Abstract
In screening for novel antibiotics, an attractive element of novelty can be represented by screening previously underexplored groups of microorganisms. We report the results of screening 200 strains belonging to the actinobacterial genus Actinoallomurus for their production of antibacterial compounds. When grown under just one condition, about half of the strains produced an extract that was able to inhibit growth of Staphylococcus aureus. We report here on the metabolites produced by 37 strains. In addition to previously reported aminocoumarins, lantibiotics and aromatic polyketides, we described two novel and structurally unrelated polyethers, designated α-770 and α-823. While we identified only one producer strain of the former polyether, 10 independent Actinoallomurus isolates were found to produce α-823, with the same molecule as main congener. Remarkably, production of α-823 was associated with a common lineage within Actinoallomurus, which includes A. fulvus and A. amamiensis. All polyether producers were isolated from soil samples collected in tropical parts of the world.
1. Introduction
Antimicrobial resistance among bacterial pathogens is becoming a major threat to human health and well-being. While different approaches can be deployed to mitigate and delay the insurgence and spread of antibiotic resistance, it is also clear that we will need a constant supply of new antibiotics, especially new chemical classes not affected by current resistance mechanisms. However, new chemical classes of antibiotics have been extremely difficult to discover from combinatorial and chemical libraries and microbial products still represent a major source of drug leads as antibiotics [1].
One of the main issues with antibiotic discovery based on microbial products is the probability of rediscovering known metabolites. This requires introducing one or more elements of novelty in the screening with respect to past efforts [2,3]. An attractive element of novelty can be represented by using novel strains, for example a taxonomic group that has not witnessed extensive analyses of its secondary metabolites, since taxonomic diversity can be seen as a surrogate for chemical diversity [4]. The main idea behind this concept is that organisms that have been subjected to different evolutionary pressures have developed unique biology to survive and, for some taxa, secondary metabolites are an important part of their biology. However, since production of secondary metabolites is not distributed equally among all species, it is important to select a taxon with a high potential to produce bioactive compounds in order to increase the probability of finding new compounds with a reasonable screening effort. Following this rationale, we initiated over a decade ago a project aimed at finding taxonomically divergent filamentous Actinobacteria, which led to the discovery of several novel taxa, including new suborders, families and genera [5,6,7,8]. One of the new taxa, originally designated as “alpha” [5], turned out to coincide with the genus Actinoallomurus (family Thermomonosporaceae), formally described in 2009 [9] with new entries added since [10,11,12,13,14,15]. With proper isolation methods, strains belonging to the genus Actinoallomurus could be effectively retrieved from a variety of soil samples, enabling the creation of a consistent collection of about 1000 isolates [2,12].
Strains belonging to the genus Actinoallomurus have been shown to produce a variety of metabolites [12,15,16,17,18,19] originating from different biosynthetic pathways. In this study, we explored 200 randomly picked Actinoallomurus isolates from the NAICONS strain collection. Together with the metabolites previously described [16,17,18,19], we analyzed the antibacterial compounds produced by 37 strains. This set of compounds includes two novel polyethers, as described here.
2. Results and Discussion
2.1. The Screened Set
The selected Actinoallomurus strains were isolated from a variety of samples collected in different continents, and representing diverse environments such as densely vegetated areas, sulfur-enriched craters of volcanic origin, and plant rhizosphere. The geographic distribution of the screened strains is listed in Table 1.

Table 1.
Geographic origin of the analyzed strains.
Three extract types were prepared from the strains―see Material and Methods―and evaluated for their ability to inhibit growth of Staphylococcus aureus and of a ΔtolC mutant of Escherichia coli. Overall, 104 and 17 strains produced at least one extract with activity against S. aureus and E. coli, respectively. All extracts with activity against E. coli were also active against S. aureus. The highest activity was observed in the mycelium and the ethyl acetate extracts at comparable frequency (57 and 46, respectively), and only in one case was the ethyl acetate exhaust extract more active. Except perhaps for an under-representation of active strains isolated from Asian samples, there was no apparent effect of the continent of origin on the frequency of anti-staphylococcal activity (Table 1). Positive extracts were analyzed as described under Material and Methods, leading to preliminary information on the chemical identity of the identified compounds. We report below the characterization of the molecules identified from 37 of the active strains.
2.2. Coumermycins, Spirotetronates, Lantibiotics and Diketopiperazines
Coumarin antibiotics target bacterial DNA gyrase and one member of this family, novobiocin, has been used to treat bacterial infections in humans caused by Gram-positive bacteria [20]. Coumermycins are other member of this family with higher antibacterial activity than novobiocin [20]. We have previously reported that the Actinoallomurus sp. K275, belonging to the Alp18 phylotype, produced several members of the coumermycin complex [12]. In the course of screening the 200 strains, two additional coumermycin producers were identified: strains ID145250 and ID145519, belonging to the Alp22 phylotype. The main coumermycin congeners produced by these three strains were A2, D1 and A1, respectively. Relevant data are shown in Supplementary Materials Figure S1. Coumermycins have been reported mostly from Streptomyces spp. [20].
Tetronate-containing polyketide natural products represent a large and diversified family of microbial metabolites with different bioactivities [21]. Halogenated spirotetronates designated NAI-414 A and B were previously described as the main products of Actinoallomurus sp. ID145414 [16]. During our screening, two strains (ID145260 and ID145814) were found to produce a molecule with m/z [M−H]− 839, an isotopic pattern compatible with the presence of two chlorines and a UV spectrum with maxima at 228 and 268 nm (see Supplementary Materials Figure S2). These properties closely resemble those reported for NAI-414 and actually match those reported for pyrrolosporin, a compound structurally related to NAI-414 but containing an additional unsaturation in the polyketide backbone. Pyrrolosporin was previously reported as a metabolite from a Micromonospora sp. [22,23].
Ribosomally synthesized and post-translationally modified peptides represent a rapidly expanding family of microbial metabolites, with lantibiotics as one of better known representatives [24]. Actinoallomurus sp. ID145699 was previously reported to produce the chlorinated lantibiotic NAI-107 and its brominated variant in Br-supplemented medium [19]. In the course of our screening, we identified strain ID145640 as an additional NAI-107 producer, with the known variations at Trp4 (hydrogen or chlorine) but just zero or one hydroxylation at Pro14 (Supplementary Materials Figures S3 and S4). Previously, NAI-107 was reported as the product of two independent Microbispora isolates [25,26].
Diketopiperazines represent a broad family of cyclized dipeptides produced by a large variety of microorganisms [27]. It is thus no surprise that one of the strains in the present work, Actinoallomurus sp. ID145219, produced two compounds with activity against S. aureus that were identified as cyclo-Phe-Leu and cyclo-Phe-Phe (see Supplementary Materials Figure S5). The structures of the metabolites mentioned in Section 2.2 are illustrated in Figure 1.
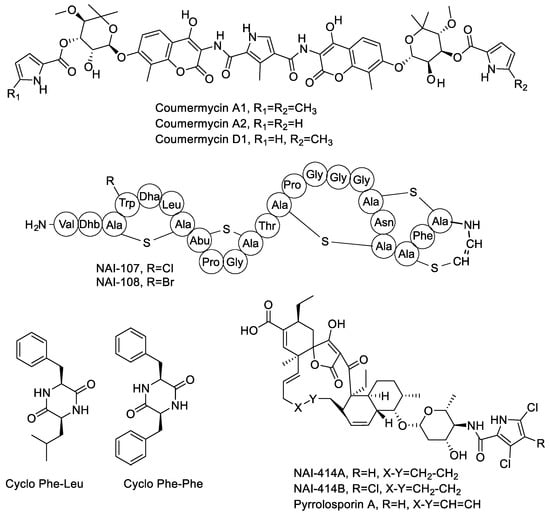
Figure 1.
Chemical structures of molecules produced by Actinoallomurus and described in Section 2.2.
2.3. Aromatic Polyketides
During the course of our screening, we frequently encountered strains producing aromatic polyketides. Identified products included the allocyclinones, hyper-halogenated angucyclinones detected from twelve independent strains belonging to three different phylotypes [17]; the paramagnetoquinones, highly paramagnetic tetracenes produced by three independent strains belonging to three different phylotypes [18]; three producers of the related dihydrobenzo-(alpha)-naphthacenequinones pradimicin (strains ID145114 and ID145318) and benanomicin (strain ID145226) (see Supplementary Materials Figure S6). Pradimicin and benanomicin present a common polyketide core decorated with a disaccharide unit and differ for the presence/absence of an N-methyl on the aminated-sugar. Both compounds were previously reported as products of Actinomadura spp., with pradimicin produced by a confirmed species of the genus, Actinomadura hibisca [28]. Benanomicin had already been reported as a metabolite of Actinoallomurus strain K15 [12].
Overall, this brief survey of aromatic polyketides indicates that Actinoallomurus spp. are capable of producing decaketides (i.e., paramagnetoquinones), undecaketides (i.e., allocyclinones, presumably undergoing oxidative ring cleavage after polyketide formation) and dodecaketides (i.e., pradimicin and benanomicin). The structures of these metabolites are shown in Figure 2.
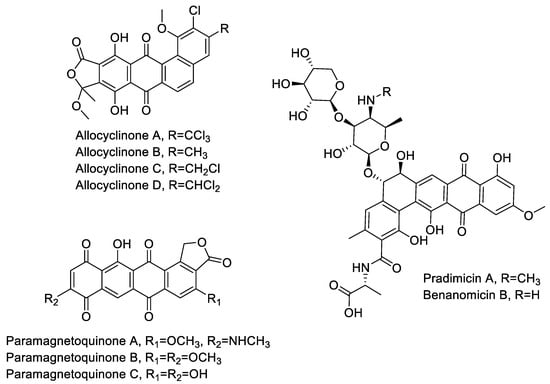
Figure 2.
Chemical structures of aromatic polyketides produced by Actinoallomurus and described in Section 2.3.
2.4. Polyethers
The extracts from several strains presented large inhibition halos against S. aureus but little or no activity against the E. coli ΔtolC strain. Upon resolution by high performance liquid chromatography (HPLC), the active fractions showed a retention time of 5–11 min and, with one exception, had no ultraviolet (UV) absorption. Mass spectrometry (MS) analysis indicated the presence of m/z signals consistent with the formation NH4+ and Na+ adducts but with no detectable H+ adducts. It should be noted that the extraction procedure and the liquid chromatography (LC)-MS eluent do not contain ammonium or sodium ions. Hence, the observation of NH4+ and Na+ adducts suggests a high cation-binding ability of the active molecules. Moreover, the fragmentation pattern showed losses of 44 amu (free carboxylic acid) and 62 amu (decarboxylation and dehydratation). As demonstrated below, we identified three distinct polyether families within twelve strains: one new compounds, designated α-823, produced by ten independent isolates; an additional new polyether, designated α-770, and the previously reported octacyclomycin, produced by one strain each. Table 2 lists the identified polyether-producing Actinoallomurus isolates, along with their origins, accession number of the 16S rRNA gene sequences and the m/z value of the most abundant congener.

Table 2.
Polyether-producing Actinoallomurus strains.
Several strains produced a likely polyether with major m/z signals [M+NH4]+ 932 and [M+Na]+ 937 (see Figure 4a,b for representative example). The metabolite produced by all these strains appeared identical (Table 2) and those from strain ID145823 were analyzed in detail. The strain produced a complex of related molecules (Figure 4a; Supplementary Materials Table S1) with similar HPLC retention times (they all eluted at ≥90% acetonitrile; see Supplementary Materials Figure S7), appearing as both [M+NH4]+ and [M+Na]+ adducts, and with similar fragmentation patterns (Supplementary Materials Figure S7). The deduced molecular formulae indicate that the congeners varied in methyl group(s) and oxygen(s) (Supplementary Materials Table S1). The structure of the major congener, designated α-823, was elucidated by a combination of NMR (Supplementary Materials Figures S8–S13) and HR-ESI-MS (Figure S14) and MS/MS analyses. The molecular formula was defined as C48H82O16Na (calculated 937.5495 [M+Na]+, found 937.5510 [M+Na]+). The analysis of 1H-monodimentional spectrum revealed the presence of four singlet and six doublet methyl signals, along with four methoxy groups. Moreover, several diastereotopic methylene signals were observed using 2D-HSQC (bi-dimensional Heteronuclear Single Quantum Coherence) experiments, indicating CH2 inserted into rigid structures or close to stereocenters. COSY (COrrelated SpectroscopY) and TOCSY (TOtal Correlated SpectroscopY) analyses, along with HMBC (Heteronuclear Multiple Bond Correlation, resulted in the structure shown in Figure 3. α-823 consists of a C30 chain with three substituted tetrahydrofuranes and three substituted tetrahydropyranes. Tetrahydrofurane C carries a deoxysugar (Figure 4). Structurally, α-823 closely resembles the polyether SF-2361, produced by an Actinomadura sp. [29,30]. Despite an identical molecular formula, α-823 carries a methyl at C-6, while a methyl group in SF-2361 has been assigned to C-2. Indeed, in the α-823 spectrum C-2 is a free methylene with δH signal at 2.18–2.54 ppm and δC at 44.7 ppm (due to the proximity to a hemiacetal and a carboxylic acid), while C-6 carries no proton (δC at 78.6 ppm) and shows HMBC correlations with a methoxy at 3.38 ppm and a singlet methyl at 1.16 ppm.
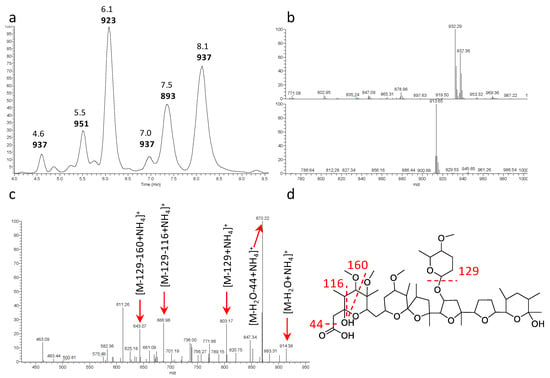
Figure 3.
Analysis of α-823. (a) Base peak chromatogram of the 4.0–10.0 min portion with retention times and m/z [M+MH4]+ values. Data obtained with a partially purified extract of Actinoallomurus sp. ID145823 (see Table S1 and Figure S7 for the congeners comparison); (b) mass spectrometry (MS) at 8.1 min in positive (above) and negative (below) ionization mode; (c) MS2 analysis of m/z [M+NH4]+ 932; (d) putative fragmentation pathway for α-823.
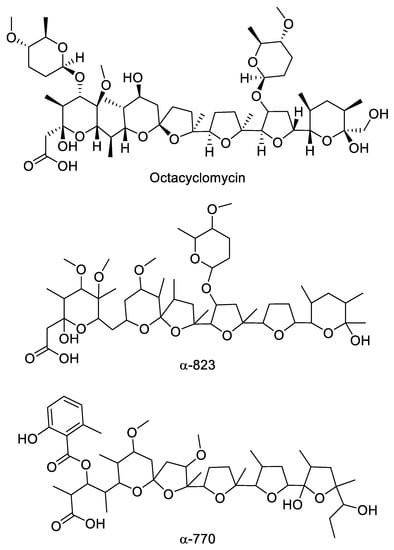
Figure 4.
Chemical structure of polyethers produced by Actinoallomurus and described in Section 2.4.
Strain ID145817 was found to produce a bioactive compound eluting at 9.0 min with no UV adsorption. Upon MS analysis, it showed m/z signals [M+NH4]+ 1034 and [M+Na]+ 1039, corresponding to the NH4+ and Na+ adduct of a molecule of 1016 amu, with major fragments at 990–995 and 972–977 (Supplementary Materials Figure S15). Additionally, a Δm/z of 129 suggested the elimination of a deoxysugar. All these properties are consistent with the metabolite produced by strain ID145817 being identical to octacyclomycin, a di-glycosylated polyether previously reported from a Streptomyces sp. [31]. NMR analysis of the purified compound confirmed this hypothesis showing signals identical to those reported in literature for octacyclomycin (data not shown). The structure of octacyclomycin is reported in Figure 4. Octacyclomycin, SF-2361 and α-823 derive from a C30 chain with identical sequence of tetrahydrofuranes and tetrahydropyranes, but differ for the number and position of methyls, oxygens and glycosyl moieties.
Strain ID145770 was found to produce an active peak eluting at 6.9 min and showing the polyether-diagnostic m/z signals [M+NH4]+ 852 and [M+Na]+ 857, corresponding to a molecule of 834 amu (Figure 5a,b). Unlike the other polyethers, however, this peak showed a UV signal with maximum at 314 nm (Figure 5a) and a Δm/z 135 upon MS fragmentation, consistent with presence of a methylsalicylate moiety (Figure 5c), a chromophore previously found in the polyether cationomycin [32]. The active molecule was produced along with a related, Δm/z +14 species, consistent with the presence of an extra methyl group atom, as listed in Table S3 and shown in Figure S16. The structure of the major congener α-770 was elucidated by a combination of NMR (Figures S17–S22), HR-ESI-MS (Figure S23) and MS/MS analyses (Figure 5). The molecular formula was defined as C45H70O14Na (calculated 857.4658 [M+Na]+, found 857.4695 [M+Na]+). The analysis of 1H- and HSQC spectra revealed the presence of 2 methoxy along with 10 methyl groups, with four of them devoid of multiplicity. Moreover, several diastereotopic methylene signals were observed, indicating CH2 inserted into rigid structures or close to stereocenters. The 2D-NMR-experiments allowed assigning the carbons in a structure consisting four substituted tetrahydrofuranes and one substituted tetrahydropyrane, as shown in Figure 4. The overall structure of α-770 is similar to that of cationomycin, produced by an Actinomadura sp. [32]. Both polyethers consists of a C27 chain with identical positioning of the methyl groups, suggesting they a common origin from incorporation of the same sequence of propionate and acetate precursors [33]; and both polyethers carry a 6-methylsalicylate moiety linked in an ester bond the C-3 hydroxyl. However, despite these similarities and the small mass difference (16 amu), α-770 and cationomycin differ significantly in the hydroxyl and methoxy decorations. Indeed, 2D-HMBC correlations established that α-770 lacks the hydroxyls at positions 15 and 5′, which are present as methoxy groups in cationomycin, as well as the hydroxyl at position 3. In contrast, α-770 carries methoxys at positions 11 and 21, while cationomycin has no hydroxyls at those positions.
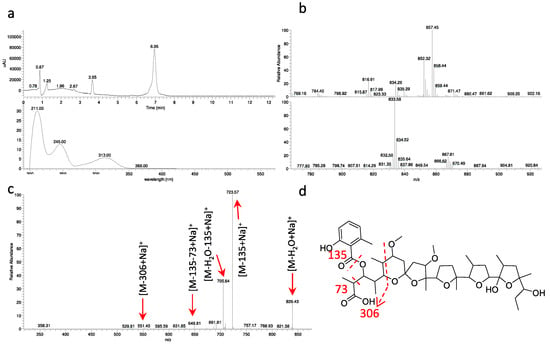
Figure 5.
Analysis of α-770. (a) UV chromatogram at 230 nm and UV spectrum of 6.9-min peak. Data obtained with a partially purified extract of Actinoallomurus sp. ID145770; (b) MS at 6.9 min in positive (above) and negative (below) ionization mode; (c) MS2 of m/z [M+NH4]+ 852; (d) putative fragmentation pathway for α-770.
The carbon chain of polyethers is assembled by type I PKSs, followed by ring formation by dedicated epoxidases [33]. The carbon chains of α-770 and α-823 are likely to derive from trideca- and pentadeca-ketide precursors, respectively. In addition, the 6-methylsalicylate unit of cationomycin has been show to result from acetate incorporation [34], consistent with the involvement of a type III PKS system.
The antimicrobial activity of polyethers is strictly connected to their ability to insert into cellular membranes and alter transport of metal cations, which leads to changes in the osmotic pressure inside the cytoplasm and cell death [30,35]. However, polyethers generally lack cellular selectivity. The antibacterial activities of α-823 and α-770 are reported in Table 3. They show potent activities against most Gram-positive bacteria, with minimal inhibitory concentrations (MICs) well below 1 µg/mL for α-770, the most active of the three compounds, with 2–4 times lower MICs than salinomycin against most of the tested strains. The polyether α-823 was generally 4–16 times less active than α-770, except for an increased activity against Mycobacterium smegmatis. No activities were detected against Gram-negative strains (not shown), except for Moraxella catarrhalis.

Table 3.
MICs (Minimal Inhibitory Concentrations) of α-770 and α-823 in comparison with salinomycin.
Some polyethers (e.g., salinomycin, monensin) are commercially used as coccidiostatic agents and salinomycin has also been evaluated as anticancer agents [36]. Recently, salinomycin and other ionophores have been shown to have transmission blocking activity against the etiological agent of malaria [37]. When tested against one chloroquine-sensitive and one chloroquine-resistant strain of Plasmodium falciparum, α-770 and α-823 showed inhibitory activity in the 2–10 nM range comparable to those of salinomycin [37]. It should be noted that, although there are over 120 reported polyethers in the literature, their mechanism of action has been studied on a limited number of molecules [30] and we are not aware of studies aimed at making polyethers selective towards a particular cell type.
The 16S rRNA gene sequences of the polyether-producing Actinoallomurus strains of Table 2 were determined and compared to those of all described Actinoallomurus species. The results are shown in Figure 6. All the ten α-823 producers cluster together in a compact branch that includes the type strains Actinoallomurus fulvus and A. amamiensis: specifically, the identical 16S rRNA gene sequences from strains ID145265, -145554, -145603 and -145830 are 100% identical to that from A. fulvus; the 16S rRNA genes sequences from ID145802 and -145828 are 99.9% and 100% identical, respectively, to that from A. amamiensis; while the identical 16S rRNA gene sequences from strains ID145804, -145811 and -145816 and that from strain ID145823 are less related to those of described species (Figure 6). The octacyclomycin producer ID145817 is less closely related (≤99.1% identity) to A. bryophytorum and A. yoronensis, although all these strains belong to a related phylogenetic branch. The α-770 producer, instead, is distantly related to A. spadix (98.7% identity) and belongs to an unrelated branch that includes, among others, strain ID145113, the producer of the aromatic polyketide paramagnetoquinone [18]. Remarkably, all polyether-producing Actinoallomurus strains were isolated from soil samples of tropical origin (Table 2), notwithstanding that 64% of the screened strains were of non-tropical origin (mostly from Europe; see Table 1). These observations suggest that the branch including A. fulvus, A. amamiensis and the α-823 producers might consist of cosmopolitan strains and that polyether production might be mostly associated with Actinoallomurus strains from tropical environment. Previous studies have established a correlation between different classes of secondary metabolites and geographic origin [38,39], although we are unaware of previous reports on biogeography of polyether production.
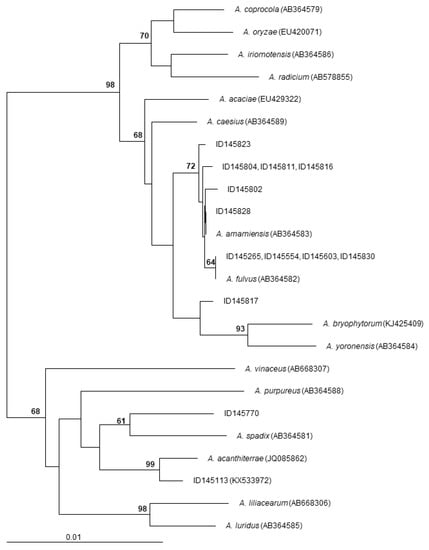
Figure 6.
Neighbor-joining tree showing the phylogenetic position of polyether-producing Actinoallomurus strains. Type-strains of all described Actinoallomurus species are included. The sequence of the paramagnetoquinone producer Actinoallomurus sp. ID145113 is also included. The tree is based on 1309 unambiguously aligned positions in the 16S rRNA gene sequences. Numbers at the nodes are bootstrap values based on 100 resamplings; only values higher than 60 are shown. Scale bar represents 1 inferred substitutions per 100 nucleotides. The tree was rooted using Streptosporangium roseum 16S rRNA gene sequence (X89947) as outgroup.
3. Materials and Methods
3.1. Bacterial Strains and Media
Actinoallomurus strains are from the NAICONS strain library. Each strain was propagated on S1-5.5 plates (60 g/L oatmeal, 18 g/L agar, 1 mL/L Trace Elements Solution) at 30 °C for two to three weeks. From these plates, the grown mycelium was used to inoculate AF-A medium (10 g/L dextrose monohydrate, 4 g/L soybean meal, 1 g/L yeast extract, 0.5 g/L NaCl, 1.5 g/L 2-(N-morpholino) ethanesulfonic acid, pH adjusted to 5.6) in shake-flasks. After 8 days in a rotatory shaker (200 rpm) at 30 °C, cultures were harvested and extracted (see Section 3.2).
PCR amplifications with the eubacterial primers F27 and R1492 and phylogenetic analyses of the 16S rRNA gene sequences were performed as previously described [40]. The 16S rRNA gene sequences have been deposited in GenBank, as listed in Table 2.
3.2. Preparation of Extracts
Three different extracts were prepared from each culture. The culture was centrifuged at 16,000 rcf for 5 min and the resulting pellet was resuspended in 0.4 vol ethanol, while the supernatant was used for ethyl acetate extraction (see below). After shaking 1 h at 55 °C, the suspension was centrifuged once more (16,000 rcf, 5 min) and the supernatant transferred to a new tube, dried under vacuum and resuspended in 10% DMSO at 0.2× the original culture volume. This extract was designated as the mycelium extract.
The supernatant from the first centrifugation step above was extracted with 0.5 vol ethyl acetate. After mixing and phase separation, the organic phase was transferred to a new tube and the aqueous phase extracted again with further 0.5 vol ethyl acetate. The two organic phases were combined, dried and resuspended at 5× the original concentration in 10% DMSO. This extract was designated as EtAc extract. The exhausted aqueous phase was also retained and tested as such.
3.3. Antibacterial Assays
The screening was performed by agar diffusion, using plates containing 15 mL of Müller-Hinton Agar and inoculated with 5 × 105 CFU/mL of the indicator strain. Strains used in this assay were Staphylococcus aureus ATCC 6538P and Escherichia coli L4242, a ΔtolC derivative of MG1061. After spotting 20 μL of the resuspended extract, plates were incubated 18–20 h at 37 °C. After HPLC fractionation, bioactive fractions were identified using the same methodology.
MIC determinations of purified compounds were performed by broth micro dilution in sterile 96-well polystyrene microtiter plates according to CLSI guidelines, using Müller Hinton broth (Difco Laboratories) containing 20 mg/L CaCl2 and 10 mg/L MgCl2 for all strains except for Streptococcus spp., which were grown in Todd Hewitt broth. Strains were inoculated at 5 × 105 CFU/mL and incubated at 37 °C for 20−24 h. Strain with an L prefix are from the NAICONS pathogens library.
3.4. Analytical Procedures
For monitoring metabolites production analytical HPLC was performed on Shimadzu Series 10 spectrophotometer (Kyoto, Japan), equipped with a reverse-phase column, LiChrospher RP-18, 5 μm, 4.6 × 125 mm (Merck, Darmstadt, Germany). Phase A was 0.1% trifluoroacetic acid (TFA), phase B acetonitrile, and the flow rate was 1 mL/min. Resolution was achieved with a linear gradient from 10% to 36% phase B in 5 min; from 36% to 50% phase B in 7 min; and from 50% to 80% phase B in 1 min; followed by a 4-min isocratic step at 80% phase B and column re-equilibration. UV detection was at 230 and 270 nm. LC-MS analyses were performed on a Dionex UltiMate 3000 coupled with an LCQ Fleet (Thermo scientific) mass spectrometer equipped with an electrospray interface (ESI) and a tridimensional ion trap. The column was an Atlantis T3 C18 5 μm × 4.6 mm × 50 mm maintained at 40 °C at a flow rate of 0.8 mL/min. Phases A and B were 0.05% TFA in water and acetonitrile, respectively. The elution was with a 14-min multistep program that consisted of 10, 10, 95, 95, 10 and 10% phase B at 0, 1, 7, 12, 12.5 and 14 min, respectively. UV-VIS signals (190–600 nm) were acquired using the diode array detector. The m/z range was 110–2000 and the ESI conditions were as follows: spray voltage of 3500 V, capillary temperature of 275 °C, sheath gas flow rate at 35 units and auxiliary gas flow rate at 15 units.
High resolution MS spectra were recorded at Unitech OMICs (University of Milano, Italy) using a Triple TOF® 6600 (Sciex) equipped with an ESI source. The experiments were carried out by direct infusion in positive ionization mode. The ESI parameters were the following: curtain gas 25 units, ion spray voltage floating 5500 v, temperature 50 °C, ion source gas1 10 units, ion source gas2 0 units, declustering potential 80 v, syringe flow rate 10 µL/min, accumulation time 1 s.
Mono- and bi-dimensional NMR spectra were measured in CDCl3 at 298K using an AMX 400 MHz spectrometer. Chemical shifts are reported relative to CDCl3 (δ 7.26 ppm).
3.5. Purification of Polyethers
α-823: Nine parallel 100-mL cultures of Actinoallomurus sp. ID145823 in AF-A medium were harvested at seven days and filtered through paper under reduced pressure to separate the mycelium from the clear broth. The latter (860 mL) was extracted three times with 450 mL ethyl acetate while the mycelium was treated with 100 mL acetone, kept on a rotary shaker 1 h and centrifuged. The combined organic phases were dried under reduced pressure and dissolved in 2 mL dichloromethane. The sample was resolved on a 12 g direct-phase Flash column RediSep RF (Teledyne Isco) by using a CombiFlash RF Teledyne Isco medium-pressure chromatography system. The column was previously conditioned at 100% dichloromethane and then eluted at 15 mL/min with a 20-min linear gradient from 0 to 10% methanol. Fractions were analyzed by LC-MS and those with the highest purity were pooled and dried, obtaining 14 mg of purified α-823. Five mg were dissolved in CDCl3 for NMR analysis.
Octacyclomycin: Two parallel 100-mL cultures of Actinoallomurus sp. ID145817 in AF-A medium were harvested at seven days and filtered through paper under reduced pressure to separate the mycelium from the clear broth. The latter (170 mL) was extracted three times with 80 mL ethyl acetate while the mycelium was treated with 100 mL ethanol, kept 1 h on a rotary shaker and centrifuged. The combined organic phases were dried under reduced pressure and dissolved in 2 mL dichloromethane. The sample was resolved by medium-pressure chromatography as described above for α-823. Fractions were analyzed by LC-MS and processed as above. Four mg of purified octacyclomycin were obtained.
α-770: Two parallel 100-mL cultures of Actinoallomurus sp. ID145770 in medium M8 [41] were harvested at seven days. Mycelium was harvested by centrifugation (10 min at 4000 rpm), treated with 20 mL ethanol, kept 1 h on a rotary shaker and centrifuged. The organic phase was recovered, dried under reduced pressure, dissolved in 2 mL dichloromethane and resolved by medium-pressure chromatography as described for α-823, except that the flow rate was set at 30 mL/min. Fractions 11–14, which showed activity against S. aureus, were analyzed by LC-MS and the ones containing similar signals were pooled, dried and dissolved in dichloromethane. A further purification step was performed by preparative thin layer chromatography on silica gel (Analtech Preparative Silica Gel GF with UV254 2000 μm; Sigma-Aldrich, St Louis, MO, USA) in dichloromethane:methanol 9:1. The spot at Rf 0.9 was dried and dissolved in CDCl3 for NMR analysis. Four mg of purified α-770 were obtained.
4. Conclusions
When grown under one routine condition in shake-flasks and only looking at metabolites with growth inhibitory activity towards S. aureus, we have been able to show that Actinoallomurus strains can express several types of biosynthetic pathways: type I (for making polyethers and spirotetronates), type II (for aromatic polyketides) and type III (for the 6-methylsalicylate moiety of the polyether α-770) polyketide synthases; ribosomally synthesized and post-translationally modified peptides (lantibiotic); aminocoumarins; and short non-ribosomal peptide synthase derived products (diketopiperazines).
Some of the observed metabolites, e.g., the previously reported aromatic polyketide allocyclinones [17] and the polyether α-823, seem to be relatively frequent metabolites in the screened Actinoallomurus strains. Other metabolites represent rarer discovery events, with identical or close matches in several Actinobacteria genera. Indeed, the genus Actinoallomurus resulted from a reclassification of Actinomadura spp. within the family Thermomonosporaceae, order Streptosporangiales [9]. Some of the compounds described here (e.g., pradimicin and benanomicin) and the α-770- and α-823-related polyethers cationomycin and SF-2361, respectively, were previously reported from Actinomadura spp. Others of the described metabolites are produced by distantly related taxa, such as NAI-107 by Microbispora spp. (family Streptosporangiaceae, order Streptosporangiales), pyrrolosporin by a Micromonospora sp. (order Micromonosporales), in addition to the Streptomyces-produced coumermycin and octacyclomycin.
Supplementary Materials
The following are available online at http://www.mdpi.com/2079-6382/7/2/47/s1, Figure S1: LC-MS, UV analysis of ethyl acetate extract of Actinoallomurus sp. ID145519, Figure S2: LC-MS, UV analysis of ethyl acetate extract of Actinoallomurus sp. ID145814, Figure S3: LC-MS, UV analysis of mycelium extract of Actinoallomurus sp. ID145640, Figure S4: Comparison between mycelium extract of Actinoallomurus sp. ID145640 and NAI-107 standard, Figure S5: LC-MS, UV analysis of ethyl acetate extract of Actinoallomurus sp. ID145219, Figure S6: LC-MS, UV analysis of ethyl acetate extract of Actinoallomurus sp. ID145114, Figure S7: Fragmentation patterns of the eight different congeners of α-823, Figure S8: 1H-NMR spectrum of α-823, Figure S9: 1H-COSY NMR spectrum of α-823, Figure S10: 1H-TOCSY NMR spectrum of α-823, Figure S11: 1H-13C HSQC NMR spectrum of α-823, Figure S12: 1H-13C HMBC NMR spectrum of α-823, Figure S13: Major COSY and HMBC correlations of α-823, Figure S14: HRESI-MS spectrum of α-823, Figure S15: LC-MS, UV analysis of mycelium extract of Actinoallomurus sp. ID145817, Figure S16: Fragmentation patters of the two different congeners of α-770, Figure S17: 1H NMR spectrum of α-770, Figure S18: 1H-COSY NMR spectrum of α-770, Figure S19: 1H-TOCSY NMR spectrum of α-770, Figure S20: 1H-13C HSQC NMR spectrum of α-770, Figure S21: 1H-13C HMBC NMR spectrum of α-770, Figure S22: Major COSY and HMBC correlations of α-770, Figure S23: HRESI-MS spectrum of α-770, Table S1: Different α-823 congeners detected in the extract from Actinoallomurus sp. ID145823, Table S2: 1H and 13C NMR data for α-823 in CDCl3, Table S3: α-770 congeners detected in the extract from Actinoallomurus sp. ID145770, Table S4: 1H and 13C NMR data for α-770 in CDCl3.
Author Contributions
M.S. and S.D. conceived and designed the experiments; M.I., A.T., J.C.S.C., G.D.G., C.B. and S.I.M. performed the experiments; M.I., A.T., J.C.S.C., G.D.G., C.B., S.I.M., M.S. and S.D. analyzed the data; M.I., A.T. and S.D. wrote the paper.
Acknowledgments
Portion of this work was part of a PhD dissertation of J.C.S.C. at the University of Warwick, UK. This work was partially supported by the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 289285 and by grants from Regione Lombardia. We are grateful to Carlo Mazzetti, Mirko Ornaghi, Roberta Pozzi and Matteo Simone for their early contributions to this project, and to Donatella Taramelli and Silvia Parapini for the anti-plasmodium activity tests. We also thank the Unitech OMICs platform at the University of Milano for HRMS analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Monciardini, P.; Iorio, M.; Maffioli, S.; Sosio, M.; Donadio, S. Discovering new bioactive molecules from microbial sources. Microb. Biotechnol. 2014, 7, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Jaspars, M.; Challis, G. Microbiology: A talented genus. Nature 2014, 506, 38–39. [Google Scholar] [CrossRef] [PubMed]
- Donadio, S.; Busti, E.; Monciardini, P.; Bamonte, R.; Mazza, P.; Sosio, M.; Cavaletti, L. Sources of polyketides and non-ribosomal peptides. Ernst Schering Res. Found. Workshop 2005, 51, 19–41. [Google Scholar]
- Busti, E.; Cavaletti, L.; Monciardini, P.; Schumann, P.; Rohde, M.; Sosio, M.; Donadio, S. Catenulispora acidiphila gen. nov., sp. nov., a novel, mycelium-forming actinomycete, and proposal of Catenulisporaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 1741–1746. [Google Scholar] [CrossRef] [PubMed]
- Cavaletti, L.; Monciardini, P.; Schumann, P.; Rohde, M.; Bamonte, R.; Busti, E.; Sosio, M.; Donadio, S. Actinospica robiniae gen. nov., sp. nov. and Actinospica acidiphila sp. nov.: Proposal for Actinospicaceae fam. nov. and Catenulisporinae subord. nov. in the order Actinomycetales. Int. J. Syst. Evol. Microbiol. 2006, 56, 1747–1753. [Google Scholar] [CrossRef] [PubMed]
- Monciardini, P.; Cavaletti, L.; Ranghetti, A.; Schumann, P.; Rohde, M.; Bamonte, R.; Sosio, M.; Mezzelani, A.; Donadio, S. Novel members of the family Micromonosporaceae, Rugosimonospora acidiphila gen. nov., sp. nov. and Rugosimonospora africana sp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2752–2758. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Ishida, Y.; Nozawa, Y.; Otoguro, M.; Suzuki, K. Transfer of Actinomadura spadix Nonomura and Ohara 1971 to Actinoallomurus spadix gen. nov., comb. nov., and description of Actinoallomurus amamiensis sp. nov., Actinoallomurus caesius sp. nov., Actinoallomurus coprocola sp. nov., Actinoallomurus fulvus sp. nov., Actinoallomurus iriomotensis sp. nov., Actinoallomurus luridus sp. nov., Actinoallomurus purpureus sp. nov. and Actinoallomurus yoronensis sp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 1867–1874. [Google Scholar] [PubMed]
- Thamchaipenet, A.; Indananda, C.; Bunyoo, C.; Duangmal, K.; Matsumoto, A.; Takahashi, Y. Actinoallomurus acaciae sp. nov., an endophytic actinomycete isolated from Acacia auriculiformis A. Cunn. ex Benth. Int. J. Syst. Evol. Microbiol. 2010, 60, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Indananda, C.; Thamchaipenet, A.; Matsumoto, A.; Inahashi, Y.; Duangmal, K.; Takahashi, Y. Actinoallomurus oryzae sp. nov., an endophytic actinomycete isolated from roots of a Thai jasmine rice plant. Int. J. Syst. Evol. Microbiol. 2011, 61, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, R.; Simone, M.; Mazzetti, C.; Maffioli, S.; Monciardini, P.; Cavaletti, L.; Bamonte, R.; Sosio, M.; Donadio, S. The genus Actinoallomurus and some of its metabolites. J. Antibiot. 2011, 64, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Koyama, R.; Matsumoto, A.; Inahashi, Y.; Omura, S.; Takahashi, Y. Isolation of actinomycetes from the root of the plant, Ophiopogon japonicus, and proposal of two new species, Actinoallomurus liliacearum sp. nov. and Actinoallomurus vinaceus sp. nov. J. Antibiot. 2012, 65, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Fukuda, A.; Inahashi, Y.; Omura, S.; Takahashi, Y. Actinoallomurus radicium sp. nov., isolated from the roots of two plant species. Int. J. Syst. Evol. Microbiol. 2012, 62, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Inahashi, Y.; Iwatsuki, M.; Ishiyama, A.; Matsumoto, A.; Hirose, T.; Oshita, J.; Sunazuka, T.; Panbangred, W.; Takahashi, Y.; Kaiser, M.; et al. Actinoallolides A-E, new anti-trypanosomal macrolides, produced by an endophytic actinomycete, Actinoallomurus fulvus MK10-036. Org. Lett. 2015, 17, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Mazzetti, C.; Ornaghi, M.; Gaspari, E.; Parapini, S.; Maffioli, S.; Sosio, M.; Donadio, S. Halogenated spirotetronates from Actinoallomurus. J. Nat. Prod. 2012, 75, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.C.S.; Maffioli, S.; Bernasconi, A.; Brunati, C.; Gaspari, E.; Sosio, M.; Wellington, E.; Donadio, S. Allocyclinones, hyperchlorinated angucyclinones and common metabolites from Actinoallomurus. J. Antibiot. 2017, 70, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.; Cruz, J.; Simone, M.; Bernasconi, A.; Brunati, C.; Sosio, M.; Donadio, S.; Maffioli, S.I. Antibacterial paramagnetic quinones from Actinoallomurus. J. Nat. Prod. 2017, 80, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.C.S.; Iorio, M.; Monciardini, P.; Simone, M.; Brunati, C.; Gaspari, E.; Maffioli, S.I.; Wellington, E.; Sosio, M.; Donadio, S. Brominated variant of the lantibiotic NAI-107 with enhanced antibacterial potency. J. Nat. Prod. 2015, 78, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, B.; Kahlich, R.; Laufer, S.; Li, S.M.; Heide, L.; Gleiter, C.H. Mass spectrometric pathway monitoring of secondary metabolites: Systematic analysis of culture extracts of Streptomyces species. Anal. Biochem. 2004, 335, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Vieweg, L.; Reichau, S.; Schobert, R.; Leadlay, P.F.; Süssmuth, R.D. Recent advances in the field of bioactive tetronates. Nat. Prod. Rep. 2014, 31, 1554–1584. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.S.; Hesler, G.A.; Gustavson, D.R.; Berry, R.L.; Tomita, K.; MacBeth, J.L.; Ross, J.; Miller, D.; Forenza, S. Pyrrolosporin A, a new antitumor antibiotic from Micromonospora sp. C39217-R109-7. I. Taxonomy of producing organism, fermentation and biological activity. J. Antibiot. 1996, 49, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.R.; Colson, K.L.; Klohr, S.E.; Lee, M.S.; Matson, J.A.; Brinen, L.S.; Clardy, J. Pyrrolosporin A, a new antitumor antibiotic from Micromonospora sp. C39217-R109-7. II. Isolation, physic-chemical properties, spectroscopic study and X-ray analysis. J. Antibiot. 1996, 9, 865–872. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.D. Antibiotics from Microbispora. U.S. Patent 6,551,591, 22 April 2003. [Google Scholar]
- Lazzarini, A.; Gastaldo, L.; Candiani, G.; Ciciliato, I.; Losi, D.; Marinelli, F.; Selva, E.; Parenti, F. Antibiotic 107891, Its Factors A1 and A2, Pharmaceutically Acceptable Salts and Compositions, and Use Thereof. U.S. Patent 7,351,687, 1 April 2008. [Google Scholar]
- Belin, P.; Moutiez, M.; Lautru, S.; Seguin, J.; Pernodet, J.L.; Gondry, M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. 2012, 29, 961–979. [Google Scholar] [CrossRef] [PubMed]
- Oki, T.; Kinoshi, M.; Tomatsu, K.; Tomita, K.; Saitoh, K.; Tsunakawa, M.; Nishio, M.; Miyaki, T.; Kawaguchi, H. Pradimicin, a novel class of potent antifungal antibiotics. J. Antibiot. 1999, 11, 1701–1704. [Google Scholar] [CrossRef]
- Tadashi, N.; Yukio, T.; Yuko, K.; Mamoru, I.; Taneto, T.; Shinji, M.; Masaji, S.; Michio, K. Novel antibiotic substance SF-2361, production and used thereof. Japan Patent 61260888, 19 November 1986. [Google Scholar]
- Kevin, D.A., II; Meujo, D.A.F.; Hamann, M.T. Polyether ionophores: Broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expert Opin. Drug Discov. 2009, 4, 109–146. [Google Scholar] [CrossRef] [PubMed]
- Funayama, S.; Nozoe, S.; Tronquet, C.; Anraku, Y.; Komiyama, K.; Omura, S. Isolation and structure of a new polyether antibiotic, octacyclomycin. J. Antibiot. 1992, 45, 1686–1691. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, G.; Kobayashi, K.; Sakurai, T.; Isono, K. Cationomycin, a new polyether ionophore antibiotic produced by Actinomadura nov. sp. J. Antibiot. 1981, 34, 1513–1514. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Oguri, H.; Watanabe, K.; Oikawa, H. Biosynthetic machinery of ionophore polyether lasalocid: Enzymatic construction of polyether skeleton. Curr. Opin. Chem. Biol. 2013, 17, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Ubukata, M.; Uzawa, J.; Isono, K. Biosynthesis of cationomycin: Direct and indirect incorporation of 13C-acetate and application of homoscalar correlated 2-D carbon-13 NMR and double quantum coherence. J. Am. Chem. Soc. 1984, 106, 2213–2214. [Google Scholar] [CrossRef]
- Rutkowski, J.; Brzezinski, B. Structures and properties of naturally occurring polyether antibiotics. BioMed Res. Int. 2013, 2013, 162513. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, F.; Wong, E.T.; Fonkem, E.; Hsieh, T.C.; Wu, JM.; Wu, E. Salinomycin: A novel anti-cancer agent with known anti-coccidial activities. Curr. Med. Chem. 2013, 20, 4095–4101. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, S.; Corbett, Y.; Ilboudo, D.P.; Misiano, P.; Dahiya, N.; Abay, S.M.; Habluetzel, A.; Grande, R.; Gismondo, M.R.; Dechering, K.J.; et al. Salinomycin and other ionophores as a new class of antimalarial drugs with transmission-blocking activity. Antimicrob. Agents Chemother. 2015, 59, 5135–5144. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.B.; Ternei, M.A.; Brady, S.F. Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. USA 2014, 111, 3757–3762. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Pregitzer, C.C.; Lemetre, C.; Ternei, M.A.; Maniko, J.; Hover, B.M.; Calle, P.Y.; McGuire, K.L.; Garbarino, J.; Forgione, H.M.; et al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 14811–14816. [Google Scholar] [CrossRef] [PubMed]
- Monciardini, P.; Sosio, M.; Cavaletti, L.; Chiocchini, C.; Donadio, S. New PCR primers for the selective amplifcation of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol. 2002, 42, 419–429. [Google Scholar] [PubMed]
- Donadio, S.; Monciardini, P.; Sosio, M. Chapter 1. Approaches to discovering novel antibacterial and antifungal agents. Methods Enzymol. 2009, 458, 3–28. [Google Scholar] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).