Abstract
Carbapenemase-producing Pseudomonas aeruginosa strains present a specific geographical distribution regarding the type of carbapenemase-encoding genes that they harbor. For more than twenty years, VIM-type enzymes were the only major carbapenemases that were detected among P. aeruginosa isolates in Greece until the emergence of NDM-1-encoding P. aeruginosa in early 2023. In the present study, we present the rapid reversal of the carbapenemase-producing P. aeruginosa epidemiology from blaVIM- to blaNDM-harbouring isolates that occurred in our hospital since then. Between January 2023 and February 2024, 139 isolates tested positive for carbapenemase production with the NG-Test CARBA 5 immunochromatographic assay. Eight isolates were processed with the Hybrispot antimicrobial resistance direct flow chip molecular assay, and the first NDM-producing isolate was further analyzed through whole genome sequencing and bioinformatics analysis. Multiple resistance genes were detected by molecular techniques in accordance with the extensively drug-resistant phenotype. The isolate that was subjected to whole-genome sequencing belonged to the P. aeruginosa high-risk clone ST308, and the blaNDM was located in the chromosome in accordance with previously reported data. During the study period, NDM-producing isolates were increasingly detected, and only five months after their emergence, they overcame VIM producers. Our results indicate the potential of this new clone to spread rapidly and predominate within healthcare institutions, further restricting the already limited treatment options.
1. Introduction
Carbapenem-resistant Pseudomonas aeruginosa (CR-PA), a major pathogen worldwide, is listed as a high-priority pathogen by the World Health Organization (WHO) Bacterial Priority Pathogens List due to its high level of antibiotic resistance and the severe infections it causes, particularly in healthcare settings [1]. In Europe, large difference are observed in the percentages of CR-PA among countries, ranging from 5% to over 50. Regarding Greece, P. aeruginosa’s resistance is quite high for carbapenems, at 48.9% compared to the EU population-weighted mean of 16.5% [2].
Carbapenem resistance in P. aeruginosa is multi-factorial [3]. Different mechanisms may be involved and may even coexist in non-susceptible isolates, like the loss of the OprD porin, the presence of inducible AmpC type β-lactamases, the overexpression of efflux pumps, and the production of carbapenemases. Among them, carbapenemases are by far the most effective because they confer high-level resistance to all or almost all β-lactams, including ceftazidime, cefepime, piperacillin–tazobactam, and carbapenems. Moreover, since carbapenem-resistance genes are often located in mobile genetic elements, they can be easily disseminated between bacterial cells and species [4].
P. aeruginosa presents a remarkable ability to acquire and host carbapenemase-encoding genes and, interestingly, most of the clinically important carbapenemases that can be found in carbapenem-resistant Gram negatives were reported for the first time in P. aeruginosa isolates worldwide. Moreover, P. aeruginosa strains that encode specific carbapenemase genes, like IMP, KPC, VIM, and NDM, present a characteristic geographical distribution and are commonly related with the so-called high-risk clones [5].
NDM is closely related to Enterobacterales and Acinetobacter spp. and has been much less frequently detected in P. aeruginosa [6]. For many years, carbapenemase-producing P. aeruginosa in Greece were almost exclusively harboring genes encoding for VIM-type carbapenemases [7,8,9,10]. Recently, however, there has been a rapid reversal to NDM-encoding P. aeruginosa in our hospital soon after the first isolation in 2023, and such isolates are quickly overcoming the VIM producers.
The present study was designed to monitor the carbapenem-resistant P. aeruginosa epidemiology from February 2023 to February 2024 within our hospital. Additionally, whole genome sequence analysis was conducted on the first NDM-encoding isolate (91845) that was recovered in May 2023 from a blood culture.
2. Results
2.1. Carbapenem Resistant P. aeruginosa Isolates
During the study period, 211 carbapenem-resistant P. aeruginosa isolates were recovered from various hospital wards. Among them, 139 isolates originating from 73 male and 66 female patients were found to be positive for carbapenemase production.
2.2. Detection of Resistance Determinants by the NG-Test CARBA 5 and the Hybrispot Antimicrobial Resistance Direct Flow Chip (AMR)
Overall, the NG-Test CARBA 5 detected 72 VIM producers and 66 NDM producers. One isolate (214929) was found to be positive for both carbapenemases (Table 1).

Table 1.
Carbapenemase-producing P. aeruginosa isolates included in the study. VIM: Verona integron-encoded metallo-β-lactamase; NDM: New Delhi metallo-β-lactamase; BAL: bronchoalveolar lavage; CVC: central venous catheter ICU A: intensive care unit A; INT B: internal medicine department B; NB: neurology department B; INT A: internal medicine department A; BH: hematology department; CARD: cardiology department; SURGC: surgery department C; SURGB: surgery department B; NS: neurosurgery; EMERG: emergency department; OPHT: ophthalmology department; NEPHR: nephrology department; CARDSURG: cardiosurgery.
There was a 100% concordance of the NG-Test CARBA 5 results with those reported by the Hybrispot. Moreover, Hybrispot revealed the presence of additional resistance determinants to other antibiotic classes in the eight isolates tested using this method (Table 2).

Table 2.
Hybrispot results reported for eight P. aeruginosa isolates. VIM: Verona integron-encoded metallo-β-lactamase; NDM: New Delhi metallo-β-lactamase.
Interestingly, after their emergence in P. aeruginosa in our hospital, NDM-encoding genes have been increasingly detected. Moreover, NDM-producing isolates rapidly overcame the VIM producers within this species, as shown in Figure 1.
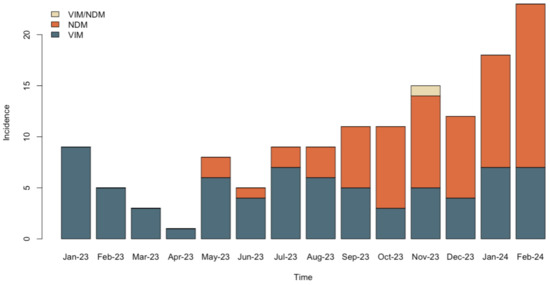
Figure 1.
Reversal of the carbapenemase-producing P. aeruginosa epidemiology from VIM to NDM in the hospital setting.
2.3. Genomic Characterization of NDM-1-Producing Isolate 91845
The NDM-1-producing isolate was genomically characterized for the presence of antimicrobial resistance genes, the presence of virulence genes, and MLST. In addition, the isolate was comparatively analyzed against the most closely related isolates, and the genomic origin of the blaNDM-1 gene, either chromosomal or plasmid, was investigated.
High-depth sequencing of isolate 91845 resulted in 155.95 M reads that were filtered to meet high-quality base calling criteria (Q30 for 96.62% of 150 bp reads). The assembled genome contains 92 contigs with an N50 value of 246.540 bp and a total genome size of 6,919,402 bp. The most closely related isolate, based on the genome-wide nucleotide identity, is a recently isolated clinical strain in Greece (GCA_030504675_1, 99.948% identity), followed by the reference blaNDM-1 human isolate from Singapore (CP020704, 99.93% identity). Two clinical isolates from the neighboring countries Italy (RefSeq assembly: GCF_013276295_1) and Serbia (RefSeq assembly: GCF_022559565.11) were identified as being closely related, with 98.99% and 98.93% identity similarity values, respectively.
2.4. MLST, Antimicrobial Resistance and Virulence Genes
Isolate 91845 belongs to the P. aeruginosa high-risk clone ST308 that has been previously reported to carry carbapenemase genes. The antimicrobial resistance analyses resulted in the following AMR genes: aac(3)-Id, aac(6′)-Ib-cr, aac(6′)-Ib3, aac(6′)-Il, aadA11, aph(3″)-Ib, aph(3′)-IIb, aph(6)-Id, blaNDM-1, blaOXA-10, blaOXA-488, blaPAO, catB7, crpP, dfrB5, floR, fosA, msr(E), qacE, qnrVC1, rmtF, sul1, and sul2. Ten virulence genes were identified: xcpP, exoU, flgC, pchB, mbtH-like, lasI, pilG, pscS, and pscF, hcp1 (Table 3).

Table 3.
Antimicrobial resistance and virulence genes found in P. aeruginosa isolate 91845.
2.5. Genomic Origin of the blandm Gene
blaNDM-1 was detected in a 2323 bp contig which also 0included an IS91 family transposase (Figure 2). No plasmid replicons have been detected in the assembled genome by PlasmidFinder, while PlasmidSPAdes did not detect blaNDM-1 in the 27 variable-length plasmid constructs. These results are in line with previous evidence that ST308 P. aeruginosa strains including blaNDM-1 are located in chromosomal regions. However, PLASme and MOB-suite classified the 2323 bp contig as a plasmid sequence originating from Enterobacterales. The prediction is considered as being high confidence, except for a 10 bp ambiguous regions identified by PLASme. In support of this finding, MOB-suite detected an AD312 plasmid (CP034849) originated from Escherichia coli that includes the blaNDM-1-containing contig as well as five other contigs of the same type (contigs 18, 39, 50, 58, and 105).
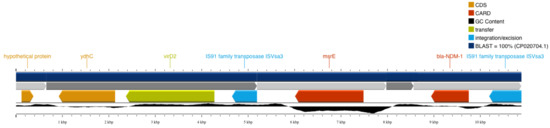
Figure 2.
The 2323 bp contig including blaNDM-1 and the neighboring IS91 family transposase.
To further assess whether these AD312-type plasmid contigs resemble known plasmids, blastn searches against complete plasmids were applied, yet with no strong evidence of similar plasmids. To further investigate the potential chromosomal origin of the blaNDM-1 gene, genome-wide pairwise alignment analysis against the reference CP020704 (Singapore) genome was performed. The genomic region neighboring blaNDM-1 in CP020704 has strong homology with the disjoint contigs 50, 79, 80, and 105 of our isolate. Figure 2, showing the 100% blast-based similarity with chromosomal regions, further supports the chromosomal origin of the blaNDM-1 gene. Interestingly, the highly homologous contigs 50 and 105 have been also identified as AD312 plasmids by MOB-suite.
3. Discussion
The New Delhi metallo-β-lactamase was named after its first report back in 2008 from Sweden regarding a patient previously hospitalized in New Delhi, India [11]. Following this initial detection, it soon became obvious that NDM-1 was already widely disseminated in the Indian subcontinent as well as in other countries, although to a lesser extent [12,13,14]. NDMs are categorized as Ambler group B carbapenemases together with other clinically important metallo-enzymes, such as VIM and IMP. Metallo-β-lactamases bear zinc in their active center and commonly hydrolyze all β-lactams except aztreonam and cefiderocol, whereas they are not inhibited by the β-lactamase inhibitors or by boronic acid. NDMs, however, present some additional unique characteristics. They hydrolyze aztreonam and they commonly present negative modified Hodge test results [15]. Resistance to cefiderocol in P. aeruginosa is still rare, but for the time being it is related only to some NDM-1-encoding lineages [16]. In Greece, NDM-1-encoding genes were introduced in late 2011 [17] and, since then, they have been well-established together with KPC, VIM, and OXA-48 carbapenemases among Enterobacterales and especially Klebsiella pneumoniae, as shown also by recent data from our hospital [18,19].
P. aeruginosa is a species that is commonly related to carbapenemases including KPC [20] and most of the clinically important metallo-β-lactamases [21,22,23,24]. Interestingly, there is a clear geographical distribution of isolates harboring different enzymes around the world, with KPCs being mainly located in Latin America, VIM and IMP presenting a global distribution, and NDMs being mostly located in India and Australia [25]. This specific distribution is associated with the spread of specific high-risk clones [5]. In Europe, the first report of NDM-encoding P. aeruginosa was reported from Serbia [26]. In 2010, at the Military Medical Academy, seven carbapenem-resistant P. aeruginosa isolates were recovered. Molecular investigation proved that two of them were the first NDM-producing P. aeruginosa worldwide. The first NDM-positive isolate was recovered from a urine sample of a 61-year-old Serbian woman with an intra-abdominal abscess, while the second one was detected in a wound of a 63-year-old woman who underwent laparotomy for intestinal carcinoma. Both patients had no previous travel history and died shortly after their admission.
The first NDM-1 in Pseudomonas aeruginosa clinical isolates from India were reported between 2011–July 2012. A total of 4 out of 200 P. aeruginosa clinical isolates investigated were NDM producers. The first was isolated from a central venous catheter culture of a 66-year-old man with a necrotizing soft tissue infection of the left lower limb. The second was also isolated from a venous catheter culture and was detected in a male patient with scarring after gallbladder removal surgery. Colistin was used successfully for both patients. The third isolate was found in the urine sample of a 56-year-old man with pyelonephritis. In this case, the treatment included colistin and amikacin. The fourth isolate was detected in a pus sample from a male patient with a compound comminuted fracture of the tibia that developed a surgical site infection [27].
In mid-2012, a case of a patient from Slovakia with NDM-1 P. aeruginosa was reported [28]. The strain was isolated from a 44-year-old patient who was hospitalized in the intensive care unit of the University Hospital in Bratislava, due to acute respiratory failure. The patient’s condition worsened and respiratory support was necessary, as he developed pneumonia. The causative agent of ventilator-associated pneumonia was Acinetobacter baumannii, according to the result of a bronchoalveolar lavage culture. During his hospitalization in the ICU, NDM-producing P. aeruginosa and Enterococcus faecalis were isolated from his blood culture. Ιn 2012, NDM-producing P. aeruginosa was also found in France. This strain was detected in a urine culture of a 63-year-old woman who was admitted to the military hospital in Bégin because of complicated pyelonephritis. It is noteworthy that the patient was hospitalized in Serbia three months before her admission [29]. A few months later, in late May 2013, the first NDM-producing P. aeruginosa was reported from Italy [30]. A 40-year-old man diagnosed with acute lymphoblastic leukemia in first remission was admitted to a hematology unit in Rome in order to undergo stem cell transplantation. Interestingly, the patient had been hospitalized in December 2012 in Belgrad, Serbia. Fifteen days after transplantation, the patient was febrile and neutropenic, and the blood cultures taken on that day were positive for P. aeruginosa. Two days later, the patient rapidly deteriorated and was admitted to the intensive care unit, where he died a few hours later due to septic shock. The isolate was afterwards found to be an NDM producer. The first autochthonous Italian NDM-encoding P. aeruginosa was isolated in August 2019. At that time, a 77-year-old female patient, who lived in a rehabilitation center, was admitted to Bari’s hospital because she suffered from chronic respiratory failure. During her hospitalization, she developed a urinary tract infection, and P. aeruginosa was isolated in the urine culture. The patient did not receive any antibiotics, and three days later she became febrile. At that point, P. aeruginosa was also isolated in her blood cultures. The patient’s condition deteriorated rapidly, and she finally died of sepsis in November 2019 [31].
After their emergence in Greece [32] and for more than 20 years, carbapenemase-producing P. aeruginosa harbored exclusively VIM-type enzymes, most often those of the VIM-2 family [33]. In 2023, NDM-producing P. aeruginosa were reported for the first time from Larisa [34]. The first isolate was recovered on 16 May 2023 from the bronchial secretions of a female patient with previous hospitalizations in various Greek ICUs before her admission at the University Hospital of Thessaly in April 2023. Eight more cases with P. aeruginosa producing NDM carbapenemases were detected in that Hospital during the same time period. There are many important similarities between our characterized isolate 91845 and those reported from Larisa. First, all characterized isolates in Greece belong to the epidemic high-risk international clone ST308. Second, they all present a high identity percentage with the reference blaNDM-1 human isolate from Singapore and, third, the NDM-encoding gene seems to be located in the chromosome.
In the past, another case of rapid reversal in carbapenemase epidemiology for yet another species, namely K. pneumoniae, has also occurred in Greece. More specific, carbapenem-resistant Klebisella pneumoniae KPC changed mostly to the VIM type rapidly after the introduction of ceftazidime/avibactam into clinical practice as a novel agent that effectively inhibited the action of KPC enzymes [35]. In P. aeruginosa, the reason is clearly not the introduction of a new antimicrobial and may rather be attributed to the ST308 potential for rapid spread.
The predominance of NDM-producing versus VIM-producing P. aeruginosa is worsening the already difficult situation regarding the treatment of the respective infections. VIM-type enzymes hydrolyze almost all β-lactams except aztreonam and cefiderocol, whereas NDM-type enzymes hydrolyze β-lactams including aztreonam. Even though cefiderocol is not yet introduced in Greece, the spread of NDM-producing P. aeruginosa could influence its success rates a priori. Indeed, resistance to cefiderocol in P. aeruginosa has been observed up to now only among some NDM-producing isolates. Ceftolozane/tazobactam, ceftazidime/avibactam, imipenem/relebactam, and meropenem/vaborbactam are not active against both VIM and NDM carbapenemases because the metallo-β-lactamases are not inactivated by the novel β-lactamase inhibitors. When active, amikacin and fosfomycin could be used to treat effectively urinary tract infections [36,37]. In our hospital epidemiology, however, resistance to these drugs among carbapenemase-producing P. aeruginosa is common. Thus, not surprisingly, genes encoding for aminoglycoside-modifying enzymes and fosfomycin resistance have been detected by the Hybrispot and NGS methods in our study. Polymyxin resistance is rare for carbapenemase-producing P. aeruginosa, and this applies in our case as well. Our isolates are susceptible to colistin by both Vitek2 and the broth microdilution method. Colistin, however, is a formerly abandoned antibiotic that presents nephrotoxicity issues, and its actual clinical usefulness has been widely questioned [38].
Our data suggest that the departments that were first affected by NDM-producing P. aeruginosa were, in chronological order, the internal medicine department A, ophthalmology, neurosurgery, and surgery department B. These departments are located far from one another inside the hospital; nevertheless, connections between them (by moving personnel for, e.g., physiotherapy) can neither be excluded nor verified retrospectively. Additionally, our institution is a tertiary care hospital admitting patients from numerous acute- and long-term care facilities, and as our colleagues from Larisa, Greece, suggested, NDM-producing P. aeruginosa was already circulating in Northern Greece [34]. That being said, we expect to have had multiple index patients; therefore, it would not seem safe to draw conclusions upon intrahospital dissemination.
The present study has some limitations that should be noted. Due to economic restrictions, only one of the study’s isolates was sequenced. The molecular characterization of all isolates would have shed more light on the dynamics of the blaNDM spread among P. aeruginosa in our hospital. More molecular data analysis could evidence a probable clonal expansion and maybe provide more information about the interactions and interplay between VIM-producing and NDM-producing strains, since the first NDM + VIM producer has already been recovered in our hospital (isolate 214929). Depending on how the present situation may develop, this could be an interesting issue for future research.
4. Materials and Methods
4.1. Hospital Setting and Patient Data
The study was conducted in AHEPA University Hospital, which is located in Northern Greece and has a 700-bed capacity. All isolates included in this study were collected as part of the standard of care protocol. Data of the patients were retrieved from the hospital’s electronic database.
4.2. Study Sample
All carbapenem-resistant (resistant to both imipenem and meropenem) P. aeruginosa isolates recovered from all hospital wards between January 2023 and February 2024 were tested for carbapenemase production with the NG-Test CARBA 5 (NG-Biotech Laboratoires, Guipry-Messac, France) immunochromatography assay. In cases of multiple isolations per patient, only one isolate was included in the study. Moreover, the results of the first NDM-1 producer (91845), the first NDM and VIM producer (214929), and of six more isolates presenting weak lines in immunochromatography were confirmed by the HybriSpot antimicrobial resistance direct flow chip (AMR) (Máster Diagnóstica, Granada, Spain) molecular assay. Additionally, the first NDM-1-producing P. aeruginosa (91845) isolated from a blood culture was subjected to whole-genome sequencing (WGS) and bioinformatic analysis.
4.3. Bacterial Identification and Susceptibility Testing
Bacterial identification was carried out by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry technology (Bruker Daltonics, Bremen, Germany), and antimicrobial susceptibility testing was performed using the Vitek2 system (bioMérieux, Marcy l’Étoile, France). Additionally, confirmatory susceptibility testing was accomplished, where applicable, by using the MICRONAUT-S MDR MRGN-screening system (Bruker Daltonics GmbH & Co. KG, Bremen, Germany). For antimicrobial susceptibility results interpretation purposes, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2023 clinical breakpoints (v 13.0) were applied.
4.4. NG-Test CARBA 5
The NG-Test CARBA 5 (NG-Biotech Laboratoires, Guipry-Messac, France) is a rapid visual multiplex immunochromatographic assay for the qualitative detection and differentiation of five major carbapenemases from carbapenem-resistant bacterial colonies. The assay consists of one cassette that includes specific areas for the detection of KPC, OXA-48-like, VIM, IMP, and NDM enzymes together with a specific control (C) area. The assay was performed according to the manufacturer’s instructions, and results were interpreted visually at 15 min after incubation in room temperature.
4.5. Hybrispot Antimicrobial Resistance Direct Flow Chip (AMR)
The Hybrispot antimicrobial resistance direct flow chip (AMR) (Máster Diagnóstica, Granada, Spain) is a microarray-based assay for the in vitro detection of multiple antibiotic resistance genes. More specifically, the assay allows the simultaneous detection of 54 antibiotic resistance genetic markers associated with multi-drug-resistant organisms, such as carbapenem-resistant Gram-negative bacteria, extended-spectrum beta-lactamase (ESBL) producers, vancomycin-resistant Enterococci (VRE), and methicillin-resistant Staphylococcus aureus (MRSA). The assay’s markers represent the major gene families including SHV, CTX-M, GES, SME, KPC, NMC/IMI, SIM, GIM, SPM, NDM, VIM, IMP, OXA23-like, OXA24-like, OXA48-like, OXA51-like, OXA58-like, MecA, VanA, and VanB. Additionally, AMR is able to identify P. aeruginosa, Acinetobacter baumannii, E. coli, Klebsiella pneumoniae, and S. aureus.
4.6. Whole-Genome Sequencing and Bioinformatics Analysis
Whole-genome sequencing was performed with theDNBSEQ-G99 high-throughput Sequencing Set based on MGI’s core DNBSEQTM sequencing technology (MGI Tech, Shenzhen, China). Raw sequencing reads were quality-checked and trimmed using fastp [39]. The draft genome was assembled using skesa 2.5.1 [40].
The antimicrobial resistant determinants, the sequence type, and plasmid replicons were detected by Staramr version 0.9.1 [41], which integrates diverse molecular profiling tools including CGE’s multilocus sequence typing [42], Resfinder [43], and PlasmidFinder [44]. To further determine whether a contig originates from a chromosome or a plasmid source of the blaNDM-containing contig, plasmid prediction and construction tools, such as PlasmidSPAdes [45], PLASme [46], and MOB-suite [47], were applied. LASTZ [48] identified highly homologous sequences against reference bla-NDM-1 assembly from the reference Singapore isolate and constructed the pairwise alignment mappings between the two isolates using Proksee [49]. Plasmid sequences were further annotated based on the presence of AMR genes by CARD Resistance Gene Identifier v.5.2.1 [50] and protein families that are linked to the integration/excision, replication/recombination/repair, transfer, and stability/transfer/defense processes of the plasmids bymobileOG-db [51]. FastANI [52] was used to calculate the genome-wide average nucleotide identity against closely related blaNDM-containing P. aeruginosa strains.
5. Conclusions
An ongoing spread of NDM-producing P. aeruginosa is taking place in Greece, where VIM-encoding P. aeruginosa strains are already endemic. The intensification of the existing infection control policies seems to be the only realistic measure to tackle the widespread dissemination of yet another potential threat to public health.
Author Contributions
E.P. contributed to the conception, supervision, drafted the manuscript, and critically revised the manuscript; G.M. contributed to the conception, design, data acquisition and interpretation, drafted the manuscript, and critically revised the manuscript; N.V. contributed to data acquisition and interpretation and drafted the manuscript; A.M. contributed to the laboratory investigation, data acquisition, data interpretation, and drafted the manuscript; A.T. contributed to data acquisition and interpretation; C.K. contributed to data acquisition; A.D. contributed to data acquisition; P.M. contributed to data acquisition and interpretation; L.S. contributed to supervision, interpretation, and critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The publication of the present data was approved by the AHEPA University Hospital bioethics committee (protocol number: 297/14.6.2024).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- WHO. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Antimicrobial Resistance Surveillance in Europe 2023–2021 Data; European Centre for Disease Prevention and Control and World Health Organization: Stockholm, Sweden, 2023.
- Meletis, G.; Exindari, M.; Vavatsi, N.; Sofianou, D.; Diza, E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia 2012, 16, 303–307. [Google Scholar]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbio. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Ismail, S.; Mahmoud, S. First detection of New Delhi metallo-β-lactamases variants (NDM-1, NDM-2) among Pseudomonas aeruginosa isolated from Iraqi hospitals. Iran. J. Microbiol. 2018, 10, 98–103. [Google Scholar] [PubMed]
- Giakkoupi, P.; Petrikkos, G.; Tzouvelekis, L.S.; Tsonas, S.; Legakis, N.J.; Vatopoulos, A.C.; WHONET Greece Study Group. Spread of integron-associated VIM-type metallo-beta-lactamase genes among imipenem-non-susceptible Pseudomonas aeruginosa strains in Greek hospitals. J. Clin. Microbiol. 2003, 41, 822–825. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsakris, A.; Poulou, A.; Kristo, I.; Pittaras, T.; Spanakis, N.; Pournaras, S.; Markou, F. Large dissemination of VIM-2-metallo-{beta}-lactamase-producing Pseudomonas aeruginosa strains causing health care-associated community-onset infections. J. Clin. Microbiol. 2009, 47, 3524–3529. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meletis, G.; Vavatsi, N.; Exindari, M.; Protonotariou, E.; Sianou, E.; Haitoglou, C.; Sofianou, D.; Pournaras, S.; Diza, E. Accumulation of carbapenem resistance mechanisms in VIM-2-producing Pseudomonas aeruginosa under selective pressure. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Pappa, O.; Kefala, A.M.; Tryfinopoulou, K.; Dimitriou, M.; Kostoulas, K.; Dioli, C.; Moraitou, E.; Panopoulou, M.; Vogiatzakis, E.; Mavridou, A.; et al. Molecular Epidemiology of Multi-Drug Resistant Pseudomonas aeruginosa Isolates from Hospitalized Patients in Greece. Microorganisms 2020, 24, 1652. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Cantόn, R.; Akόva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. European Network on Carbapenemases. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef]
- Johnson, A.P.; Woodford, N. Global spread of antibiotic resistance: The example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 2013, 62, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, K.; Toleman, M.; Walsh, T.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Tychala, A.; Meletis, G.; Mantzana, P.; Kassomenaki, A.; Katsanou, C.; Daviti, A.; Kouroudi, L.; Skoura, L.; Protonotariou, E. Replacement of the Double Meropenem Disc Test with a Lateral. Flow Assay for the Detection of Carbapenemase-Producing Enterobacterales and Pseudomonas aeruginosa in Clinical Laboratory Practice. Antibiotics 2023, 12, 771. [Google Scholar] [CrossRef]
- Oliver, A.; Rojo-Molinero, E.; Arca-Suarez, J.; Beşli, Y.; Bogaerts, P.; Cantón, R.; Cimen, C.; Croughs, P.D.; Denis, O.; Giske, C.G.; et al. Pseudomonas aeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: Update from ESGARS-ESCMID/ISARPAE Group. Clin. Microbiol. Infect. 2024, 30, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Voulgari, E.; Gartzonika, C.; Vrioni, G.; Politi, L.; Priavali, E.; Levidiotou-Stefanou, S.; Tsakris, A. The Balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J. Antimicrob. Chemother. 2014, 69, 2091–2097. [Google Scholar] [CrossRef]
- Protonotariou, E.; Meletis, G.; Pilalas, D.; Mantzana, P.; Tychala, A.; Kotzamanidis, C.; Papadopoulou, D.; Papadopoulos, T.; Polemis, M.; Metallidis, S.; et al. Polyclonal Endemicity of Carbapenemase-Producing Klebsiella pneumoniae in ICUs of a Greek Tertiary Care Hospital. Antibiotics 2022, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G.; Malousi, A.; Tychala, A.; Kassomenaki, A.; Vlachodimou, N.; Mantzana, P.; Metallidis, S.; Skoura, L.; Protonotariou, E. Probable Three-Species In Vivo Transfer of blaNDM-1 in a Single Patient in Greece: Occurrence of NDM-1-Producing Klebsiella pneumoniae, Proteus mirabilis, and Morganella morganii. Antibiotics 2023, 20, 1206. [Google Scholar] [CrossRef]
- Villegas, M.V.; Lolans, K.; Correa, A.; Kattan, J.N.; Lopez, J.A.; Quinn, J.P.; Colombian Nosocomial Resistance Study Group. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 2007, 51, 1553–1555. [Google Scholar] [CrossRef]
- Watanabe, M.; Iyobe, S.; Inoue, M.; Mitsuhashi, S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1991, 35, 147–151. [Google Scholar] [CrossRef]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef]
- Toleman, M.A.; Simm, A.M.; Murphy, T.A.; Gales, A.C.; Biedenbach, D.J.; Jones, R.N.; Walsh, T.R. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: Report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 2002, 50, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Song, W.; Park, M.J.; Jeong, S.; Lee, N.; Jeong, S.H. Molecular Characterization of the First Emerged NDM-1-Producing Pseudomonas aeruginosa Isolates in South Korea. Microb. Drug Resist. 2021, 27, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The Intriguing Carbapenemases of Pseudomonas aeruginosa: Current Status, Genetic Profile, and Global Epidemiology. Yale J. Biol. Med. 2022, 22, 507–515. [Google Scholar]
- Jovcic, B.; Lepsanovic, Z.; Suljagic, V.; Rackov, G.; Begovic, J.; Topisirovic, L.; Kojic, M. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 2011, 55, 3929–3931. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, A.; Praharaj, A.K.; Kumar, M.; Grover, N. Emergence of NDM-1 in the Clinical Isolates of Pseudomonas aeruginosa in India. J. Clin. Diagn. Res. 2013, 7, 1328–1331. [Google Scholar] [CrossRef] [PubMed]
- Kulkova, N.; Babalova, M.; Sokolova, J.; Krcmery, V. First report of New Delhi metallo-β-lactamase-1-producing strains in Slovakia. Microb. Drug Resist. 2015, 21, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Janvier, F.; Jeannot, K.; Tessé, S.; Robert-Nicoud, M.; Delacour, H.; Rapp, C.; Mérens, A. Molecular characterization of blaNDM-1 in a sequence type 235 Pseudomonas aeruginosa isolate from France. Antimicrob Agents Chemother. 2013, 57, 3408–3411. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Fortini, D.; Galetti, R.; Garcia-Fernandez, A.; Nardi, G.; Orazi, D.; Capone, A.; Majolino, I.; Proia, A.; Mariani, B.; et al. Isolation of NDM-1-producing Pseudomonas aeruginosa sequence type ST235 from a stem cell transplant patient in Italy, May 2013. Eur. Surveill. 2013, 18, 20633. [Google Scholar] [CrossRef]
- Loconsole, D.; Accogli, M.; Monaco, M.; Grosso, M.; De Robertis, A.L.; Morea, A.; Capozzi, L.; Del Sambro, L.; Simone, A.; De Letteriis, V.; et al. First detection of autochthonous extensively drug-resistant NDM-1 Pseudomonas aeruginosa ST235 from a patient with bloodstream infection in Italy, October 2019. Antimicrob. Resist. Infect. Control 2020, 9, 73. [Google Scholar] [CrossRef]
- Tsakris, A.; Pournaras, S.; Woodford, N.; Palepou, M.F.; Babini, G.S.; Douboyas, J.; Livermore, D.M. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 2000, 38, 1290–1292. [Google Scholar] [CrossRef]
- Mavroidi, A.; Tsakris, A.; Tzelepi, E.; Pournaras, S.; Loukova, V.; Tzouvelekis, L.S. Carbapenem-hydrolysing VIM-2 metallo- beta-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 2000, 46, 1041–1042. [Google Scholar] [CrossRef]
- Tsilipounidaki, K.; Gkountinoudis, C.; Florou, Z.; Fthenakis, G.; Miriagou, V.; Petinaki, E. First Detection and Molecular Characterization of Pseudomonas aeruginosa blaNDM-1 ST308 in Greece. Microorganisms 2023, 11, 2159. [Google Scholar] [CrossRef]
- Papadimitriou-Olivgeris, M.; Bartzavali, C.; Lambropoulou, A.; Solomou, A.; Tsiata, E.; Anastassiou, E.D.; Fligou, F.; Marangos, M.; Spiliopoulou, I.; Christofidou, M. Reversal of carbapenemase-producing Klebsiella pneumoniae epidemiology from blaKPC- to blaVIM-harbouring isolates in a Greek ICU after introduction of ceftazidime/avibactam. J. Antimicrob. Chemother. 2019, 1, 2051–2054. [Google Scholar] [CrossRef] [PubMed]
- López Montesinos, I.; Gómez-Zorrilla, S.; Palacios-Baena, Z.R.; Prim, N.; Echeverria-Esnal, D.; Gracia, M.P.; Montero, M.M.; Durán-Jordà, X.; Sendra, E.; Sorli, L.; et al. Aminoglycoside or Polymyxin Monotherapy for Treating Complicated Urinary Tract Infections Caused by Extensively Drug-Resistant Pseudomonas aeruginosa: A Propensity Score-Adjusted and Matched Cohort Study. Infect. Dis. Ther. 2022, 11, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Samonis, G.; Maraki, S.; Karageorgopoulos, D.E.; Vouloumanou, E.K.; Falagas, M.E. Synergy of fosfomycin with carbapenems, colistin, netilmicin, and tigecycline against multidrug-resistant Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa clinical isolates. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 695–701. [Google Scholar] [CrossRef]
- Montero, M.; Horcajada, J.P.; Sorlí, L.; Alvarez-Lerma, F.; Grau, S.; Riu, M.; Sala, M.; Knobel, H. Effectiveness and safety of colistin for the treatment of multidrug-resistant Pseudomonas aeruginosa infections. Infection 2009, 37, 461–465. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4. 0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. PlasmidSPAdes: Assembling plasmids from whole genome sequencing data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Shang, J.; Ji, Y.; Sun, Y. PLASMe: A tool to identify PLASMid contigs from short-read assemblies using transformer. Nucleic Acids Res. 2023, 51, 83. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, A.M.; Liljeberg, P.; Plosila, J.; Tenhunen, H. LastZ: An Ultra Optimized 3D Networks-on-Chip Architecture. In Proceedings of the 2011 14th Euromicro Conference on Digital System Design: Architectures, Methods and Tools, DSD 2011, Oulu, Finland, 31 August–2 September 2011; pp. 173–180. [Google Scholar]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, 517–525. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. MobileOG-db: A manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, 99122. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).