Abstract
Escherichia coli O157:H7, Salmonella and Staphylococcus aureus are common foodborne pathogens. We determined the prevalence of E. coli O157:H7 and Salmonella in feces and milk and the prevalence of S. aureus in milk from dairy cattle and camels in the Borana pastoral community in the Southern Oromia Region of Ethiopia. Paired individual cow composite (pooled from all quarters in equal proportions) milk and fecal samples were collected from cows (n = 154) and camels (n = 158). Samples were cultured on bacterial isolation and identification media. E. coli O157:H7 and Salmonella isolates were further tested for susceptibility against nine antimicrobial drugs. Different risk factors associated with hygienic milking practices were recorded and analyzed for their influence on the prevalence of these bacteria in milk and feces. The prevalence of E. coli O157:H7 and Salmonella in feces were 3.9% and 8.4%, respectively, in cows, and 0.6% and 2.5%, respectively, in camels. E. coli O157:H7 and Salmonella were detected in the composite milk samples of 2.6% and 3.9% of the cows, respectively, and 0% and 1.3% of the camels, respectively. S. aureus was detected in composite milk samples of 33.4% of the cows and 41.7% of the camels. All E. coli O157:H7 (n = 11) and Salmonella (n = 25) isolates from both animal species and sample types were resistant to at least one antimicrobial drug. Multidrug resistance was observed in 70% (7/10) of the E. coli O157:H7 fecal and milk isolates from cows and 33.3% (2/6) of the Salmonella fecal and milk isolates from camels. The prevalence of these bacteria in feces and milk was not affected by risk factors associated with milking practices. Given the very close contact between herders and their animals and the limited availability of water for hand washing and udder cleaning, these bacteria are most likely present in all niches in the community. Improving community awareness of the need to boil milk before consumption is a realistic public health approach to reducing the risk of these bacteria.
1. Introduction
Milk and milk products play a significant role in human health and well-being [1,2]. However, milk-borne pathogens cause human diseases ranging from gastrointestinal disturbances such as diarrhea and vomiting to systemic and even life-threatening illnesses [3,4,5,6]. The presence of milk-borne pathogens in milk has both public health and economic importance [7,8]. The economic losses incurred by the dairy industry can be associated with reduced consumer confidence impacting the market for dairy products [9,10], product recalls, or the effects of some pathogens on animal productivity. The microbiological quality of dairy products in relation to foodborne pathogens is of great concern worldwide and is especially true in developing countries where dairy products are commonly handled under inadequate hygienic conditions and frequently consumed raw [8,11]. Milk-borne pathogens, including Salmonella, E. coli O157:H7 and Staphylococcus aureus can infect humans following the consumption of non-pasteurized milk and milk products [7,12]. Lack of routine milk pasteurization practices coupled with poor hygienic milk handling and processing under traditional livestock production systems is common in many developing countries [11,13,14,15,16]. In Ethiopia, a recent review of the available literature [17] indicated medians of 6% and 10% prevalences of Salmonella and E. coli O157:H7, respectively, in raw cow milk.
Most studies on E. coli O157:H7 in livestock species have been conducted on samples collected from different parts of beef cattle, sheep and goats at abattoirs or slaughterhouses and retail meat from different livestock species and other food samples [18,19,20,21,22,23]. The overall prevalence of E. coli O157:H7 in meat and other sample types was low, usually below 10%, but most of them had high antimicrobial resistance patterns, including multidrug resistance phenotypes [18,19,20,21,22,23]. Studies on Salmonella in Ethiopia have focused on testing the presence of Salmonella in different livestock species and foods of animal origin (meat and its minced products, raw eggs and raw milk), animal feces and human stool and their antimicrobial susceptibility profiles [24,25,26,27,28]. The prevalence of Salmonella is low in ruminants (cattle, sheep and goats) but high in pigs [26]. The prevalence of Salmonella in food of animal origin ranges from 3 to 10%, and antimicrobial drug resistance has also been observed against almost all tested antibiotics that are commonly used in both veterinary and human health sectors [24,25,27,28]. Staphylococcus aureus is the most common and frequently isolated bacteria responsible for mastitis, with variable prevalence in cows, and udder quarters, from different parts of Ethiopia [11,29,30,31,32,33].
Although E. coli O157:H7, Salmonella and S. aureus have been extensively studied in the highlands of Ethiopia [11,13,18,34,35,36,37], their statuses are not well understood in the pastoral settings where large herds of livestock are raised in extensive systems. Borana is an expansive savanna grassland in the Southern Oromia State of Ethiopia. It is characterized by an arid to semi-arid climate where the community’s livelihood mainly depends on livestock production. Milk is commonly consumed by the Borana pastoral community [38,39]. In this community, milking cows and processing milk are conducted using local traditional methods that are affected by various socio-cultural practices and beliefs [15,40]. Information on the occurrence of foodborne pathogens such as E. coli O157:H7 and Salmonella and the major milk-borne pathogen S. aureus in dairy animals and their milk is limited in these pastoral communities. The objective of this study was to determine the prevalence of E. coli O157:H7, Salmonella and S. aureus in dairy cows and camels raised under the pastoral livestock production system in Borana. Antimicrobial resistance of E. coli O157:H7 and Salmonella isolates were also determined.
2. Results
2.1. Description of the Study Animals
Dairy Cows: Paired fecal and milk samples were collected from 154 lactating cows belonging to 96 herds in 13 villages from 4 districts in the Borana zone (Figure 1). On average, 1.6 cows were sampled per herd, with a median of 1 and a range of 1–8 cows per herd. Only 1 cow was sampled per herd in two-thirds of the herds (66.7%; n = 96); 2 cows were sampled in 17.7% of the herds; 3 cows were sampled in 11.5% of the herds; and in the remaining four herds (4.2%), 4, 5, 6 or 8 cows were sampled per herd. Almost all study cows (98.7%, n = 154) were sampled from herds that raised mixed livestock species, with two-thirds (66.9%) of the sampled cows being from herds that raised 4 livestock species (cattle, camels, goats and sheep; Table 1. The study cows were seven years old on average, with the majority (57.8%) of the cows in good condition at the time of sampling. On average, the study cows were 11.5 months in lactation, with a mean parity number of 2.6 (Table 1).
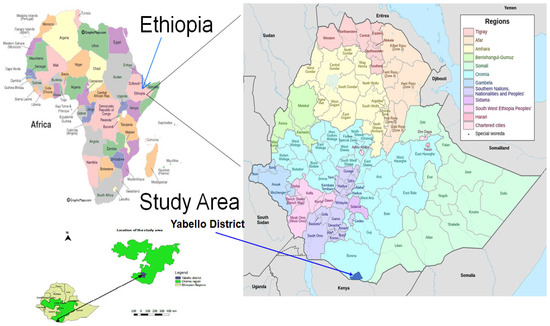
Figure 1.
Geographical location of the study area.

Table 1.
Description of the study cow population sampled for paired fecal and milk samples in the Borana pastoral community.
With regards to milking practices, over half (56.5%) of the study cows were milked into a locally made container called an “Okole”, (Figure 2E,F) which is a bucket made from the fresh skin of a giraffe or cow [41]. Another milk collection container was “Welki”, which is made locally from wood (Figure 2B–D). The rest of the cows and camels were milked into commercially available plastic buckets. Nearly all cows (91%) had relatively clean udders, and most were milked with no hand washing (80%), udder preparation (93%), or container cleaning (77%). Most cows (68%) were restrained by a rope tied to the hocks during milking (Figure 2G), and calves were allowed to suckle in more than half of the cows (58%) or restrained by a person (Figure 2I). Cows were milked primarily by women (84%; Table 1). The fecal consistency of the study cows was mostly normal or fluid and almost all cows had no teat lesions (Table 1).

Figure 2.
Locally made milk collection and storage containers. (A) Gorfa is a milk container that is handmade using traditional techniques. It is made from very tightly woven strands of vegetable or sisal fibers bunched together and wrapped at regular intervals with either one or two other fibers and decorated with cowry shells. Prior to use for milk storage, it is cleaned with water and smoked with glowing embers of local trees (Ejersa, scientific name Olea europaea subsp. cuspidate; Daanse, scientific name Faurea speciose; and Birreessa, scientific name Terminalia brownie) usually used for smoking milk containers [41]. The container is light and extremely durable and the inside has a black encrusted patina which makes it waterproof and ideal as a liquid container; (B–D) Welki is a temporary milk collection container locally made from wood and used when milking. It is available in different sizes, including small (B), medium (C) and large (D); (E,F) Okole is temporatry milk collection container locally made from skin of cattle and available in different sizes including large (E) and small (F); (G) a rope tied across both hind limbs at the hock joint using a milker’s knot to prevent the cow from kicking during milking; (H) a pastoralist woman kneel down on her leg and hold Welki (temporary milk collection container) tightly between her thighs and milking the cow quickly with both hands; (I) a person restraining the calf while the cow is milked. This picture was taken during study tim.
Dairy Camels: paired fecal and milk samples were collected from 158 lactating camels belonging to 91 herds in 10 villages from 4 districts in the Borana zone (Table 2). On average, 1.7 camels were sampled per herd, with a median of 1 and a range of 1–4 camels per herd. Only one camel was sampled per herd in over half of the herds (53.9%; n = 91); two camels were sampled in 24.2% of the herds; three camels were sampled in 16.5% of the herds; and four camels were sampled per herd in the remaining five herds (5.5%). Almost all study camels (98.7%, n = 158) were sampled from herds that raised mixed livestock species, with the majority of the camels (86.1%) sampled being from herds that raised four livestock species (cattle, camels, goats and sheep; Table 2). The study camels were nine years old, on average, with most of the camels being in good (46%) or medium (41%) body condition at the time of sampling. On average, the study camels were 10 months in lactation, with a mean parity number of 3.1 (Table 2).

Table 2.
Description of the study camel population sampled for paired fecal and composite milk samples in the Borana pastoral community.
With regards to hygienic milking practices, commercially obtained plastic containers and two locally made milk collection containers, Welki (Figure 2B–D) and Okole (Figure 2E,F), were the most common milking utensils used in the community to milk the study camels. The majority of the camels (84%) had relatively clean udders, and most camels were milked with no hand washing (89%), udder preparation (96.2%) or container cleaning (87%). The overwhelming majority of the study camels (89%) were manually restrained during milking and calves were allowed to suckle in half (58%) of the camels sampled. Unlike cows, camel milking was performed by at least two people helping each other, with family members being involved in the task (Figure 3 and Table 2). The fecal consistencies of the study camels were almost equally divided between normal and hard, and almost all study camels (98%) had no teat lesions (Table 2).
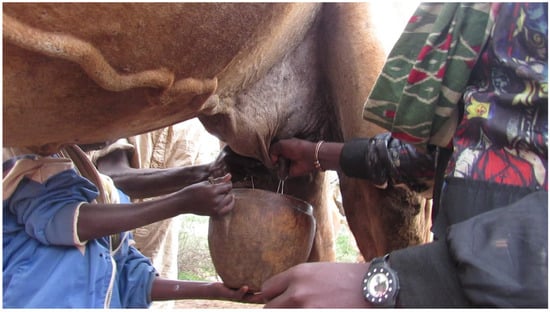
Figure 3.
A camel milked into a Welki by three persons. A camel can be milked by two or three persons from a standing position depending on availability of person to help. If two persons are milking, one person holds the milk collection container with one hand and milks the animal with the other hand, while the second person milks the camel with both hands [42]. If three persons are milking, one person holds the milk collection container and the two persons milk the camel as shown in this figure. Milk let-down takes a shorter time and milkers milk the camel quickly and collect milk within a short time. The picture was taken during study time.
2.2. Prevalence of E. coli O157:H7, Salmonella and Staphylococcus aureus in the Feces and Milk of Lactating Cows and Camels in the Borana Pastoralist Community
The prevalence of E. coli O157:H7 and Salmonella were 3.9% and 8.4%, respectively, in cow feces (Figure 4). E. coli O157:H7 and Salmonella were detected in 2.6% (4/154) and 3.9% (6/154), respectively, of composite milk samples from cows (Figure 4). All the E. coli O157:H7 and 77% (10/13) of the Salmonella-positive cows were from Yabello district. Cows that were positive for both pathogens were found in various villages in the positive districts, with most cases detected in one village (Dida Yabello). Most cases occurred in cows sampled from mixed herds raising the four livestock species: cattle, camels, goats and sheep.
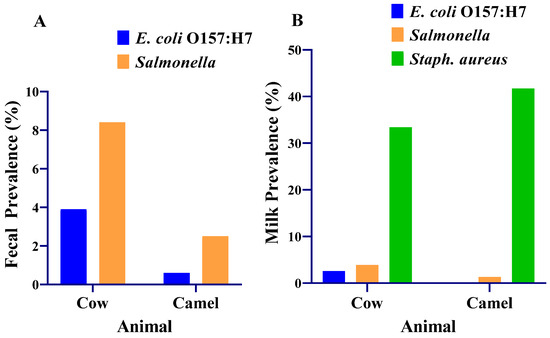
Figure 4.
Prevalence of E. coli O157:H7, Salmonella and Staphylococcus aureus in the milk and feces of lactating cows and camels in the Borana pastoralist community. (A) Fecal prevalence of E. coli O157:H7 and Salmonella; (B) milk prevalence of E. coli O157:H7, Salmonella and S. aureus. Staphylococcus aureus was cultured from milk samples collected from 119 cows and 130 camels.
The prevalence of E. coli O157:H7 and Salmonella were 0.6% and 2.5%, respectively, in the feces of camels. E. coli O157:H7 was not detected in any of the composite camel milk samples tested. Salmonella was detected in 1.3% (2/158) of composite camel milk (Figure 4).
The prevalence of Salmonella was significantly higher (p = 0.025) in the feces of cows than in the feces of camels. The prevalences of E. coli O157:H7 in feces were not different between cows and camels (p = 0.064). The prevalences of E. coli O157:H7 and Salmonella (p = 0.169) in composite milk did not differ significantly (p = 0.058) between the cows and the camels.
S. aureus was detected in composite milk samples of 33.4% of the cows and 41.7% of the camels. The prevalence of S. aureus was significantly higher (p = 0.026) in composite milk samples from camels (41.7%) than in composite milk samples from cows (33.4%).
2.2.1. Association between Risk Factors and E. coli O157:H7, Salmonella and S. aureus Prevalence in Cow Feces and Milk
Prevalence of E. coli O157:H7 in fecal samples: Study district was significantly associated with the prevalence of both E. coli O157:H7 (p = 0.02) and Salmonella (p = 0.019) in the feces of cows (Table 3).

Table 3.
Effects of various risk factors on the prevalence of E. coli O157:H7 and Salmonella in the feces of dairy cows in the Borana pastoralist community.
As shown in Table 3, the prevalence of E. coli O157:H7 in feces was not significantly affected by age (p = 0.575), body condition score (p = 0.641), stage of lactation at sampling (p = 0.575), or parity number (p = 0.407) of the cow. Similarly, E. coli O157:H7 in the feces was not significantly affected by the type of container used for milking (p = 0.414), the person milking the cow (p = 1.00), whether or not hands were washed before milking (p = 0.575), whether or not the milk container was washed before milking (p = 0.338), by the type of restraining method used during milking (p = 1.00), or whether or not calves were allowed to suckle during milking (p = 0.693). E. coli O157:H7 was not significantly affected by the fecal consistency (p = 0.398) or the presence of teat lesions (p = 0.182).
The 6 positive fecal samples were obtained from cows that were 5 (1 sample), 6 (3), 8 (1) and 10 (1) years old. Five of the six positive samples were obtained from cows in good body condition. Five positive fecal samples were obtained from cows in late lactation (12 months in lactation) and a single positive sample was obtained from an early lactating cow (2 months in lactation). Five of the six positive fecal samples were obtained from cows between 1 and 3 parities, while four of the positive fecal samples were obtained from cows milked into plastic containers and the remaining two samples were obtained from cows milked into Okole. We noted that all positive fecal samples were obtained from cows that were milked without hand washing. Four of the positive fecal samples were obtained from cows restrained by ropes tied to the hocks during milking. Five of the six positive fecal samples were obtained from cows with normal fecal consistency. Similarly, five of the six positive fecal samples were obtained from cows with no teat lesions.
Prevalence of Salmonella in feces: The prevalence of Salmonella in cow feces (Table 3) was not significantly affected by the age (p = 0.665), body condition score (p = 0.536), stage of lactation at sampling (p = 0.545), or parity number (p = 0.645) of the cow. Similarly, the prevalence of Salmonella in the feces was not significantly affected by the type of container used for milking (p = 0.939), the person milking the cow (p = 0.452), whether or not hands were washed before milking (p = 0.468), whether or not the milk container was washed before milking (p = 0.3), or by the type of restraining method used during milking (p = 1.00). Calf suckling before milking significantly reduced the prevalence of Salmonella in cow feces (p = 0.042); the prevalence of Salmonella in the feces was 14.1% (n = 64) in calf-suckled cows versus 4.4% (n = 90) in cows milked without calf suckling. The prevalence of Salmonella in the feces was not significantly affected by fecal consistency (p = 0.077) or the presence of teat lesions (p = 0.100).
Salmonella was detected in cows aged between 5 and 9 years old, with most (46.2%, n = 13) detected in 8-year-old cows. Positive samples were obtained from cows in good or medium body condition. Most positive samples (10/13) were obtained from cows in late lactation (12–24 months in lactation), with the remaining 3 positive samples coming from early lactating cows (1–6 months in lactation). The positive fecal samples were obtained from cows with parities between 1 and 4. Most positive fecal samples were obtained from cows milked into Okole (7 positives) or plastic containers (5 positives), while the one remaining positive sample was from a cow milked into a jerrycan. All positive cows were milked by women. We noted that 12 of the 13 positive fecal samples were obtained from cows that were milked without washing hands and without cleaning containers. Nine of the positive fecal samples were obtained from cows restrained using ropes tied to the hocks during milking, while the remaining four were from cows manually restrained. Nine of the thirteen positive fecal samples were obtained from cows with normal fecal consistency, three were from cows with fluid fecal consistency and the remaining one positive sample was from a cow with a hard fecal consistency. All Salmonella-positive fecal samples were obtained from cows with no teat lesions.
E. coli O157:H7 in composite milk: Overall, four composite milk samples from the dairy cows were positive for E. coli O157:H7. E. coli O157:H7 positivity was not significantly (p > 0.05) associated with any of the risk factors included in the analysis (Table 4). However, there were some notable observations within the categories of risk factors.

Table 4.
Effects of various risk factors on the detection of E. coli O157:H7 and Salmonella in composite milk samples from dairy cows in the Borana pastoralist community.
District was not significant (p = 0.089), but all four positive E. coli O157:H7 composite milk samples came from Yabello only. Village was not significant (p = 0.154), with E. coli O157:H7 occurring in only two of the villages: Dharito (three of the four positive composite milk samples) and Colqasa (one positive sample). All four E. coli O157:H7-positive composite milk samples were obtained from mixed herds that raised all four livestock species (cattle, camels, goats, sheep), although the factor was not significant (p = 0.78). The age of the cow was not significantly associated with E. coli O157:H7 positivity (p = 0.623), although three of the four positive composite milk samples were from 8-year-old cows. All four positive composite milk samples were obtained from cows in good BCS and lactating for 12 months, with no significant effects of BCS (p = 0.383) or stage of lactation (p = 0.904) on the detection of E. coli O157:H7 in composite cow milk. The effect of parity was not significant (p = 0.415); one positive milk sample each was obtained from cows in their 1st and 3rd parity, while the remaining two positive samples were from cows in their 4th parity. Two each of the four positive composite milk samples were obtained from cows milked into Okole or plastic containers, with no significant effect of milking utensils (p = 0.807). The person milking the cow did not have any significant effect (p = 1.00), with all composite milk samples being obtained from cows milked by women. Hand washing (p = 0.584) and container cleaning (p = 0.575) before milking were not significant, although all four E. coli O157:H7-positive composite milk samples were obtained from cows milked without hand washing or container cleaning. The type of restraint was not significant (p = 1.00); three positive milk samples were obtained from cows milked following restraint with a rope. Three of the positive composite milk samples were obtained from cows that were milked after calf suckling (p = 0.642), from cows with fluid fecal consistency (p = 0.359) and from cows with no teat lesions (p = 0.125), although these factors were not significant. All four positive composite milk samples were obtained from cows that were milked without udder preparation (p = 1.00) and from cows that had relatively clean udders (p = 1.00).
Salmonella in composite milk: Overall, six Salmonella-positive composite milk samples were obtained from the cows sampled. Except for village (p < 0.001) and parity number (p = 0.002), all other factors were not significantly associated (p > 0.05) with the detection of Salmonella from the composite milk samples of dairy cows (Table 4). Although district was not significant (p = 0.382), Salmonella was detected in composite milk samples from cows in two of the three districts (Dubuluk and Yabello), with three positive samples each.
Staphylococcus aureus in composite milk: Most of the risk factors were not significantly associated (p > 0.05) with the prevalence of S. aureus in cows (Table 5). In the composite milk samples collected from cows, only village (p = 0.051) and fecal consistency (p = 0.002) were significantly associated with S. aureus prevalence.

Table 5.
Effects of various risk factors on the detection of Staphylococcus aureus in composite milk samples from dairy cows and camels in the Borana pastoralist community.
2.2.2. Association between Risk Factors and E. coli O157:H7, Salmonella and S. aureus Prevalence in Camel Feces and Milk
Prevalence of E. coli O157:H7 in fecal samples: The effects of the various studied risk factors on the prevalence of E. coli O157:H7 and Salmonella in camel feces are presented in Table 6. The single fecal sample that was positive for E. coli O157:H7 was obtained from a 10-year-old camel (with no age effect; p = 1.00) with a medium body condition score (p = 0.538), in her 7th month of lactation (p = 0.032) and her third parity (p = 1.00), who was milked into an Okole (p = 0.608) by a man and a woman (p = 0.31). The cow was milked after handwashing (p = 0.108) and cleaning the milking utensil (p = 0.133), and she was manually restrained (p = 1.00). Her calf was allowed to suckle (p = 1.00), her feces had a hard consistency (p = 0.513) and she had no teat lesions (p = 1.00).

Table 6.
Effects of the various risk factors on the detection of Salmonella in fecal and composite milk samples from camels in the Borana pastoralist community.
Prevalence of Salmonella in fecal samples: All the risk factors analyzed were not significantly associated (p > 0.05) with the prevalence of Salmonella in fecal samples of lactating camels (Table 6). However, it is worth mentioning the following observed trends within the categories of each risk factor. Two of the four positive fecal samples were from 6- and 8-year-old camels, with no significant age effect (p = 0.4). Three positive fecal samples were obtained from camels in good BCS, with the remaining one positive sample coming from a camel with poor BCS; however, BCS was not significantly associated with the prevalence of Salmonella in feces (p = 0.234). The stage of lactation was not significant (p = 0.073); one Salmonella-positive feces sample was obtained from an early lactating camel (2 months in lactation), while three Salmonella-positive feces samples were obtained from late lactating camels (4–5 and 7 months in lactation). The parity number was not significant (p = 0.605); two Salmonella-positive camels were in their first parity, while the remaining two camels were in their 2nd and 4th parities. The milking container used was not significantly associated with the prevalence of Salmonella in feces (p = 0.714); two Salmonella-positive camels were milked into an Okole or a plastic container, while the other two camels were milked into Welki. The prevalence of Salmonella in fecal samples was not associated with the person(s) milking the camel (p = 0.131); all Salmonella-positive camels were milked by different people. Handwashing and container cleaning before milking were not significantly associated with the prevalence of Salmonella in fecal samples (p = 1.00), although all Salmonella-positive camels were milked without hand washing or container cleaning. Three of the four positive samples were obtained from manually restrained camels, although the restraint type was not significantly associated with the prevalence of Salmonella in fecal samples (p = 0.369). Three of the four camels were milked without calf suckling, although calf suckling was not significantly associated with the prevalence of Salmonella in fecal samples (p = 0.620). Fecal consistency was not significantly associated with the prevalence of Salmonella in feces (p = 1.00); two Salmonella-positive samples were obtained from camels with hard and normal fecal consistency each. Teat lesion was not significantly associated with the prevalence of Salmonella in feces (p = 1.00), although all four Salmonella-positive camels had no teat lesions. Although udder preparation did not have any significant effect on the prevalence of Salmonella in feces (p = 1.00), all Salmonella-positive camels did not undergo udder preparation prior to milking. Three of the Salmonella-positive camels had relatively clean udders, although this was not significant (p = 0.502).
Salmonella and S. aureus in composite milk: Salmonella was detected in two composite milk samples from the camels, but its detection was not significantly associated with any of the risk factors analyzed (p > 0.05; Table 6). Most of the risk factors were not significantly associated with the prevalence of S. aureus in camels (p > 0.05; Table 5), and district was the only risk factor that was significantly associated with S. aureus prevalence in composite milk samples from camels (p = 0.036).
2.3. Antimicrobial Resistance of E. coli O157:H7 and Salmonella
Antimicrobial susceptibility testing was performed for 11 E. coli O157:H7 isolates (10 fecal and milk samples from cows and one fecal sample from a camel) and 25 Salmonella isolates (19 fecal and milk samples from cows and 6 fecal and milk samples from camels). Antimicrobial susceptibility test results for E. coli O157:H7 and Salmonella isolates from milk and fecal samples for nine antimicrobial agents are shown in Table 7. Inhibition zone diameters for the antimicrobials on the test panel are provided in Table 7 and Supplementary Table S1. The isolates showed varying degrees of susceptibility to the antimicrobial agents tested. All E. coli O157:H7 isolates were susceptible to nalidixic acid, gentamicin, ciprofloxacin and chloramphenicol. Antimicrobial resistance of E. coli O157:H7 isolates was observed against ampicillin (100% of the isolates), streptomycin (73%), tetracycline (64%) and trimethoprim (18.2%). All Salmonella isolates were susceptible to nalidixic acid, gentamicin, ciprofloxacin and trimethoprim. On the other hand, Salmonella isolates were resistant to ampicillin (100% of the isolates), streptomycin (28%), kanamycin (4%) and tetracycline (12%) (Table 8).

Table 7.
Antimicrobial concentrations (µg/disk), interpretive categories and zone diameter (mm) breakpoints for Enterobacteriaceae.

Table 8.
Antimicrobial susceptibility test results for E. coli O157:H7 and Salmonella isolates from milk and fecal samples collected from lactating cows and camels under pastoral production system.
All E. coli O157:H7 isolates from fecal and milk samples from cows were resistant to at least one antimicrobial agent. Multidrug resistance (MDR), defined as resistance to ≥3 antimicrobial classes [43], was observed in 70% (7/10) of the E. coli O157:H7 isolates from fecal and milk samples of cows. The single E. coli O157:H7 isolate from camel feces was resistant only to ampicillin. Salmonella isolates from fecal and milk samples from cows were resistant to ampicillin alone (79%) or co-resistant to one or two drugs in two other antimicrobial classes (21%). While MDR was not observed in the cow isolates, 33.3% (2/6) of the Salmonella isolates from camel milk and feces showed MDR (Table 9).

Table 9.
Antimicrobial resistance profiles of E. coli O157:H7 and Salmonella isolates from cow and camel fecal and milk samples from the Borana pastoral community.
3. Discussion
The present study was conducted as part of a milk hygiene improvement research project in the Borana pastoral communities [13,44] to determine the prevalence of E. coli O157:H7 and Salmonella (in both feces and milk) and S. aureus (in composite milk only) in lactating cows and camels. Studies focusing on the prevalence of these pathogens in lactating dairy animals are scarce [45,46] and the available ones were mainly conducted in the central highlands of Ethiopia and focused primarily on animals destined for slaughter at abattoirs [18,47,48,49,50].
The 3.9% prevalence of E. coli O157:H7 in fecal samples from cows is comparable to the prevalence observed in cattle feces from abattoirs in Ethiopia (4.7%) [50] and Qatar (5%) [51]. On the other hand, a lower prevalence (1.9%) of E. coli O157:H7 was reported in central Ethiopia [18]. Compared to the present study, a higher prevalence (10.7%) of E. coli O157:H7 in cattle feces was reported in Riyadh, Saudi Arabia [52]. The same study [52] also reported a 2.4% prevalence of E. coli O157:H7 in camel feces, which is higher than the 0.6% prevalence observed in our study. Similar to the present study, low prevalences (1% [51] and 0.6% [53] of E. coli O157:H7 in camels were also reported elsewhere. The absence of E. coli O157:H7 in camel milk in the present study is contrary to a previous study from Qatar, which reported a high occurrence of E. coli O157:H7 (34%, n = 50) in camel fecal samples [51].
In the present study, the 8.4% prevalence of Salmonella in fecal samples from cows is higher than the 2.3% prevalence [46] but nearly similar to the 7.7% prevalence reported in dairy farms in Addis Ababa [45]. Farm-level contamination of cow milk with Salmonella in the present study (3.9%) is similar to the 3.1% prevalence in a previous report from central Ethiopia [45]. However, making these valid comparisons can be difficult given that most of the previous studies mostly involved cattle bound for slaughter after transportation from their initial production sites. Stress due to transportation can increase pathogen shedding. In the present study, on-farm samples were collected from animals raised under natural conditions in an extensive livestock production system.
The results of the present study showed that considerable proportions of the raw milk sampled from cows and camels at the farm level (primary production) were positive for Salmonella and E. coli O157:H7. Under such circumstances, the pathogens can present public health risks given that raw milk consumption is common in the area [15] and that attitudinal changes from this practice were not sustained after public education [13]. Further, the risk is potentiated by the ability of E. coli O157:H7 to survive harsh conditions, such as the low pH of dairy products [54]. We noted that under such subsistence farming, milk production is primarily for household consumption, with little sold to meet the financial needs of the family. There are no refrigeration or pasteurization facilities in this area and, as such, milk is consumed raw, posing a significant risk to consumers, especially children.
Risk factors such as pre-milking teat washing, milkers’ hand washing, presence of trauma/injury on teats, pre-cleaning of milk collection containers, milkers (male or female), animal body condition and fecal consistency were collected and their effects on the prevalence of these bacteria were analyzed in fecal and milk samples (Table 1 and Table 2). None of these risk factors significantly influenced the prevalence of these pathogens in fecal and milk samples. Given the fact that none of these hygienic milking practices were used in these areas before and that producers lack enough water for cleaning and have limited experience with hygienic milking procedures, it is not surprising to find no effects of these risk factors. In the absence of basic access to clean water and toilets in pastoral communities, and widely practiced raw milk consumption and close human–animal contact, the prevalence of these pathogens in feces, milk and other niches in these pastoral communities may not vary. However, there are no widespread milk-borne illnesses due to these pathogens. It is not clear whether this is due to adaptation to these pathogens due to frequent exposure early on or whether it is due to other mechanisms.
The current study on the antimicrobial susceptibility of E. coli O157:H7 and Salmonella revealed varying degrees of susceptibility to the antimicrobial agents tested. The degrees of susceptibility of E. coli O157:H7 and Salmonella isolates to specific antimicrobials varied from 0% to 100%. All isolates, from both cows and camels, were 100% susceptible to nalidixic acid, gentamicin and ciprofloxacin, which is in agreement with previous studies in Ethiopia [50]. The finding that all isolates were 100% resistant to ampicillin is in line with a previous study [50] and may indicate the widespread use of this antimicrobial in pastoral communities, mainly for the treatment of mastitis in dairy animals [11]. A similar study [45] in central Ethiopia also indicated resistance of Salmonella isolates to commonly used antimicrobials, including ampicillin (100%), streptomycin (66.7%), nitrofurantoin (58.3%), kanamycin and tetracycline (33.3%).
In conclusion, E. coli O157:H7 and Salmonella were detected in the milk and feces of a considerable number of lactating cows. Similarly, S. aureus was detected in milk of lactating cows and camels. The presence of these pathogens in cow milk indicates that they were shedding through milk from infected gland or contaminated either by infected cows or unhygienic conditions during milking and handling at the level of primary production. This is particularly important in causing potential health effects in people who commonly consume raw milk and milk products. Moreover, the occurrence of multidrug-resistant E. coli O157:H7 and Salmonella in the feces and milk of lactating cows can pose a significant public health risk. Therefore, relevant intervention programs and the creation of awareness on best practices for milk handling as well as control and surveillance programs for antimicrobial usage in animals can be implemented to minimize the contamination of milk and milk products with antimicrobial-resistant pathogens.
As a limitation, we did not conduct whole genome sequencing and comparative analyses of the bacterial isolates obtained from milk and feces to determine whether the isolates were genetically identical but contaminating different samples or whether they were genetically different. Additionally, further detailed investigations are required to understand short-term and long-term health-related problems or impact caused by frequent exposure of public especially children at early age in life to these foodborne pathogens in this pastoral community.
4. Materials and Methods
4.1. Study Area
The study was conducted in selected villages in four districts (Yabello, Surupha, Dubuluk and Elweya) of the Borana zone, Oromia (Figure 1). These villages were selected based on their high milk production potential and ease of accessibility via cars. Borana zone is located in the lowlands of the Southern part of Oromia, Ethiopia. Yabello is the capital city of Borana zone and is about 570 km from Addis Ababa (Figure 1). The Borana pastoral area has a semi-arid to arid climate with dry and rainy seasons and bimodal rainfall distribution consisting of a long rainy season from March to May and a short rainy season from September to November. Despite usually expecting two rainy seasons, rainfall is increasingly becoming erratic and highly variable, resulting in frequent droughts and variability in livestock and livestock products off-take. The Borana community comprises both pastoral (those who only raise livestock) and agropastoral (those who grow some crops and also raise livestock) communities. Livestock production is a major source of livelihood for the community. Borana pastoralists historically raise only cattle; however, due to recent increasing erratic rainfall and drought problems, they have diversified their herds by additionally raising more drought-resilient livestock, including camels, goats and sheep [39]. The study area is typical of other pastoral settings where communities heavily depend on animal production usually raised comingled together or mixed species (cattle, sheep, goats and camels) [13,15]. People, domestic animals and wild animals share spaces and drinking water and live in close contact, which may favor the cross-species transmission of many infectious diseases, including the foodborne pathogens targeted in this study. Moreover, this study area is close to the border with Northern Kenya and Somalia and there is frequent cross-border contact between herders through grazing lands, livestock trade business as well as animal and human drugs smuggled across borders. Additional description of the study area is available elsewhere [13,15].
4.2. Study Design and Sample Size Calculation
A cross-sectional study was conducted in April 2018 to determine the prevalence of E. coli O157:H7 and Salmonella in the feces and milk and S. aureus in milk of dairy cows and camels. The study population comprised healthy-looking lactating cows and camels managed under a traditional/extensive husbandry system in the study area. Convenience sampling was used to select an individual animal from each herd. Paired fecal and milk samples were collected from each animal to determine the apparent prevalence of the target pathogens. The number of animals required to estimate prevalence was calculated using the following formula, which has been described elsewhere [55]:
where N is the required sample size, d is absolute precision (d = 0.05), and p is expected prevalence. The prevalence (p) used in the sample size calculation was a pooled prevalence of 7.47% obtained from a meta-analysis of Salmonella in ruminants in Ethiopia [26].
Accordingly, 106 camels and 106 cows were needed, assuming equal sample sizes for E. coli O157:H7, Salmonella and S. aureus. However, to account for herd-level clustering of bacterial infection and contamination, the target sample size was adjusted for an intra-cluster correlation coefficient (ρ) of 0.2 [56], and about 1–8 animals were sampled per herd. The study’s design effect (deff), calculated as deff = 1 + (m − 1)ρ, where m = 3 is the cluster size and ρ = 0.2 is the correlation coefficient, was 1.4. Therefore, the sample size obtained using a simple random sampling formula was adjusted by multiplying it by the deff, resulting in 149 cows and 149 camels. In the end, to account for any potential sample losses, paired fecal and milk samples were collected from 154 lactating cows and 158 camels.
4.3. Milk and Fecal Sample Collection and Transportation
Fecal samples (~15 g) were collected rectally from individual animals using a gloved hand while the animals were restrained. A 30 mL sample of composite milk (pooled milk from all quarters) was collected from each animal. Prior to sample collection into sterile tubes, milk was collected from each animal either into commercially obtained plastic containers or locally made milk collection containers (Okole, made from cattle hide, or Welki, made from wood) (Figure 2). Samples were collected either early in the morning (around 5 a.m.) before the animals were released to pasture or after 5 p.m. in the evening when animals returned to their housing. Composite milk and fecal samples were collected in sterile bottles labeled with unique animal identifier numbers consisting of animal species, herd and sampling date. “Okole” is a bucket made from the fresh skin of a giraffe or cow. Samples were kept at +4 °C and transported to the microbiology laboratory at the International Livestock Research Institute (ILRI) in Addis Ababa, Ethiopia. Samples were stored at −20 °C until processed for microbiological analysis. During field sampling, data on potential risk factors associated with milking and hygienic practices such as the containers used for milk collection, the presence of trauma on teats or udders, whether milker(s) washed their hands, the udders and milk collection containers before milking, the animal restraining methods used during milking, the sex (male or female) and total number of milkers, fecal consistency on the day of milk collection and overall body condition of each animal were also collected.
4.4. Bacterial Isolation and Identification
4.4.1. Salmonella spp. and E. coli O157:H7
Isolation and identification of the bacteria was done using standard techniques recommended by the International Organizations for Standardization [57] with some modifications [58]. Samples were pre-enriched by mixing 10 g of feces or 10 mL of milk with 90 mL of buffered peptone water (BPW; Oxoid, Basingstoke, UK) in Whirl-Pak filter bags (Thomas Scientific, Houston, TX, USA). The mixture was homogenized in a laboratory blender (Oxoid). Pre-enrichments were incubated at 25 °C for 2 h, then at 42 °C for 6 h and held at +4 °C until they were processed the next day for isolation of E. coli O157:H7 and Salmonella.
Pre-enrichment broth (1 mL) was added to 20 µL of anti-E. coli O157:H7 immunomagnetic separation (IMS) beads for E. coli O157:H7 isolation (Dynabeads anti-E. coli O157:H7; Applied Biosystems, Foster, CA, USA) or 20 µL of Salmonella-specific IMS beads for Salmonella isolation (Dynal, Lake Success, NY, USA) as previously described [59,60]. Briefly, E. coli O157:H7- and Salmonella-specific IMS beads were re-suspended by gently vortexing the mixture to ensure that the pellet was completely suspended. Twenty microliters (20 µL) of re-suspended paramagnetic beads was transferred to Eppendorf tubes (Oxoid) and 1 mL of the enriched culture was added into the Eppendorf tubes. Each tube was vortexed for 10–30 min at room temperature. Tubes were then transferred to a manual magnetic particle concentrator (MPC-S; Oxoid) with a magnetic strip in place, inverted several times and left to separate for 3 min. The supernatant was aspirated and discarded. The magnetic strip was removed and 1 mL of phosphate buffered saline with Tween 20 (PBS-T; Sigma chemical Co., Saint Louis, MO, USA) was added to each tube. The beads were re-suspended by inverting MPC several times with the tubes still in place. The magnetic strip was replaced and the above steps were repeated three times. To prevent cross-contamination, separate sterile micropipette tips were used for each sample.
The final bead–bacteria complexes (50 µL) were plated on CHROMagar O157 plates (CHROMAgar-O157:H7; DRG International, Mountainside, NJ, USA) supplemented with novobiocin (5 mg/L) and potassium tellurite (2.5 mg/L; Sigma chemical Co) and incubated at 37 °C overnight for the isolation of E. coli O157:H7. Following incubation, presumptive E. coli O157:H7 colonies with a mauve-pink color on the CHROMAgar plates were picked and inoculated on nutrient agar slants and incubated at 37 °C for 18 h. Slants were stored at +4 °C until biochemical tests were performed.
For the isolation of Salmonella, bacteria–bead complexes were eluted into 3 mL of Rappaport Vassiliadis soya peptone broth (RVS; Oxoid) and incubated at 42 °C for 18 h. After incubation, a loopful of RVS broth enrichment culture was plated onto xylose lysine deoxycholate (XLD) agar (Oxoid) supplemented with 4.6 mL/L tergitol), 15 mg/L novobiocin and 5 mg/L cefsulodin (XLDtnc; Sigma chemical Co.) and incubated at 37 °C for 18 h. A suspected Salmonella colony based on characteristic appearance on the XLD plate was inoculated on nutrient agar slants and incubated at 37 °C for 18 h. Slants were stored at +4 °C until biochemical tests were performed.
For biochemical tests, colonies were re-streaked on nutrient agar (Oxoid) plates and incubated at 37 °C for 24 h. E. coli O157:H7 and Salmonella isolates were biochemically tested using triple sugar iron agar (TSI; Oxoid), L-lysine decarboxylation test, indole production, citrate utilization test and methyl red (MR) and Voges Proskauer (VP) tests. Pure colonies from nutrient agar plates were picked and inoculated in biochemical test tubes containing TSI agar, lysine decarboxylase broth, Simon’s citrate agar and tryptone broth, and incubated at 37 °C for 24 h (more than 24 h incubation was needed for the citrate utilization test) [57]. Colonies producing an alkaline slant with acid (yellow color) butt on TSI with hydrogen sulfide and gas production, formation of purple/pink color of L-lysine decarboxylation broth, color change in Simon’s citrate agar from green to blue, positive for MR test, negative for VP test and negative for tryptophan utilization (yellow–brown ring) indicating the absence of indole production were considered Salmonella positive. Isolates positive for indole, negative for citrate utilization, negative for VP and positive for L-lysine decarboxylation were presumptively considered to be E. coli O157:H7. E. coli O157:H7 isolates were further confirmed using a latex agglutination test with the O157:H7 antigen (Remel, Lenexa, KS, USA), following the manufacturer’s instruction.
4.4.2. Staphylococcus aureus
S. aureus was isolated from milk samples according to ISO 6888-1 [61] using Baird-Parker agar (Oxoid). The methodology was modified to follow only qualitative detection of the pathogen. Egg emulsion was prepared locally from fresh chicken eggs with intact shells purchased from a local market in Addis Ababa. The eggs were cleaned with a brush using a liquid detergent and rinsed under running water. The eggshells were disinfected by immersing them in 70% ethanol for 30 s and then air drying. Each egg was broken under aseptic conditions (in the biosafety hood) and the yolk separated from the white via repeated transfer of the yolk from one half of the shell to the other. The yolk was placed in a sterile flask and sterile water was added at four times the volume and mixed thoroughly. The mixture was heated in a water bath set at 47 °C for 2 h and then kept at +3 °C ± 2 °C for 18–24 h to allow for precipitate formation. The supernatant liquid was aseptically collected into a fresh sterile flask for use. The emulsion was stored at +3 °C ± 2 °C for a maximum of 72 h.
Sixty-three grams of agar (Oxoid) was added to one liter of distilled water and boiled to dissolve the medium; this was then autoclaved at 121 °C for 15 min. After the agar was cooled to 50 °C, 50 mL of egg yolk emulsion and 3.5% potassium tellurite solution (Oxoid) were aseptically added proportionally. The mixture of molten agar was added to sterile petri dishes and allowed to solidify and then kept under sterile conditions until use. Immediately before use, the surface of the plate was dried and 0.1 mL aliquot of milk was spread using a sterile wire loop. The plate was incubated at 37 °C for 24 h and checked for typical S. aureus colonies. Negative plates were incubated for up to 48 h. Each plate was examined for typical S. aureus colonies, which appear as black colonies surrounded by a clear zone. Typical colonies were selected and sub-cultured on tryptone soya yeast extract agar (TSYEA) and incubated at 37 °C for 24–48 h for purity. The presumptive pure colony was inoculated on TSAYE agar and incubated at 37 °C overnight. Finally, the pure colony was inoculated in TSAYE broth, incubated overnight and then stored at −80 °C in sterile 85% glycerol at a proportion of 500 µL culture and 500 µL glycerol at the Forage and Feed Development Lab at ILRI.
4.5. Antimicrobial Susceptibility Testing for E. coli O157:H7 and Salmonella Isolates
Antimicrobial susceptibility testing was performed according to the Clinical and Laboratory Standards Institute [62] using the Kirby–Bauer disk diffusion method. The antimicrobial disks were obtained from HIMEDIA (Mumbai, India); their list, disk concentration and CLSI interpretation breakpoints are shown in Table 1. These antimicrobials were selected based on their availability in the study area and the possibility of use by herders in the study areas. Pure colony grown on nutrient agar was transferred to a 5 mL tryptone soya broth (TSB; Oxoid) and incubated at 37 °C for 18 h until growth reached 0.5 McFarland turbidity standards (Oxoid). A sterile cotton swab was dipped into the suspension and swabbed uniformly in three directions over the surface of Mueller–Hinton agar plates (Oxoid) and kept at room temperature for 30 min to allow drying. Antibiotic disks were placed on the inoculated plates using sterile forceps by gently pressing onto the agar to ensure firm contact on the surface and incubated at 37 °C for 24 h. After incubation, the diameters of the zone of inhibition were measured using a caliper and compared with CLSI [62] zone size interpretative guidelines for the family of Enterobacteriaceae as sensitive, intermediate or resistant (Table 1).
4.6. Data Analysis
Data were recorded in Microsoft Excel (Redmond, WA, USA) and cleaned for any entry errors. Data were analyzed in STATA, version 16 (StataCorp LLC, College Station, TX, USA). Descriptive statistics such as frequencies were used to estimate the prevalence of the pathogens in both composite milk and feces samples. Univariate analysis of the association between pathogen presence and potential risk factors was conducted using Fisher’s exact or chi-squared tests. A p-value < 0.05 (hereafter simply presented as P) was interpreted as a statistically significant association.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13010026/s1, Table S1: Inhibition zone diameter of antimicrobial disks used to determine the antimicrobial susceptibility of E. coli and Salmonella isolates from dairy cows and camels in the Borana pastoral community in South Oromia, Ethiopia.
Author Contributions
K.A., O.K.D., G.E.A. and D.G. contributed to the conceptualization and design of the study and to manuscript writing and editing; G.E.A. performed the statistical analyses; D.H. wrote the first draft of the manuscript; D.H. and H.D. conducted the microbiological tests and the antimicrobial susceptibility testing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the United States Agency for International Development Bureau for Food Security under Agreement #AID-OAA-L-15-00003 as part of the Feed the Future Innovation Lab for Livestock Systems. Any opinions, findings, conclusions, or recommendations expressed here are those of the authors alone. This study was also supported by the CGIAR Research Program on Livestock and the CGIAR Research Program on Agriculture for Nutrition and Health. We thank all donors and organizations that globally support CGIAR’s work through their contributions to the CGIAR Trust Fund. The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or U.S. Government determination or policy. The mention of trade names or commercial products in this publication by United States Department of Agriculture (USDA) author (GEA) is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Institutional Review Board Statement
The study was approved by the Research Ethics Review Committee of the College Veterinary Medicine and Agriculture of Addis Ababa University (Ref: VM/ERC/27/05/10/2018), ILRI’s Animal Care and Use Committee (ref: ILRI-IACUC2018-04) and ILRI’s Committee on the use of human subjects in research (ILRI-IREC2016-20).
Informed Consent Statement
Verbal consent was obtained from the animal owners at the time of sample collection.
Data Availability Statement
The data can be obtained upon request from the corresponding author.
Acknowledgments
We thank Silvia Alonso for the contribution on project administration and management, support on study design and implementation and comments on the earlier version of the manuscript. We also thank the pastoral livestock community for their willingness to be part of this study.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Thorning, T.K.; Raben, A.; Tholstrup, T.; Soedamah-Muthu, S.S.; Givens, I.; Astrup, A. Milk and dairy products: Good or bad for human health? An assessment of the totality of scientific evidence. Food Nutr. Res. 2016, 60, 32527. [Google Scholar] [CrossRef] [PubMed]
- Givens, D.I. MILK Symposium review: The importance of milk and dairy foods in the diets of infants, adolescents, pregnant women, adults, and the elderly. J. Dairy Sci. 2020, 103, 9681–9699. [Google Scholar] [CrossRef] [PubMed]
- Hetzel, M.; Bonfoh, B.; Farah, Z.; Traore, M.; Simbe, C.F.; Alfaroukh, I.O.; Schelling, E.; Tanner, M.; Zinsstag, J. Diarrhoea, vomiting and the role of milk consumption: Perceived and identified risk in Bamako (Mali). Trop. Med. Int. Health 2004, 9, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.P.; Boor, K.J.; Murphy, S.C.; Murinda, S.E. Food safety hazards associated with consumption of raw milk. Foodborne Pathog. Dis. 2009, 6, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, S.; Hoffmann, S.; White, A.; Ahn, J.W.; McQueen, R.B.; Scallan Walter, E. Cost of Hospitalizations for Leading Foodborne Pathogens in the United States: Identification by International Classification of Disease Coding and Variation by Pathogen. Foodborne Pathog. Dis. 2021, 18, 812–821. [Google Scholar] [CrossRef]
- Diab, M.S.; Tarabees, R.; Elnaker, Y.F.; Hadad, G.A.; Saad, M.A.; Galbat, S.A.; Albogami, S.; Hassan, A.M.; Dawood, M.A.O.; Shaaban, S.I. Molecular Detection, Serotyping, and Antibiotic Resistance of Shiga Toxigenic Escherichia coli Isolated from She-Camels and In-Contact Humans in Egypt. Antibiotics 2021, 10, 1021. [Google Scholar] [CrossRef]
- Scallan Walter, E.J.; Griffin, P.M.; Bruce, B.B.; Hoekstra, R.M. Estimating the Number of Illnesses Caused by Agents Transmitted Commonly Through Food: A Scoping Review. Foodborne Pathog. Dis. 2021, 18, 841–858. [Google Scholar] [CrossRef]
- Alegbeleye, O.O.; Guimarães, J.T.; Cruz, A.G.; Sant’Ana, A.S. Hazards of a ‘healthy’ trend? An appraisal of the risks of raw milk consumption and the potential of novel treatment technologies to serve as alternatives to pasteurization. Trends Food Sci. Technol. 2018, 82, 148–166. [Google Scholar] [CrossRef]
- Dhanashekar, R.; Akkinepalli, S.; Nellutla, A. Milk-borne infections. An analysis of their potential effect on the milk industry. Germs 2012, 2, 101–109. [Google Scholar] [CrossRef]
- Petrovski, K.R.; Trajcev, M.; Buneski, G. A review of the factors affecting the costs of bovine mastitis. J. S. Afr. Vet. Assoc. 2006, 77, 52–60. [Google Scholar] [CrossRef]
- Balemi, A.; Gumi, B.; Amenu, K.; Girma, S.; Gebru, M.; Tekle, M.; Rius, A.A.; D’Souza, D.H.; Agga, G.E.; Kerro Dego, O. Prevalence of Mastitis and Antibiotic Resistance of Bacterial Isolates from CMT Positive Milk Samples Obtained from Dairy Cows, Camels, and Goats in Two Pastoral Districts in Southern Ethiopia. Animals 2021, 11, 1530. [Google Scholar] [CrossRef] [PubMed]
- Costard, S.; Espejo, L.; Groenendaal, H.; Zagmutt, F.J. Outbreak-Related Disease Burden Associated with Consumption of Unpasteurized Cow’s Milk and Cheese, United States, 2009–2014. Emerg. Infect. Dis. 2017, 23, 957–964. [Google Scholar] [CrossRef]
- Amenu, K.; Agga, G.E.; Kumbe, A.; Shibiru, A.; Desta, H.; Tiki, W.; Dego, O.K.; Wieland, B.; Grace, D.; Alonso, S. MILK Symposium review: Community-tailored training to improve the knowledge, attitudes, and practices of women regarding hygienic milk production and handling in Borana pastoral area of southern Ethiopia. J. Dairy Sci. 2020, 103, 9748–9757. [Google Scholar] [CrossRef] [PubMed]
- Kamana, O.; Ceuppens, S.; Jacxsens, L.; Kimonyo, A.; Uyttendaele, M. Microbiological Quality and Safety Assessment of the Rwandan Milk and Dairy Chain. J. Food Prot. 2014, 77, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Amenu, K.; Wieland, B.; Szonyi, B.; Grace, D. Milk handling practices and consumption behavior among Borana pastoralists in southern Ethiopia. J. Health Popul. Nutr. 2019, 38, 6. [Google Scholar] [CrossRef] [PubMed]
- Majalija, S.; Tumwine, G.; Kiguli, J.; Bugeza, J.; Ssemadaali, M.A.; Kazoora, H.B.; Muwanguzi, E.N.; Nantima, N.; Tuyiragize, R. Pastoral community practices, microbial quality and associated health risks of raw milk in the milk value chain of Nakasongola District, Uganda. Pastoralism 2020, 10, 3. [Google Scholar] [CrossRef]
- Keba, A.; Rolon, M.L.; Tamene, A.; Dessie, K.; Vipham, J.L.; Kovac, J.; Zewdu, A. Review of the prevalence of foodborne pathogens in milk and dairy products in Ethiopia. Int. Dairy J. 2020, 109, 104762. [Google Scholar] [CrossRef] [PubMed]
- Abdissa, R.; Haile, W.; Fite, A.T.; Beyi, A.F.; Agga, G.E.; Edao, B.M.; Tadesse, F.; Korsa, M.G.; Beyene, T.; Beyene, T.J.; et al. Prevalence of Escherichia coli O157:H7 in beef cattle at slaughter and beef carcasses at retail shops in Ethiopia. BMC Infect. Dis. 2017, 17, 277. [Google Scholar] [CrossRef]
- Asrat, D.; Hiko, A.; Zewde, G. Occurrence of Escherichia coli O157:H7 in retail raw meat products in Ethiopia. J. Infect. Dev. Ctries. 2008, 2, 389–393. [Google Scholar] [CrossRef][Green Version]
- Haile, A.F.; Alonso, S.; Berhe, N.; Atoma, T.B.; Boyaka, P.N.; Grace, D. Prevalence, Antibiogram, and Multidrug-Resistant Profile of E. coli O157: H7 in Retail Raw Beef in Addis Ababa, Ethiopia. Front. Vet. Sci. 2022, 9, 734896. [Google Scholar] [CrossRef]
- Haile, A.F.; Alonso, S.; Berhe, N.; Bekele Atoma, T.; Boyaka, P.N.; Grace, D. Escherichia coli O157:H7 in Retail Lettuce (Lactuca sativa) in Addis Ababa City: Magnitude of Contamination and Antimicrobial Susceptibility Pattern. Front. Microbiol. 2021, 12, 694506. [Google Scholar] [CrossRef] [PubMed]
- Bekele, T.; Zewde, G.; Tefera, G.; Feleke, A.; Zerom, K. Escherichia coli O157:H7 in Raw Meat in Addis Ababa, Ethiopia: Prevalence at an Abattoir and Retailers and Antimicrobial Susceptibility. Int. J. Food Contam. 2014, 1, 4. [Google Scholar] [CrossRef]
- Abebe, E.; Gugsa, G.; Ahmed, M.; Awol, N.; Tefera, Y.; Abegaz, S.; Sisay, T. Occurrence and antimicrobial resistance pattern of E. coli O157:H7 isolated from foods of Bovine origin in Dessie and Kombolcha towns, Ethiopia. PLOS Neglected Trop. Dis. 2023, 17, e0010706. [Google Scholar] [CrossRef] [PubMed]
- Ejo, M.; Garedew, L.; Alebachew, Z.; Worku, W. Prevalence and Antimicrobial Resistance of Salmonella Isolated from Animal-Origin Food Items in Gondar, Ethiopia. BioMed Res. Int. 2016, 2016, 8. [Google Scholar] [CrossRef]
- Abate, D.; Assefa, N. Prevalence and antimicrobial resistance patterns of Salmonella isolates in human stools and animal origin foods in Ethiopia: A systematic review and meta-analysis. Int. J. Health Sci. 2021, 15, 43–55. [Google Scholar]
- Tadesse, G.; Tessema, T.S. A meta-analysis of the prevalence of Salmonella in food animals in Ethiopia. BMC Microbiol. 2014, 14, 270. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, G.; Gebremedhin, E.Z. Prevalence of Salmonella in raw animal products in Ethiopia: A meta-analysis. BMC Res. Notes 2015, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Alemayehu, D.; Molla, B.; Muckle, A. Prevalence and Antimicrobial Resistance Pattern of Salmonella Isolates from Apparently Healthy Slaughtered Cattle in Ethiopia. Trop. Anim. Health Prod. 2003, 35, 309–319. [Google Scholar] [CrossRef]
- Abebe, R.; Hatiya, H.; Abera, M.; Megersa, B.; Asmare, K. Bovine mastitis: Prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet. Res. 2016, 12, 270. [Google Scholar] [CrossRef]
- Tora, E.T.; Bekele, N.B.; Suresh Kumar, R.S. Bacterial profile of bovine mastitis in Ethiopia: A systematic review and meta-analysis. PeerJ 2022, 10, e13253. [Google Scholar] [CrossRef]
- Mekonnen, S.A.; Lam, T.J.G.M.; Hoekstra, J.; Rutten, V.P.M.G.; Tessema, T.S.; Broens, E.M.; Riesebos, A.E.; Spaninks, M.P.; Koop, G. Characterization of Staphylococcus aureus isolated from milk samples of dairy cows in small holder farms of North-Western Ethiopia. BMC Vet. Res. 2018, 14, 246. [Google Scholar] [CrossRef] [PubMed]
- Yimana, M.; Tesfaye, J. Isolation, identification and antimicrobial profile of methicillin-resistant Staphylococcus aureus from bovine mastitis in and around Adama, Central Ethiopia. Vet. Med. Sci. 2022, 8, 2576–2584. [Google Scholar] [CrossRef] [PubMed]
- Demil, E.; Teshome, L.; Kerie, Y.; Habtamu, A.; Kumilachew, W.; Andualem, T.; Mekonnen, S.A. Prevalence of subclinical mastitis, associated risk factors and antimicrobial susceptibility of the pathogens isolated from milk samples of dairy cows in Northwest Ethiopia. Prev. Vet. Med. 2022, 205, 105680. [Google Scholar] [CrossRef] [PubMed]
- Gutema, F.D.; Rasschaert, G.; Agga, G.E.; Jufare, A.; Duguma, A.B.; Abdi, R.D.; Duchateau, L.; Crombe, F.; Gabriel, S.; De Zutter, L. Occurrence, Molecular Characteristics, and Antimicrobial Resistance of Escherichia coli O157 in Cattle, Beef, and Humans in Bishoftu Town, Central Ethiopia. Foodborne Pathog. Dis. 2021, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Getaneh, A.M.; Gebremedhin, E.Z. Meta-analysis of the prevalence of mastitis and associated risk factors in dairy cattle in Ethiopia. Trop. Anim. Health Prod. 2017, 49, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, S.A.; Koop, G.; Melkie, S.T.; Getahun, C.D.; Hogeveen, H.; Lam, T. Prevalence of subclinical mastitis and associated risk factors at cow and herd level in dairy farms in North-West Ethiopia. Prev. Vet. Med. 2017, 145, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Amenu, K.; Grace, D.; Nemo, S.; Wieland, B.M. Bacteriological quality and safety of ready-to-consume milk and naturally fermented milk in Borana pastoral area, southern Ethiopia. Trop. Anim. Health Prod. 2019, 51, 2079–2084. [Google Scholar] [CrossRef]
- Sadler, K.; Catley, A. Milk Matters: The Role and Value of Milk in the Diets of Somali Pastoralist Children in Liben and Shinile, Ethiopia; Feinstein International Center, Tufts University and Save the Children: Addis Ababa, Ethiopia, 2009. [Google Scholar]
- Megersa, B.; Markemann, A.; Angassa, A.; Zárate, A.V. The role of livestock diversification in ensuring household food security under a changing climate in Borana, Ethiopia. Food Secur. 2014, 6, 15–28. [Google Scholar] [CrossRef]
- Amenu, K.; Szonyi, B.; Grace, D.; Wieland, B. Important knowledge gaps among pastoralists on causes and treatment of udder health problems in livestock in southern Ethiopia: Results of qualitative investigation. BMC Vet. Res. 2017, 13, 303. [Google Scholar] [CrossRef]
- Amdhun, K. Milk Container Sanitation Regime Using Wood Smoke: Perceived Roles and Effects on Milk Quality in Borana, Ethiopia; MVSc, College of Veterinary Medicine and Agriculture, Addis Ababa University: Bishoftu, Ethiopia, 2019. [Google Scholar]
- Karumba, T. Camel’s Milk Set for Boom Times, p In AFP. 2011. Available online: https://timesofmalta.com/articles/view/Camel-s-milk-set-for-boom-times.372990 (accessed on 10 June 2023).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Amenu, K.; Alonso, S. Improving Handling Practices and Microbiological Safety of Milk and Milk Products in Borana Pastoral Communities, Ethiopia. 2019, pp. 1–2. Available online: https://livestocklab.ifas.ufl.edu/media/livestocklabifasufledu/pdf-/ETH-Amenu-Borana-milk-project-leaflet.pdf (accessed on 10 June 2023).
- Addis, Z.; Kebede, N.; Sisay, Z.; Alemayehu, H.; Wubetie, A.; Kassa, T. Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: A cross sectional study. BMC Infect. Dis. 2011, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Eguale, T.; Engidawork, E.; Gebreyes, W.A.; Asrat, D.; Alemayehu, H.; Medhin, G.; Johnson, R.P.; Gunn, J.S. Fecal prevalence, serotype distribution and antimicrobial resistance of Salmonellae in dairy cattle in central Ethiopia. BMC Microbiol. 2016, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Molla, W.; Molla, B.; Alemayehu, D.; Muckle, A.; Cole, L.; Wilkie, E. Occurrence and antimicrobial resistance of Salmonella serovars in apparently healthy slaughtered sheep and goats of central Ethiopia. Trop. Anim. Health Prod. 2006, 38, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Zewdu, E.; Cornelius, P. Antimicrobial resistance pattern of Salmonella serotypes isolated from food items and personnel in Addis Ababa, Ethiopia. Trop. Anim. Health Prod. 2009, 41, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Sibhat, B.; Molla Zewde, B.; Zerihun, A.; Muckle, A.; Cole, L.; Boerlin, P.; Wilkie, E.; Perets, A.; Mistry, K.; Gebreyes, W.A. Salmonella serovars and Antimicrobial Resistance Profiles in Beef Cattle, Slaughterhouse Personnel and Slaughterhouse Environment in Ethiopia. Zoonoses Public Health 2011, 58, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Ashenafi, B.; Paulos, D.; Abera, M.; Tefera, G.; Hailu, D.; Kasaye, S.; Amenu, K. Occurrence of Escherichia coli O157:H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol. 2017, 17, 24. [Google Scholar] [CrossRef]
- Mohammed, H.O.; Stipetic, K.; Salem, A.; McDonough, P.L.; Chang, Y.F.; Sultan, A. Risk of Escherichia coli O157:H7, Non-O157 Shiga Toxin–Producing Escherichia coli, and Campylobacter spp. in Food Animals and Their Products in Qatar. J. Food Prot. 2015, 78, 1812–1818. [Google Scholar] [CrossRef]
- Bosilevac, J.M.; Gassem, M.A.; Al Sheddy, I.A.; Almaiman, S.A.; Al-Mohizea, I.S.; Alowaimer, A.; Koohmaraie, M. Prevalence of Escherichia coli O157:H7 and Salmonella in Camels, Cattle, Goats, and Sheep Harvested for Meat in Riyadh. J. Food Prot. 2015, 78, 89–96. [Google Scholar] [CrossRef]
- Disassa, N.; Sibhat, B.; Mengistu, S.; Muktar, Y.; Belina, D. Prevalence and Antimicrobial Susceptibility Pattern of E. coli O157:H7 Isolated from Traditionally Marketed Raw Cow Milk in and around Asosa Town, Western Ethiopia. Vet. Med. Int. 2017, 2017, 7581531. [Google Scholar] [CrossRef]
- Tsegaye, M.; Ashenafi, M. Fate of Escherichia coli O157:H7 during the processing and storage of Ergo and Ayib, traditional Ethiopian dairy products. Int. J. Food Microbiol. 2005, 103, 11–21. [Google Scholar] [CrossRef]
- Thrusfield, M. Veterinary Epidemiology, 3rd ed.; Blackwell Publishing Professional: Ames, IA, USA, 2005. [Google Scholar]
- Otte, M.J.; Gumm, I.D. Intra-cluster correlation coefficients of 20 infections calculated from the results of cluster-sample surveys. Prev. Vet. Med. 1997, 31, 147–150. [Google Scholar] [CrossRef] [PubMed]
- ISO 6579-1:2017; Microbiology of the Food Chain Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella Part 1: Detection of Salmonella spp. ISO: Geneva, Switzerland, 2017.
- Agga, G.E.; Arthur, T.M.; Hinkley, S.; Bosilevac, J.M. Evaluation of Rectoanal Mucosal Swab Sampling for Molecular Detection of Enterohemorrhagic Escherichia coli in Beef Cattle. J. Food Prot. 2017, 80, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Reynolds, K.; Bauer, N., Jr.; Keen, J.E. Shiga-toxigenic Escherichia coli O157:H7 infections among livestock exhibitors and visitors at a Texas County Fair. Vector Borne Zoonotic Dis. 2005, 5, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Keen, J.E.; Wittum, T.E.; Dunn, J.R.; Bono, J.L.; Durso, L.M. Shiga-toxigenic Escherichia coli O157 in agricultural fair livestock, United States. Emerg. Infect. Dis. 2006, 12, 780–786. [Google Scholar] [CrossRef]
- ISO 6888-1; Microbiology of the Food Chain-Horizontal Method for the Enumeration of Coagulase-Positive (Staphylococcus aureus and Other Species)-Part 1:Method Using Baird-Parker Agar Medium. ISO: Geneva, Switzerland, 2021.
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 8th ed.; Clinical and Laboratory Standards Insititute: Wayne, PA, USA, 2009. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).