Abstract
Staphylococcus spp. poses a significant threat to human and animal health due to their capacity to cause a wide range of infections in both. In this study, resistance genes conferring antibiotic resistance in Staphylococcus spp. and Mammaliicoccus sciuri isolates from humans and poultry in Edo state, Nigeria, were investigated. In April 2017, 61 Staphylococcus spp. isolates were obtained from urine, wounds, nasal and chicken fecal samples. Species identification was carried out by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Antimicrobial susceptibility testing was performed using the Kirby-Bauer method for 16 antibiotics. Whole-genome sequencing was used for characterization of the isolates. The 61 investigated isolates included Staphylococcus aureus, S. arlettae, M. sciuri, S. haemolyticus, and S. epidermidis. A total of 47 isolates (77%) belonged to human samples and 14 (23%) isolates were collected from poultry samples. All were phenotypically resistant to at least three antimicrobial(s). Multiple resistance determinants were detected in the human and poultry isolates analyzed. Phylogenetic analysis revealed close relatedness among the isolates within each species for S. arlettae, M. sciuri, and S. haemolyticus, respectively. This study delivered comprehensive genomic insights into antibiotic-resistant Staphylococcus species and M. sciuri isolates from human and poultry sources in Edo state, Nigeria, from a One Health perspective.
1. Introduction
Bacteria of the genus Staphylococcus can be found widely in the environment and are also isolated from humans and various animal species, including poultry. The genus Staphylococcus includes 72 validly published species (https://lpsn.dsmz.de/search?word=Staphylococcus, accessed on 13 June 2024). A previous report shows that pathogenic species include coagulase-positive staphylococci, such as S. aureus, S. intermedius, S. lutrae, and S. delphini and some strains of the species S. hyicus [1,2]. Among those, Staphylococcus aureus is one of the most important species able to cause hospital, community and farm-acquired infections among animals and human populations. S. aureus has also been reported as the third most important worldwide cause of food-borne infections [3]. Other Staphylococcus species, which are coagulase-negative (CoNS), can be implicated in nosocomial infections as well, and they are also detected as uncommon food-poisoning causative agents [4]. Various previous researches on CoNS showed the increasing medical importance of this Staphylococcus subtype in humans [5], including species such as S. haemolyticus, S. epidermidis or S. saprophyticus, and its role in the spread of antimicrobial resistance genes (ARGs) [6]. CoNS possess fewer virulence factors that participate in the pathogenesis of infection when compared with S. aureus, but recently, CoNS have emerged as common causes of nosocomial infections. In addition, increasing rates of antibiotic resistance have been detected in CoNS, in some cases even greater than for S. aureus, which limits the therapeutic options available [7]. In poultry, the most relevant CoNS with poultry comprise: S. xylosus, S. sciuri and S. cohnii [8]. Regarding staphylococci, several studies report on the isolation of these bacteria, especially on S. aureus from humans in Nigeria. However, few studies are available about Staphylococcus spp. obtained from poultry and poultry food products intended for human consumption, especially for developing countries like Nigeria.
Previous reports showed that antimicrobial resistance increases the severity of food-borne infections [9] and other diseases in humans as well as animals [10,11]. The extended use of antimicrobials in livestock, including poultry, has increased the ability of Staphylococcus spp. to acquire a plethora of resistance genes [12].
Staphylococcus spp., especially S. aureus strains, are known to produce β-lactamases and to acquire and disseminate different types of resistance genes through mobile genetic elements, plasmids, and transposons, playing an important role in the emergence of multiple drug resistance [13]. In human and veterinary medicine, methicillin-resistant S. aureus (MRSA) is considered a pathogen of relevance for public health since it is the source of infections associated with a high rate of mortality worldwide [14]. Moreover, the World Health Organization (WHO) has classified MRSA strains as “high priority 2 pathogens”. Within the definition of MRSA is included the non-susceptibility of S. aureus strains to at least one antimicrobial in three or more antibiotic categories. Presenting resistance to oxacillin or cefoxitin also induces resistance against other β-lactams [15]. The spread of antimicrobial resistance, especially among CoNS in healthy poultry, is a global health concern for human and animal health [6].
To determine the role of Staphylococcus species in disease processes and to evaluate the treatment options, it is important to correctly identify resistant strains. In this study, we assessed the prevalence of resistant Staphylococcus species in humans and poultry in Edo state, Nigeria, and characterized them using whole-genome sequence-based typing.
2. Results
2.1. Identification of Staphylococcus spp. Including S. (Mammaliicoccus) sciuri
Sixty-one isolates were confirmed as Staphylococcus spp., including eight M. sciuri isolates, by MALDI-TOF MS and/or rMLST. Whole genome-based taxonomic analysis performed using DNA-DNA hybridization (dDDH) and Tetra Correlation Search (TCS) confirmed the identity of some of the isolates. A total of 24 of the 61 confirmed isolates were identified as S. aureus, 11 as S. haemolyticus, 8 as M. sciuri, 7 as S. arlettae, 6 as S. epidermidis, 2 as S. saprophyticus, 2 as S. ureilyticus and 1 as S. xylosus (Figure 1). Moreover, 30 of the 61 identified isolates originated from nasal swabs of healthy students, 14 isolates were from healthy chicken fecal samples, 10 isolates were from urine of healthy students and 7 originated from clinical wound samples. In addition, 3 out of 21 S. aureus isolates were categorized as resistant/multidrug-resistant, with none of the isolates identified as MRSA based on the absence of mecA. The resistant/MDR isolates also included M. sciuri (100%), S. epidermidis (2/6; 33%), S. arlettae (100%), S. haemolyticus (10/11; 91%), S. ureilyticus (100%), S. xylosus (100%) and S. saprophyticus (1/2; 50%). The resistant/multidrug resistant (MDR) isolates originated from chicken fecal samples (8/14; 57%), nasal swab samples (10/30; 33%), urine samples (9/10; 90%) and clinical wound samples (n = 7, 100%).
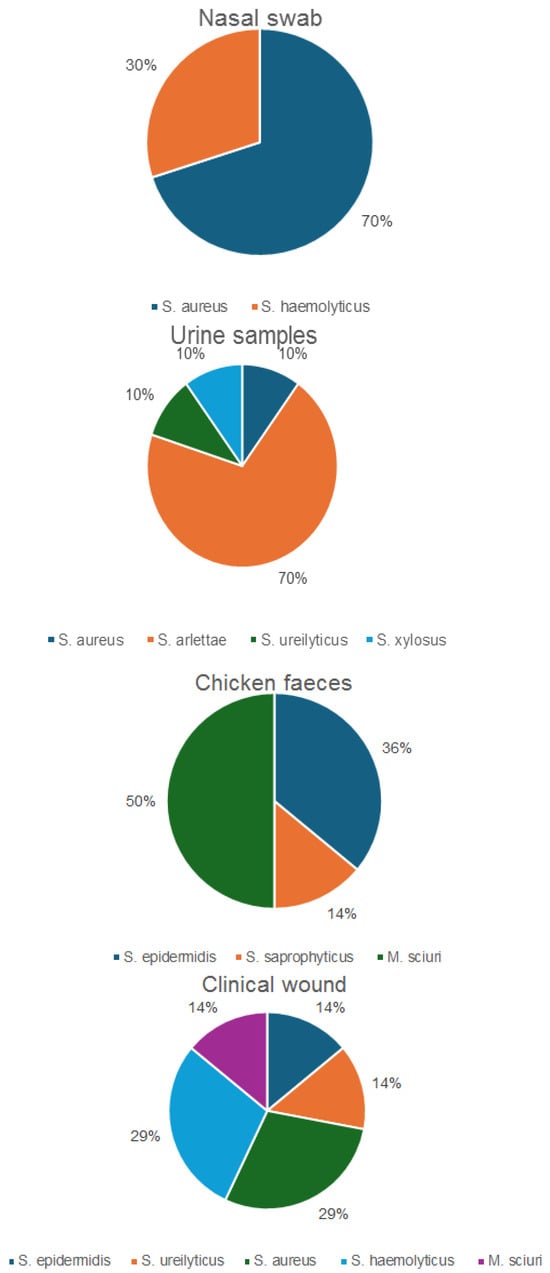
Figure 1.
Distribution of the confirmed Staphylococcus spp. isolates, including M. sciuri.
2.2. Antimicrobial Susceptibility
All the Staphylococcus spp. isolates (n= 26) tested were resistant to penicillin except M. sciuri (n = 7 out of 8 isolates, 87.5%). Moreover, 8 out of 10 isolates (80%) of S. haemolyticus were resistant to cefoxitin. Resistance to cefoxitin was also detected among M. sciuri (3 out of 8 isolates, 37.5%) and S. aureus (1 out of 3 isolates, 33%). The percentages of resistant Staphylococcus spp., including M. sciuri isolates, are shown in Table 1. All the tested isolates were susceptible to linezolid and amikacin. Only S. xylosus was 100% sensitive to trimethoprim. S. epidermidis was also 100% sensitive to erythromycin and clindamycin. All the isolates of S. arlettae, S. ureilyticus and S. epidermidis were sensitive to moxifloxacin and ciprofloxacin. S. ureilyticus, S. arlettae and S. xylosus were 100% sensitive to gentamicin (Table 1). The MIC of 12 cefoxitin-resistant isolates confirmed the resistant/multi-resistant phenotype of the isolates (Supplementary Table S1). Twenty-three multi-resistance phenotypes were observed among the resistant isolates. The resistance profiles of the isolates are shown in Table 2. Two M. sciuri isolates from chicken feces and an S. haemolyticus isolate from a wound had profiles with the highest number of antibiotics they showed resistance to (Table 2 and Table 3).

Table 1.
Percentage of resistant Staphylococcus spp., including M. sciuri isolates.

Table 2.
Resistance profile of the Staphylococcus spp., including M. sciuri isolates.

Table 3.
Resistance phenotypes in the Staphylococcus spp., including M. sciuri isolates.
2.3. Antimicrobial Resistance Genes and SCCmec Element
None of the S. aureus isolates carried the methicillin resistance gene mecA. All the isolates had the tetracycline resistance gene tet38, 67% β-lactam resistance gene blaZ coexisting with blaI, blaR1, 67% dfrG gene and 33% fosB (Supplementary Table S1). Multiple resistance determinants were detected in the human and poultry isolates analyzed, which included aac(6′)-Ie/aph(2″)-Ia, mecA, mecA1, fosB, ermC, mphC tet(38), tetK, and dfrG resistance genes. Details of the resistance genes detected in the human and poultry isolates are shown in Supplementary Table S1.
The SSCmec_type_III(3A) containing the type 3 ccr gene complex(ccrA3B3), the regulatory genes mecR1 and mecI that control the expression of methicillin resistance and the mecA gene were detected in two M. sciuri poultry isolates. The SSCmec type V, containing ccrC and mec complex C, was detected in 4 out of 10 (25%) characterized S. haemolyticus strains (Supplementary Table S1).
2.4. Multilocus Sequence Typing of the Staphylococcus spp. Isolates
Three different sequence types (ST1, ST15, ST669) were identified among the three S. aureus isolates. Two of the S. aureus isolates had different spa types, t084 and t127, respectively. One of the S. aureus isolate had an unknown spa type. Novel STs were detected in the S. epidermidis and M. sciuri isolates. One, two and four isolates of M. sciuri had the new sequence types ST222, ST223, and ST224, respectively. ST 195 was detected in an M. sciuri isolate. The two S. epidermidis isolates had a novel ST 1179. Four sequence types, ST56, ST30, ST54,and ST49, were identified among the S. haemolyticus isolates, with ST 56 (n = 5) predominating among them. No known sequence type was identified in the S. arlettae, S. ureilyticus, S. saprophyticus and S. xylosus isolates as no MLST schemes are available for them.
Four plasmid replicon types (rep5, rep7, rep16, rep24) were observed among the three S. aureus isolates. Eleven non-S. aureus isolates (one S. xylosus, six M. sciuri, three S. haemolyticus and one S. arlettae) did not have any plasmid replicon types. The S. saprophyticus isolate was characterized by the highest number of plasmid replicon types, comprising 5 out of 12 replicon types observed in this study (Supplementary Table S1). The predominant plasmid replicon type in the non-S. aureus isolates was rep7,which was detected in 11 from 31 non-S. aureus isolates examined. Six out of seven S. arlettae isolates had a replicon type (rep7). The two S. ureilyticus isolates differed in their replicon type composition, having only replicon type rep10 in common. The S. epidermidis isolates with a novel ST 1179 had identical plasmid replicon types, rep22 and rep39. Eight different replicon types (rep7, rep10, rep13, rep21, rep22, rep20, repUS22, rep24) were detected in 7 out of 10 S. haemolyticus isolates. The replicon type rep US12 was the only plasmid replicon type detected in the M. sciuri isolates.
2.5. Virulence Genes Extracted from S. aureus Genomes
The identified virulence genes in the three resistant S. aureus isolates included determinants encoding for surface cell-bound proteins of the microbial surface components recognizing adhesive matrix molecules (MSCRAMM) family recognizing adhesive matrix molecules (clfB, fnbA, isdA map), genes whose products are part of the immune evasion mechanisms (ads, coa, sbi, vWbp) and determinants encoding different toxins and extracellular enzymes (Supplementary Table S1). Genes involved in the formation of the polysaccharide matrix of biofilms (icaA-D, icaR), capsule biosynthesis (cap8) as well as regulatory proteins were also identified. The frequently occurring virulence genes found in all the S. aureus isolates were adsA, aur, cap8A-G, cap8L-P, ebp, esaAB, essAB, esaG6, esaG8, esaG9, esxA, geh, sbi, sspBC, LukD, hp, icaA-D, icaR, hld and hlgABC. The genes detected in the characterized S. aureus isolates also included sec, selk, selI-encoding staphylococcal enterotoxins, hly/hla, alpha hemolysin enterotoxin-encoding gene, lukD, leukocidin gene, hlb, β-hemolysin enterotoxin-encoding gene, genes encoding hemolysin (hld, hlgA-C), immune-modulatory factors (sak, chp), staphylococcal complement inhibitor (scn) and genes encoding invasive toxins (sak, hysA).
2.6. Genetic Comparison of the Staphylococcus spp. Isolates, Including M. sciuri Isolates
CgMLST analysis of the Staphylococcus aureus isolates (Figure 2) revealed more than 1305 alleles of difference between the three isolates. Based on the official cluster threshold (CT) of 24 allelic differences, no cluster was observed. As for the Staphylococcus haemolyticus isolates (n = 10) (Figure 3), a maximum of 710 allelic differences were identified across the MST. Three different clusters were obtained. Cluster 1 had three isolates (two isolates from nasal swabs of healthy students and one isolate from wounds) with similar plasmid incompatibility group rep 21 and ST-56. The hospital where the isolates from wounds were obtained is not in close proximity to the location where the nasal samples of the healthy individuals were collected. Cluster 2 had two isolates with sequence type ST-30 (one poultry isolate with plasmid incompatibility group rep 22 and a closely related isolate from nasal swabs of healthy students) with an allelic difference of 1.0. The farms where the poultry samples were obtained were in the same town as the healthy individuals. Cluster 3 included two closely related isolates with an allelic difference of 3.0 and identical ST-56 from nasal swabs of healthy students and wounds. Whole genome-based cgMLST analysis of the M. sciuri isolates (n = 8) revealed up to 1716 allelic differences across the MST (Figure 4). Based on the defined cluster threshold (CT) of 10 allelic differences, two different clusters were obtained. The isolates in cluster 1 (Figure 4) included one isolate from the nasal swabs of healthy students and three isolates from healthy poultry animals. The isolates with the novel sequence type ST-224 were closely related, with an allelic difference of 5.0. Cluster 2 consisted of two closely related isolates from healthy poultry (one allele). Regarding Staphylococcus arlettae, six out of seven isolates had a plasmid incompatibility group rep 7 (Figure 5). Overall, the seven isolates displayed up to 862 alleles of difference.
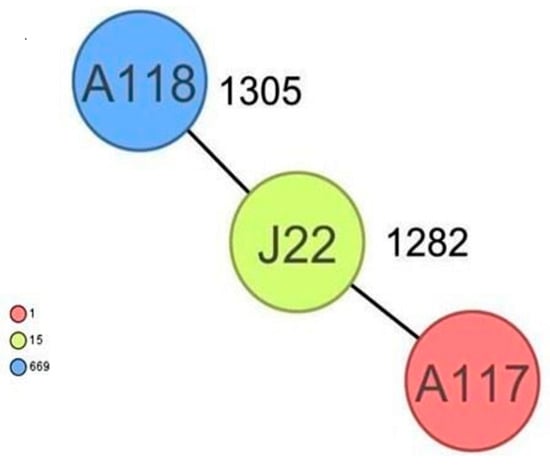
Figure 2.
Minimum spanning tree for 3 S. aureus isolates based on cgMLST of S aureus. Colors correspond to the sequence types of the isolates. Each circle represents isolates with an allelic profile based on the sequences of 1861 core genome targets.
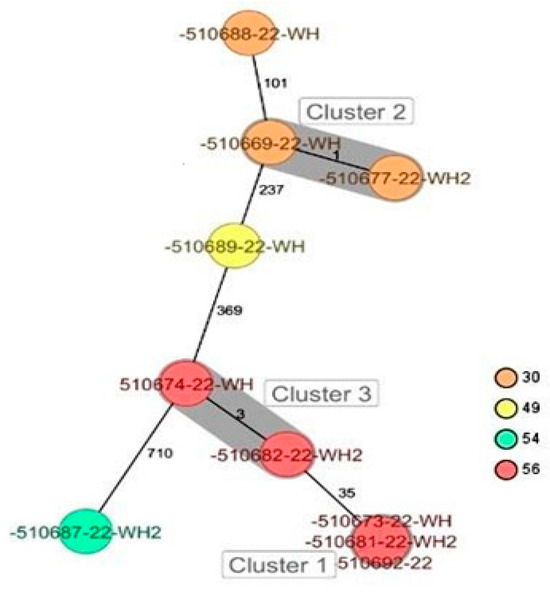
Figure 3.
Minimum spanning tree for 10 S. haemolyticus isolates based on cgMLST of S. haemolyticus. Colors correspond to the sequence types of the isolates. Each circle represents isolates with an allelic profile based on the sequences of 1721 core genome targets. Isolates with closely related genotypes were identified with a maximum of 3 allelic differences and are shaded in gray.
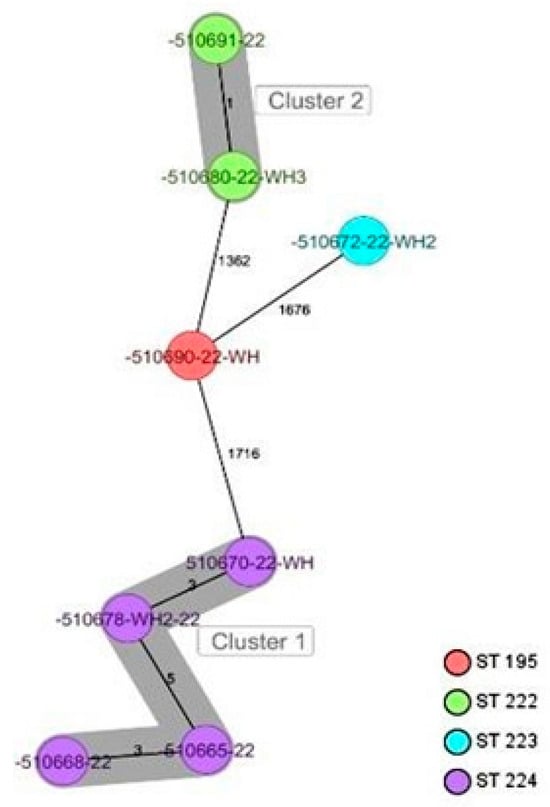
Figure 4.
Minimum spanning tree for 8 M. sciuri isolates based on cgMLST of M. sciuri. Colors correspond to the sequence types of the isolates. Each circle represents isolates with an allelic profile based on the sequences of 1923 core genome targets. Isolates with closely related genotypes were identified with a maximum of 5 allelic differences and are shaded in gray.
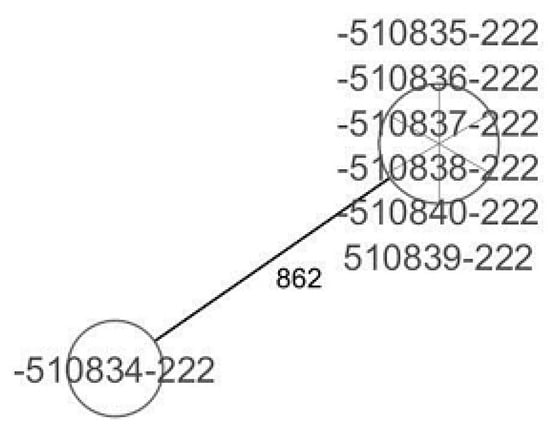
Figure 5.
Minimum spanning tree for 7 S. arlettae isolates based on cgMLST of S. arlettae. Each circle represents isolates with an allelic profile based on the sequences of 1930 core genome targets.
3. Discussion
This study aimed to identify antibiotic-resistant Staphylococcus spp. isolates from humans and poultry in Edo state, Nigeria, and to characterize the isolates using whole-genome sequencing (WGS). The results showed that MDR staphylococci are prevalent in samples originating from healthy humans, clinical human samples and poultry samples in Edo state, Nigeria. The detection of Staphylococcus spp. and M. sciuri isolates in this study is in agreement with previous reports on the presence of Staphylococcus aureus and CoNS in humans and livestock [16,17,18]. Previous reports of interspecies transmission between humans and livestock emphasize the importance of understanding host-specific antimicrobial resistance patterns for S. aureus and other Staphylococcus spp. to study their transmission to animals and humans [17,19,20]. Resistant S. aureus isolates were detected in samples from three humans (two wound swabs and one nasal swab) and none had the mecA gene (MRSA). This contrasts with other Nigerian studies and reports from other geographic regions of MRSA from human samples [21,22,23]. Two methicillin-sensitive Staphylococcus aureus (MSSA) (ST1 and ST15) identified in this study matched the findings of a previous study in this region in which these STs were also detected [24]. S. aureus ST669 has also been previously reported as MSSA [25].
The presence of virulence factors in the S. aureus isolates, such as surface proteins, biofilms, exoenzymes, exotoxins, and exfoliative toxins, is linked to the ability to cause different infections [26]. The presence of cell wall-associated adhesive molecules, such as fnb (encoding fibronectin-binding protein), detected in this study and the ability of S. aureus to successfully persist within the hospital and community is known to be responsible for the possibility of severe animal and human infections [27,28]. Another virulence determinant detected in this study was the icaA gene, encoding the N-acetylglucosamyl transferase responsible for intracellular adhesion [29,30].
Significantly, the S. haemolyticus and M. sciuri that were multidrug resistant and also positive for mecA/mecA1 in this study were isolated from both humans and poultry. Previous studies in Nigeria have found MRSA and CoNS largely among humans, with limited reports in poultry [21,31,32]. The level of antimicrobial resistance of Staphylococcus spp. to critically needed antimicrobial agents is a public health concern. The overall frequency of resistant/multidrug-resistant isolates in this study was 56%. The inappropriate use of antibacterial agents in Nigeria, both in humans and poultry, has been previously reported, which could explain the high frequency of bacterial resistance found to common antibacterial agents in this study [33,34,35]. One of the profiles that had the highest number of antibiotic resistance combinations was detected in M. sciuri isolates from chicken feces. M. sciuri has been primarily considered a bacterium associated with livestock, which can be found in large numbers in the farm environment [36,37]. Outside the farm environment, the colonizing population may be low, but they are found to readily adapt and persist in healthcare settings and thus may pose a threat to human health [38,39]. A previous report showed that the clinical relevance of M. sciuri is mainly attributed to its resistance to methicillin [40]. The consumption of poultry food/meat containing antibiotic-resistant Staphylococcus spp. may lead to food poisoning. Furthermore, the handling or ingesting of staphylococci-contaminated meat/food is a possible risk factor for colonization by methicillin-resistant staphylococci [41,42]. The results from this study show that the frequency of multidrug-resistant staphylococci, especially CoNS, in poultry is alarming and this may represent a public health problem.
Genomic analysis of the M. sciuri isolates showed close genetic relatedness between isolates recovered from humans and poultry from the same town, which suggests possible transmission between the different hosts and that the strains may not be host-specific. Regarding the plasmids, all harbored ARGs, and this agrees with previous studies on Staphylococcus spp. [17,43]. Furthermore, cgMLST analysis indicated a substantial genetic relatedness among the isolates within specific species, suggesting potential transmission pathways or shared resistance mechanisms. These findings underscore the urgent need for enhanced surveillance and targeted interventions to mitigate the spread of antimicrobial resistance in both human and veterinary medicine. Although the horizontal transmission of plasmids was not demonstrated in this study, the potential risks of transmission of plasmid-borne ARGs from M. sciuri to other Staphylococcus species has previously been reported [44,45]. This highlights the importance of genomic surveillance for ARG detection to avoid their plasmidic spread of AMR gene carriage on plasmids in S. aureus and coagulase negative staphylococci. The CoNS isolates in this study were observed to have varying plasmid content. The presence of ARGs on plasmids in CoNS isolated from human and poultry supports previous reports that CoNS may serve as reservoirs for the spread of AMR [17,46]. Dissemination of plasmids carrying multi-resistance genes will substantially limit the therapeutic efficacy of antibiotic agents and urgently warrants surveillance of staphylococci from both human and animal sources.
All the resistant S. haemolyticus in this study also carried the mecA gene. S. haemolyticus has been reported as an emerging multi-resistant nosocomial pathogen and may represent the most commonly isolated CoNS from blood cultures [47]. ST30 S. haemolyticus was detected in our work in two reservoirs (healthy humans and poultry), which suggests clonal transmission of the isolate between the two reservoirs. This clone has also been reported to be detected in blood stream and nosocomial infections, often showing vancomycin hetero-resistance thus confirming invasive characteristics of the CoNS clone [48].
SCCmec types I, II and III have previously been reported to be associated with MRSA strains associated with healthcare infections, while types IV and V are reported to be associated with livestock-associated infections [49,50]. The results of our work are in agreement with reports [51,52] that demonstrated that SCCmec type V with ccrC and mec complex C can be associated with S. haemolyticus strains isolated from certain geographical areas, including some African countries, such as Algeria and Mali. Also, SCCmec type III MRSA isolates have previously been reported to have high resistance to several antimicrobials [53], which is consistent with the results from this study. The two poultry M. sciuri isolates with the SCCmec type III(3A) element had a profile of high antimicrobial resistance.
4. Materials and Methods
4.1. Sample Collection and Processing
In April 2017, 130 samples from urine and 50 from the nose were taken from healthy students at the College of Pharmacy, Igbinedion University Okada. A healthy student was an individual who had a self-reported health status and/or an individual with no visible signs of illness after physical examination. Sampling was arbitrarily carried out, with no inclusion and exclusion criteria involved. Samples were sent immediately upon collection to the Department of Pharmaceutical Microbiology Laboratory for further analysis. In addition, previously identified Staphylococcus spp. wound isolates (n = 70) from inpatients and out-patients were obtained from the medical microbiology laboratory of the University of Benin Teaching Hospital, Edo state, Nigeria, during the same study period. Four poultry farms in Okada and Benin city, Edo state, were also visited once during April 2017 and 100 chicken fecal samples were collected and immediately sent to the Department of Pharmaceutical Microbiology Laboratory for further processing. Most samples were obtained from healthy subjects to assess the possible carriage of resistant Staphylococci spp. Samples were processed using standard microbiological techniques, as previously described [54]. A number of three to four different colonies per plate with different morphology were recovered and further investigated. Isolation of Staphylococcus spp. was achieved by inoculating samples on Mannitol salt agar plates (Oxoid, Hampshire, United Kingdom) and incubating them for 24 h at 37 °C. Sixty-one different colonies obtained from the agar plates were sub-cultured on blood agar plates (Columbia agar + 5% sheep blood) (BioMérieux, Marcy l’Etoile, France) to obtain pure colonies. Matrix-assisted laser desorption ionizationtime of flight (MALDI-TOF) mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany) analysis was used for species identification.
4.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was carried out on the 61 isolates identified by MALDI-TOF using first the KirbyBauer susceptibility testing technique [55] against 16 antibiotics: vancomycin, teicoplanin, linezolid, fusidic acid, cefoxitin, benzyl penicillin, amoxicillin-clavulanic acid, ciprofloxacin, amikacin, moxifloxacin, minocycline, gentamicin, erythromycin, clindamycin, trimethoprim, and rifampicin (Oxoid, Basingstoke Hampshire, UK).
The minimum inhibitory concentrations (MICs) for benzyl penicillin, ciprofloxacin, trimethoprim, cefoxitin, moxifloxacin, gentamicin, teicoplanin, vancomycin, amikacin, erythromycin, clindamycin, minocycline, and rifampicin were retrieved using E-test strips (BioMérieux Marcy L’Etoile, France and Liofilchem sr Zona industriale, Abruzzi, Italy). The MICs were determined for twelve isolates that were cefoxitin resistant in the Kirby-Bauer test. Those twelve cefoxitin-resistant isolates and another twenty-two arbitrarily selected isolates that were multidrug resistant, as previously defined [14], were selected for characterization by whole-genome sequencing. The results were interpreted using the European Committee on Antimicrobial Susceptibility Testing criteria [56].
4.3. Whole-Genome Sequencing
Whole-genome sequencing (WGS) was carried out as previously described [57] for 34 resistant/multidrug-resistant Staphylococcus spp. isolates. Briefly, high-molecular weight (HMW) DNA was extracted from the isolates using the MagAttract HMW DNA extraction kit (Qiagen, Hilden, Germany). Library preparations were produced using the Illumina Nextera XT DNA library preparation kit (Illumina Inc., San Diego, CA, USA) for a 2 × 300 bp paired-end sequencing on an Illumina Miseq sequencer (Miseq v3.0, Illumina Inc., San Diego, CA, USA). Samples were sequenced to achieve a minimum of 30-fold coverage. Genome assembly was carried out using SPAdes version 3.15.2 [58]. FastQC v0.11.7 was used for quality control of the assemblies. Read trimming was performed with Trimmomatic v0.36. Ribosomal multilocus sequence typing (rMLST) (https://pubmlst.org/species-id, accessed on 11 July 2022) was used to confirm the isolate species. Whole-genome-based taxonomic analysis was performed using DNADNA hybridization (dDDH) (https://tygs.dsmz.de, accessed on 11 July 2022) and Tetra Correlation Search (TCS) (http://jspecies.ribohost.com/jspeciesws, accessed on 11 July 2022) to confirm the species for some isolates. To determine the phylogenetic relationships between the isolates of the same species, the genomes were analyzed with Ridom Seqsphere+ v8.3.5 (Ridom, Münster, Germany) using core genome multilocus sequence typing (cgMLST) and conventional MLST.
A public cgMLST scheme was used for S. aureus [59], while ad hoc cgMLST schemes were generated for M. sciuri, S. haemolyticus and S. arlettae with 1923, 1721 and 1930 core genome targets, respectively. The official cluster distance threshold of 24 alleles was used to identify related S. aureus isolates. Clonal M. sciuri and S. haemolyticus isolates were identified with a cluster threshold of 10 alleles, respectively. “Good core genome targets” were defined based on the criteria described in [60]. Minimum spanning trees (MSTs) were generated to visualize the genetic relatedness between the isolates of the same species. The Spa types [61], sequence types (STs) [62], antimicrobial resistance genes and virulence genes [63] were extracted from the WGS data using Seqsphere+ v9.0.3 with the Comprehensive Antibiotic Resistance Database-Resistance Gene Identifier (CARD-RGI) [64], the NCBI AMRFinder database [65] and the VFDB database (https://dx.doi.org/10.1093%2Fnar%2Fgki008, accessed on 11 July 2022), respectively
New sequence types (STs) of S. epidermidis and M. sciuri were submitted to the respective PubMLST database (https://pubmlst.org/organisms/staphylococcus-epidermidis, accessed on 11 July 2022) [66] and (https://pubmlst.org/bigsdb?db=pubmlst_msciuri_seqdef, accessed on 11 July 2022) for curation. The staphylococcal chromosome cassette mec elements (SCCmec) typing of methicillin resistant isolates was retrieved using SCCmecFinder version 1.2 (https://cge.food.dtu.dk/services/SCCmecFinder/, accessed on 15 August 2023) [67]).
5. Conclusions
Our study provides a detailed report not only on multi-resistant S. aureus but also multi-resistant CoNS using WGS. A high frequency of multidrug-resistant CoNS was reported in this study. To the best of our knowledge, this study represents the first molecular characterization of Nigerian human and poultry CoNS isolates. CoNS isolates are increasingly recognized to cause clinically relevant nosocomial and community-acquired infections. The presence of the mecA gene and SCCmec element (a mobile genetic element) in CoNS (with varying sequence types and plasmid replicon types) from human and poultry in this study further elucidates the importance of periodical surveillance involving molecular typing and monitoring of antimicrobial resistance patterns in antibiotic stewardship programs, both in human and veterinary medicine. These would enhance the design of antibiotic prescription policies and hospital infection control strategies. Knowledge of the genetic relatedness among isolates within specific species that suggests transmission pathways/shared resistance mechanisms is important. It curtails the menace of antimicrobial drug resistance posed by these pathogens. In addition, the comparison of staphylococci genomes allowed the specific detection of virulent strains, especially CoNS, in both humans and poultry, which is important and useful in infection control.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics13080733/s1, Table S1: Antimicrobial resistance genes in Staphylococcus spp.
Author Contributions
Conceptualization: C.J.; methodology: C.J., T.O.O., J.U.A., A.A.A. and A.T.S.; investigation: C.J., B.D.-P. and A.S.; software, validation, formal analysis C.J., F.A., A.C.-R. and W.R.; writing original draft, preparation, writing, review and editing: C.J., review and editing: T.O.O., J.U.A., A.A.A., A.T.S., A.S., B.D.-P., F.A., A.C.-R. and W.R.; supervision: A.C.-R. and W.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an Ernst-Mach follow-up grant, Ernst Mach Worldwide 2021/2022 to C.J. by the Austrian Federal Ministry of Science, Research and Economy (BMWFW). This work was supported financially by the Austrian Agency for Health and Food Safety (AGES), Vienna, Austria.
Institutional Review Board Statement
The wound isolates were pre-identified; hence, research ethics approval was not required. No contact was produced with patients and the original samples from the hospital. Informed consent was not required by the institution. The isolate data were obtained from clinical records and anonymously handled. Approval from the Igbinedion university ethical committee was duly obtained for samples obtained from the healthy students. The document IUO/ethics/22/009 was initiated and obtained for the study.
Informed Consent Statement
All the participants were duly informed about the purpose and procedures of the study. Written informed consent from the students was obtained before samples were collected.
Data Availability Statement
This Whole-Genome Shotgun project has been deposited at the DDBJ/ENA/GenBank under the BioProject accession no. PRJNA863242. The version described in this paper is the first version. The raw sequence reads have been deposited in the sequence read archive (SRA).
Acknowledgments
We thank Petra Hasenberger and Silke Stadlbauer for their assistance in carrying out the work.
Conflicts of Interest
No conflicts of interest are declared.
References
- Pottumarthy, S.; Schapiro, J.M.; Prentice, J.L.; Houze, Y.B.; Swanzy, S.R.; Fang, F.C.; Cookson, B.T. Clinical isolates of Staphylococcus intermedius masquerading as methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2004, 42, 5881–5884. [Google Scholar] [CrossRef] [PubMed]
- Van Hoovels, L.; Vankeerberghen, A.; Boel, A.; Van Vaerenbergh, K.; De Beenhouwer, H. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 2006, 44, 4609–4612. [Google Scholar] [CrossRef] [PubMed]
- Achi, O.; Madubuike, C. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from retail ready to eat foods in Nigeria. J. Res. Microbiol. 2007, 2, 516–523. [Google Scholar]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed]
- Chajecka-Wierzchowska, W.; Zadernowska, A.; Nalepa, B.; Sierpińska, M.; Łaniewska-Trokenheim, L. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin—Phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015, 46, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.; Ziebuhr, W.; Becker, K. Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infect. 2019, 25, 1071–1080. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Agersal, Y.; Ahrens, P.; Jørgensen, J.C.Ø.; Madsen, M.; Jensen, L.B. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet. Microbiol. 2000, 74, 353–364. [Google Scholar] [CrossRef]
- Hashempour-Baltork, F.; Hosseini, H.; Shojaee-Aliabadi, S.; Torbati, M.; Alizadeh, A.M.; Alizadeh, M. Drug resistance and the prevention strategies in food borne Bacteria: An update review. Adv. Pharm. Bull. 2019, 9, 335–347. [Google Scholar] [CrossRef]
- Smith, R.; Coast, J. The true cost of antimicrobial resistance. BMJ 2013, 346, f1493. [Google Scholar] [CrossRef]
- Adenipekun, E.O.; Jackson, C.R.; Oluwadun, A.; Iwalokun, B.A.; Frye, J.G.; Barrett, J.B.; Hiott, L.M.; Woodley, T.A. Prevalence and Antimicrobial Resistance in Escherichia coli from Food Animals in Lagos, Nigeria. Microb. Drug Resist. 2015, 21, 358–365. [Google Scholar] [CrossRef]
- Otalu, O.; Junaidu, K.; Chukwudi, O.E.; Jarlath, U.V. Multi-drug resistant coagulase positive Staphylococcus aureus from live and slaughtered chickens in Zaria, Nigeria. Int. J. Poult. Sci. 2011, 10, 871–875. [Google Scholar] [CrossRef]
- McCallum, N.; Berger-Bachi, B.; Senn, M.M. Regulation ¨of antibiotic resistance in Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 118–129. [Google Scholar] [CrossRef]
- Tiemersma, E.W.; Bronzwaer, S.L.; Lyytikäinen, O.; Degener, J.E.; Schrijnemakers, P.; Bruinsma, N.; Monen, J.; Witte, W.; Grundmann, H. Methicillin-resistant Staphylococcus aureus in Europe, 1999–2002. Emerg. Infect. Dis. 2004, 10, 1627. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard defnitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Shittu, A.O.; Taiwo, F.F.; Frobose, N.J.; Schwartbeck, B.; Niemann, S.; Mellmann, A.; Schaumburg, F. Genomic analysis of Staphylococcus aureus from the West African Dwarf (WAD) goat in Nigeria. Antimicrob. Resist. Infect. Control 2021, 10, 122. [Google Scholar] [CrossRef]
- Egyir, B.; Dsani, E.; Owusu-Nyantakyi, C.; Amuasi, G.R.; Owusu, F.A.; Allegye-Cudjoe, E.; Addo, K.K. Antimicrobial resistance and genomic analysis of staphylococci isolated from livestock and farm attendants in Northern Ghana. BMC Microbiol. 2022, 22, 180. [Google Scholar] [CrossRef]
- Pimenta, L.K.L.; Rodrigues, C.A.; Filho, A.R.G.; Coelho, C.J.; Goes, V.; Estrela, M.; Priscila de Souza Avelino, M.A.G.; Vieira, J.D.G.; Carneiro, L. Staphylococcus spp. Causatives of Infections and Carrier of blaZ, femA, and mecA Genes Associated with Resistance. Antibiotics 2023, 12, 671. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3, e00305–e00311. [Google Scholar] [CrossRef] [PubMed]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.G.; Feil, E.J.; et al. Gene exchange drives the ecological success of a multihost bacterial pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef]
- Ibadin, E.E.; Enabulele, I.O.; Muinah, F. Prevalence of mecA gene among staphylococci from clinical samples of a tertiary hospital in Benin City, Nigeria. Afr. Health Sci. 2017, 17, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Karikari, A.B.; Frimpong, E.; Owusu-Ofori, A. Methicillin-resistant Staphylococcus aureus among patients in a teaching hospital in Ghana. Int. J. One Health 2017, 3, 46–49. [Google Scholar] [CrossRef]
- Sangeeta, R.; Lyndsey, L.; Roberta, M.; Linzy, J.; Doreene, R.H. Antimicrobial resistance and genetic diversity of Staphylococcus aureus collected from Livestock, poultry and Humans. One Health 2022, 15, 100407. [Google Scholar]
- Igbinosa, E.O.; Beshiru, A.; Igbinosa, I.H.; Ogofure, A.G.; Ekundayo, T.C.; Okoh, A.I. Prevalence, multiple antibiotic resistance and virulence profile of methicillin-resistant Staphylococcus aureus (MRSA) in retail poultry meat from Edo, Nigeria. Front. Cell Infect. Microbiol. 2023, 13, 1122059. [Google Scholar] [CrossRef]
- Washington, M.A.; Agee, W.A., III; Kajiura, L.; Staege, C.M.; Uyehara, C.F.; Barnhill, J.C. An Analysis of Staphylococcus aureus Infections at a Military Medical Center Using the PLEX-ID Combined Polymerase Chain Reaction-Mass Spectrometry System. Maj. Mil. Med. 2014, 179, 445. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.R.; Batistao, D.W.F.; Ribas, R.M.; Sousa, A.M.; Pereira, M.O.; Botelho, C.M. Staphylococcus aureus virulence factors and disease. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Mendez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; pp. 702–710. [Google Scholar]
- Rahimi, F.; Katouli, M.; Karimi, S. Bioflm production among methicillin resistant Staphylococcus aureus strains isolated from catheterized patients with urinary tract infection. Microb. Pathog. 2016, 98, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Motallebi, M.; Jabalameli, F.; Asadollahi, K.; Taherikalani, M.; Emaneini, M. Spreading of genes encoding enterotoxins, haemolysins, adhesin and bioflm among methicillin resistant Staphylococcus aureus strains with staphylococcal cassette chromosome mec type IIIA isolated from burn patients. Microb. Pathog. 2016, 97, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Lappin-Scott, H.M.; Bass, C. Biofilm formation: Attachment, growth and detachment of microbes from surfaces. Am. J. Infect. Control 2001, 29, 250–251. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 2001, 39, 2151–2156. [Google Scholar] [CrossRef]
- Parom, S.K.; Igwe, J.C.; Bolaji, R.O.; Adeshina, G.O.; Onaolapo, J.A. Molecular characterization of methicillin resistance gene among Staphylococcus aureus isolated from poultry farms in Kaduna, Nigeria. J. Biosci. Biotechnol. Discov. 2018, 3, 5–13. [Google Scholar] [CrossRef]
- Ogundipe, F.O.; Ojo, O.E.; Feßler, A.T.; Hanke, D.; Awoyomi, O.J.; Ojo, D.A.; Akintokun, A.K.; Schwarz, S.; Maurischat, S. Antimicrobial resistance and virulence of methicillin-resistant Staphylococcus aureus from human, chicken and environmental samples within live bird markets in three Nigerian cities. Antibiotics 2020, 9, 588. [Google Scholar] [CrossRef] [PubMed]
- Kantiyok, T.J.; Arowolo, O.A.R.; Asanga, S.A.; Gberindyer, F.A. Antibacterial Use and Level of Resistance in Poultry: A Retrospective Study in North-Central and South-West Nigeria. J. Poult. Res. 2018, 15, 13–17. [Google Scholar]
- Jesumirhewe, C.; Springer, B.; Allerberger, F.; Ruppitsch, W. Whole genome sequencing of extended-spectrum beta-lactamase genes in Enterobacteriaceae isolates from Nigeria. PLoS ONE 2020, 15, e0231146. [Google Scholar] [CrossRef] [PubMed]
- Ndahi, M.D.; Hendrikse, R.; Helwigh, B.; Card, R.M.; Fagbamila, I.O.; Abiodun-Adewusi, O.O.; Ekeng, E.; Adetunji, V.; Adebiyi, I.; Andersen, J.K. Determination of antimicrobial use in commercial poultry farms in Plateau and Oyo States, Nigeria. Antimicrob. Resist. Infect. Control 2023, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Piessens, V.; van Coillie, E.; Verbist, B.; Supré, K.; Braem, G.; Van Nuffel, A.; De Vuyst, L.; Heyndrickx, M.; De Vliegher, S. Distribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herds. J. Dairy Sci. 2011, 94, 2933–2944. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Vanderhaeghen, W.; Angeles Argudín, M.; Haesebrouck, F.; Butaye, P. Characterization of methicillin-resistant Staphylococcus sciuri isolates from industrially raised pigs, cattle and broiler chickens. J. Antimicrob. Chemother. 2014, 69, 2928–2934. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Dakić, I.; Djukić, S.; Lozuk, B.; Svabić-Vlahović, M. Surgical wound infection associated with Staphylococcus sciuri. Scand. J. Infect. Dis. 2002, 34, 685–686. [Google Scholar] [CrossRef] [PubMed]
- Dakić, I.; Morrison, D.; Vuković, D.; Savić, B.; Shittu, A.; Ježek, P.; Hauschild, T.; Stepanović, S. Isolation and molecular characterization of Staphylococcus sciuri in the hospital environment. J. Clin. Microbiol. 2005, 43, 2782–2785. [Google Scholar] [CrossRef] [PubMed]
- Rolo, J.; Worning, P.; Boye, J.; Sobral, R.; Bowden, R.; Bouchami, O.; Damborg, P.; Guardabassi, L.; Perreten, V.; Westh, H.; et al. Evidence for the evolutionary steps leading to mecA-mediated β-lactam resistance in staphylococci. PLoS Genet. 2017, 13, e1006674. [Google Scholar] [CrossRef]
- Bortolaia, V.; Espinosa-Gongora, C.; Guardabassi, L. Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. 2016, 22, 130–140. [Google Scholar] [CrossRef]
- Silva, V.; Caniça, M.; Ferreira, E.; Vieira-Pinto, M.; Saraiva, C.; Pereira, J.E.; Capelo, J.L.; Igrejas, G.; Poeta, P. Multidrug-Resistant Methicillin-Resistant Coagulase-Negative Staphylococci in Healthy Poultry Slaughtered for Human Consumption. Antibiotics 2022, 11, 365. [Google Scholar] [CrossRef] [PubMed]
- Mccarthy, A.J.; Lindsay, J.A. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Schwarz, S.; Cai, J.; Fan, R.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J. Antimicrob. Chemother. 2016, 71, 1474–1478. [Google Scholar] [CrossRef] [PubMed]
- Dhaouadi, S.; Soufi, L.; Campanile, F.; Dhaouadi, F.; Sociale, M.; Lazzaro, L. Prevalence of meticillin-resistant and -susceptible coagulase-negative staphylococci with the first detection of the mecC gene among cows, humans and manure in Tunisia. Int. J. Antimicrob. Agents 2020, 55, 105826. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 2013, 35, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Bathavatchalam, Y.D.; Solaimalai, D.; Amladi, A.; Dwarakanathan, H.T.; Anandan, S.; Veeraraghavan, B. Vancomycin heteroresistance in Staphylococcus haemolyticus: Elusive phenotype. Future Sci. OA 2021, 7, FSO710. [Google Scholar] [CrossRef]
- Monecke, S.; Coombs, G.; Shore, A.C.; Coleman, D.C.; Akpaka, P.; Borg, M.; Chow, H.; Ip, M.; Jatzwauk, L.; Jonas, D.; et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011, 6, e17936. [Google Scholar] [CrossRef]
- Sarkhoo, E.; Udo, E.E.; Boswihi, S.S.; Monecke, S.; Mueller, E.; Ehricht, R. The Dissemination and Molecular Characterization of Clonal Complex 361 (CC361) Methicillin-Resistant Staphylococcus aureus (MRSA) in Kuwait Hospitals. Front. Microbiol. 2021, 12, 658772. [Google Scholar] [CrossRef]
- Ruppé, E.; Barbier, F.; Mesli, Y.; Maiga, A.; Cojocaru, R.; Benkhalfat, M.; Benchouk, S.; Hassaine, H.; Maiga, I.; Diallo, A.; et al. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 2009, 53, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Bouchami, O.; Hassen, A.; Lencastre, H.; Miragaia, M. High prevalence of mec complex C and ccrC is independent of SCCmec type V in Staphylococcus haemolyticus. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 605–614. [Google Scholar] [CrossRef]
- Szczuka, E.; Krajewska, M.; Lijewska, D.; Bosacka, K.; Kaznowski, A. Diversity of staphylococcal cassette chromosome mec elements in nosocomial multiresistant Staphylococcus haemolyticus isolates. J. Appl. Genet. 2016, 57, 543–547. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ebrahim-Saraie, H.S.; Motamedifar, M.; Sarvari, J.; Alfatemi, S.M.H. Emergence of SCCmec Type I Obtained from Clinical Samples in Shiraz Teaching Hospitals, South-West of Iran. Jundishapur. J. Microbiol. 2015, 8, e16998. [Google Scholar] [CrossRef] [PubMed]
- Cheesebrough, M. District Laboratory Practice in Tropical Countries, 2nd ed.; Cambridge University Press: Cambridge, UK, 2006; pp. 178–187. [Google Scholar]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 12.0; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2022. [Google Scholar]
- Jesumirhewe, C.; Springer, B.; Allerberger, F.; Ruppitsch, W. Genetic characterization of antibiotic resistant Enterobacteriaceae isolates from bovine animals and the environment in Nigeria. Front. Microbiol. 2022, 13, 793541. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Leopold, S.R.; Goering, R.V.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Ruppitsch, W.; Pietzka, A.; Prior, K.; Bletz, S.; Fernandez, H.; Allerberger, F.; Harmsen, D.; Mellmann, A. Defining and evaluating a core genome MLST scheme for whole genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 2869–2876. [Google Scholar] [CrossRef]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Day, N.P.J.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Strauß, L.; Rufng, U.; Abdulla, S.; Alabi, A.; Akulenko, R.; Garrine, M.; Germann, A.; Grobusch, M.P.; Helms, V.; Herrmann, M.; et al. Detecting Staphylococcus aureus virulence and resistance genes: A comparison of whole-genome sequencing and DNA microarray technology. J. Clin. Microbiol. 2016, 54, 1008–1016. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017, expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- International Working Group on the Classifcation of Staphylococcal Cassette Chromosome. Classifcation of Staphylococcal Cassette Chromosome mec (SCCmec): Guidelines for Reporting Novel SCCmec Elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).