Abstract
Salmonella continues to be a major threat to public health, especially with respect to strains from a poultry origin. In recent years, an increasing trend of antimicrobial resistance (AMR) in Salmonella spp. was observed due to the misuse of antibiotics. Among the approaches advised for overcoming AMR, probiotics from the Lactobacillus genus have increasingly been considered for use as effective prophylactic and therapeutic agents belonging to the indigenous microbiota. In this study, we isolated lactobacilli from the ilea and ceca of hens and broilers in order to evaluate their potential probiotic properties. Four species were identified as Limosilactobacillus reuteri (n = 22, 45.8%), Ligilactobacillus salivarius (n = 20, 41.6%), Limosilactobacillus fermentum (n = 2, 4.2%) and Lactobacillus crispatus (n = 1, 2%), while three other isolates (n = 3, 6.25%) were non-typable. Eight isolates, including Ligilactobacillus salivarius (n = 4), Limosilactobacillus reuteri (n = 2), L. crispatus (n = 1) and Lactobacillus spp. (n = 1) were chosen on the basis of their cell surface hydrophobicity and auto/co-aggregation ability for further adhesion assays using the adenocarcinoma cell line Caco-2. The adhesion rate of these strains varied from 0.53 to 10.78%. Ligilactobacillus salivarius A30/i26 and 16/c6 and Limosilactobacillus reuteri 1/c24 showed the highest adhesion capacity, and were assessed for their ability to compete in and exclude the adhesion of Salmonella to the Caco-2 cells. Interestingly, Ligilactobacillus salivarius 16/c6 was shown to significantly exclude the adhesion of the three Salmonella serotypes, S. Enteritidis, S. Infantis and S. Kentucky ST 198, to Caco-2 cells. The results of the liquid co-culture assays revealed a complete inhibition of the growth of Salmonella after 24 h. Consequently, the indigenous Ligilactobacillus salivarius 16/c6 strain shows promising potential for use as a preventive probiotic added directly to the diet for the control of the colonization of Salmonella spp. in poultry.
1. Introduction
Non-typhoidal Salmonella strains are the leading cause of foodborne gastroenteritis [1]. Poultry products are primarily consumed worldwide and are commonly known to be reservoirs for a variety of microorganisms. Salmonella is the most frequently encountered pathogen in poultry products, as well as the most prominent inhabitant of avian gastrointestinal tracts (GIT) [2]. In developing countries, a high prevalence of Salmonella has been recorded, ranging from 13% to 39% in South America, estimated at 35% in Africa, and ranging from 35% to 50% in Asia [3]. In Lebanon, according to our recent study, the percentage of contamination of poultry meat at the retail level (supermarkets and restaurants) was estimated at 22.4% [4].
Several control strategies have been adopted to reduce or eliminate Salmonella at the farm level. It is well known that the use of antibiotic growth promoters (AGPs) and other prophylactic treatments improve animal health and productivity rates in livestock farming [5]. However, the mass use of antibiotics as feed additives has led to the emergence and spread of antimicrobial resistant pathogens and epidemic multi-drug-resistant clones and/or resistance genes in poultry reservoirs [6]. Recently, resistance to critical antibiotics, namely fluoroquinolones and expanded-spectrum β-lactam antibiotics, has spread worldwide and reached humans through the food chain [7]. Consequently, since 2006, AGP use in the animal industry has been completely banned in the EU [8] and reduced in many other countries [9]. However, in Lebanon, there are no current regulations concerning the use of AGPs in animal husbandry (personal communication with the ministry of agriculture).
Many countries have also implemented control programs to tackle Salmonella in poultry farms. Such was the case in the USA (National Poultry Improvement Plan (NPIP) for the eradication of Salmonella in eggs (1989) and meats (1994)) and the EU (Commission Regulation (EC), No. 2160/2003). These measurements ultimately led to the successful reduction of targeted Salmonella spp., but unfortunately cleared the way for the emergence of more resistant, less common serotypes, such as S. Heidelberg and S. Kentucky [10].
A promising alternative strategy against enteropathogens is the use of lactic acid bacteria (LAB) as probiotics. Probiotics are non-pathogenic live microorganisms which confer health benefits on their host when ingested in adequate quantities [11]. The use of (direct-fed microbial) probiotics as broiler growth promoters [12,13] could improve livestock health and might reduce the emergence of antimicrobial resistance (AMR) [14]. Strains of Lactobacillus spp. and Bifidobacterium spp. are the most widely studied probiotics acting against gastrointestinal microbial pathogens [15], especially against Salmonella infections in the broiler gastrointestinal tract [16,17]. Two fundamental mechanisms of inhibition of pathogenic microorganisms have been described, namely the direct cell competitive exclusion and the production of inhibitory compounds, including lactic and acetic acids, hydrogen peroxide, bacteriocin or bacteriocin-like inhibitors, and fatty and amino acid metabolites [18].
Intestinal adhesion and colonization are the first steps of the Salmonella infection process in poultry. Therefore, the adhesion ability is an essential prerequisite of, and one of the main criteria for selecting, potential probiotic strains [11]. The probiotic ability prevents the selected strains from undergoing direct elimination by peristalsis and inhibits the colonization of enteric pathogens in chickens by competitive exclusion [19]. Methods of evaluating the capacity of LAB to adhere to poultry epithelia may include in vitro analysis of, for example, cell aggregation, cell wall hydrophobicity, and adhesion to the human colorectal adenocarcinoma cell line (Caco-2) and chicken hepatocellular carcinoma cell line (LMH) [12]. Since bacterial populations of GIT are specific to their animal hosts, poultry-derived probiotics could be more effective than non-specific microbial agents [20].
This study aims to develop an effective probiotic derived from broiler and chicken GITs. In this regard, in vitro experiments were conducted to reveal the probiotic activity of native poultry-derived lactobacilli strains against the most relevant and drug-resistant Salmonella spp. (S. Enteritidis, S. Infantis and S. Kentucky ST198) in Lebanese poultry farms. The screening of lactobacilli strains for their anti-Salmonella activity, safety and surface probiotic properties was also carried out. Finally, the lactobacilli showing a great probiotic potential were selected for the further in vitro characterization of their adhesion ability and kinetics in co-culture. In fact, their adhesion and abilities to exclude and compete with Salmonella serotypes in epithelial tissues, using the Caco-2 cell line as an experimental model, were evaluated, as well as their capacity to inhibit pathogen growth in a mixed co-culture model.
2. Results
2.1. Screening of Lactobacilli and Their Anti-Salmonella Activity
In total, 210 stains (155 from the 16 trials and 55 from commercial birds) which presented as gram-positive bacilli/coccobacilli with no catalase activity were collected from broiler ceca and ileum samples. All lactobacilli were found to produce inhibition zones against the three serotypes of Salmonella based on the agar spot-on-lawn assay. The radii of their inhibition zones ranged from 1.2 to 4.4 cm (data not shown). However, the cell-free supernatants (CFSs) of all lactobacilli, neutralized to pH 6.8, did not display any antimicrobial effects against the Salmonella serotypes studied.
2.2. Genotypic Identification of Lactobacilli Isolates with Phylogenetic Relations
Lactobacilli strains (n = 48) were chosen according to their high anti-Salmonella activity in the spot-on-lawn test. The 16S rRNA gene sequence analysis identified four species: Limosilactobacillus reuteri (formerly Lactobacillus reuteri) (n = 22, 45.83%), Ligilactobacillus salivarius (formerly Lactobacillus salivarius) (n = 20, 41.66%), Limosilactobacillus fermentum (formerly Lactobacillus fermentum) (n = 2, 4.16%) and Lactobacillus crispatus (n = 1, 2%) (Figure 1). The three remaining isolates (16/i10, 14/i15, A30/c2, 6.25%) were non-typable. The most common species were Limosilactobacillus reuteri and Ligilactobacillus salivarius. The phylogenetic tree demonstrated a close relation among the same species. However, we could not obtain a better strain resolution at the subspecies level among Limosilactobacillus reuteri. To gain further insight into the genetic dissimilarities and evolutionary relationships among the lineages isolated here would require profound core-gene-based phylogenetic analyses. These analyses are not considered here, since the focus of our study is the probiotic potential of lactobacilli strains.
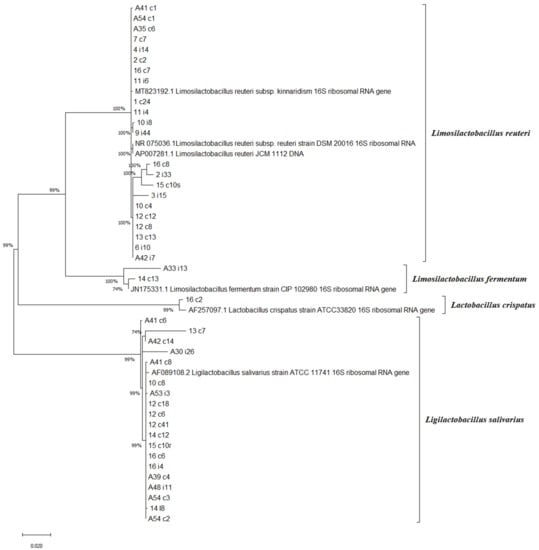
Figure 1.
A maximum-likelihood phylogenetic tree reconstructed using 16S rRNA gene sequences. The percentage of replicate trees in which the associated species clustered together in the bootstrap test (1000 replicates) are shown next to the branches [21]. Evolutionary analyses were conducted in MEGA X. Limosilactobacillus fermentum comb. nov. CIP 102980 (JN175331), Lactobacillus crispatus ATCC 33820T (AF257097), Ligilactobacillus salivarius comb. nov. ATCC 11741T (AF089108), Limosilactobacillus reuteri comb. nov. JCM 1112 (AP007281), Limosilactobacillus reuteri subsp.kinnaridis (MT823192), and Limosilactobacillus reuteri subsp. reuteri DSM20016 (NR075036) were selected as type strains. The 16S rRNA gene accession numbers are provided in parentheses.
2.3. Analysis of Surface Properties
The visual screening of the forty-eight chosen lactobacilli isolates showed that most of the strains were Agg+/Agg− (75%), while Agg+ and Agg− represented 14,6% and 10.4%, respectively (data not shown). These results were confirmed by auto-aggregation assays at 4 h. As shown in Figure 2, category I demonstrated a significant auto-aggregation percentage (≥65%) compared to category II (≤10%), while category III ranged from 10 to 65% except for three strains: one > 65% and two ≤ 10%. Auto-aggregation was determined in all the lactobacilli tested (n = 45, 90%) at 24 h.
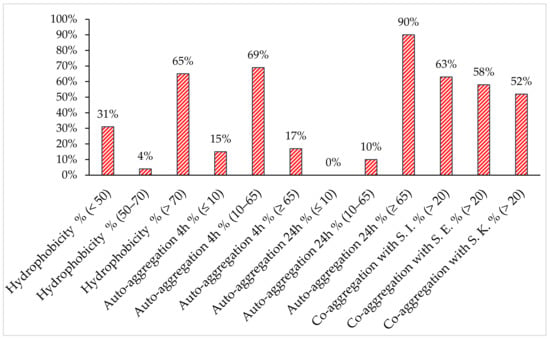
Figure 2.
Isolate distribution in defined ranges of percentage of hydrophobicity, auto-aggregation and co-aggregation with the three Salmonella spp. (S. Enteritidis (S.E.), S. Kentucky ST198 (S.K.) and S. Infantis (S.I.).
The co-aggregation properties of the lactobacilli strains with Salmonella serotypes differed among the strains and ranged from 0 to 94.6% (data not shown). They co-aggregated with S. Kentucky ST198, S. Enteritidis and S. Infantis at 52%, 58% and 63%, respectively. Otherwise, a high affinity for xylene was shown (65%) compared to non-hydrophobic isolates (31%).
2.4. Hydrophobicity and Auto/co-Aggregation Correlation
The results obtained from the analysis of the lactobacilli surfaces were subjected to principal component analysis (PCA) (Figure 3). The first PC1 and second PC2 principal components could explain 47.1% and 28.13% of the total variance, respectively. Based on the cell surface properties, eight lactobacilli strains were chosen for further adhesion assays, whose characteristics are summarized in Table 1. Ligilactobacillus salivarius A30/i26 was shown to be highly hydrophobic (98.84% ± 1.34), possessing an aggregation phenotype (Agg+) and an ability to aggregate rapidly at 4 h (76.15% ± 3.93). The most co-aggregative strains were L. crispatus 16/c2, Limosilactobacillus reuteri 12/c8 and Ligilactobacillus salivarius (16/c4, 16/c6 and 14/i8). In addition to these properties, Ligilactobacillus salivarius 16/c6 did not exhibit auto-aggregation at 4 h but only at 24 h (9.89% ± 3.63 and 95.91% ± 2.58, respectively). However, Ligilactobacillus salivarius 16/c4 displayed an aggregation phenotype (Agg+) and rapidly auto-aggregated at 4 h (76.23% ± 3.38). Both Lactobacillus spp. 16/i10 and Limosilactobacillus reuteri 1/c24 displayed high hydrophobicity levels (98.36% ± 3.63 and 91.81% ± 7.78, respectively); however, they either did not show an auto-aggregation capacity at 4 h, or only did so to a moderate degree (6.16% ± 5.53 and 13.76% ± 1.87, respectively) (Table 1).
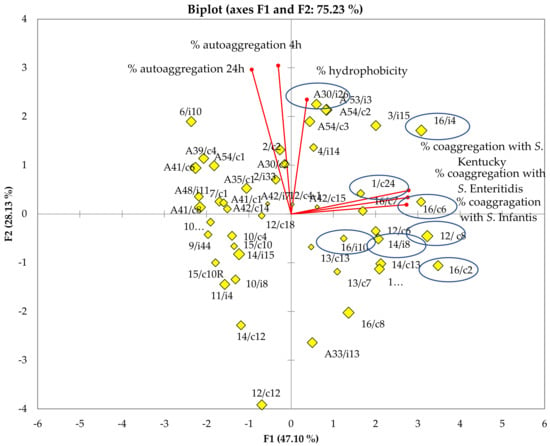
Figure 3.
Graphic representation of the principal component analysis (PCA) of surface proprieties, including hydrophobicity and auto/co-aggregation, for the 48 lactobacilli isolated stains. The selected stains are encircled.

Table 1.
Cell surface properties of the eight selected strains of lactobacilli.
There was no significant correlation between hydrophobicity, auto-aggregation, and co-aggregation among the forty-eight strains tested (Table 2). On the contrary, a significant correlation was detected (p < 0.05) between the co-aggregation results of the three Salmonella serotypes with lactobacilli isolates, since the correlation coefficient value reached up to 0.890.

Table 2.
Correlation of Pearson coefficients between hydrophobicity, auto-aggregation and co-aggregation of the 48 lactobacilli isolates. The index of Pearson was used to evaluate the correlation between the six assays, hydrophobicity, auto-aggregation and co-aggregation between the lactobacilli strains and S. Enteritidis, S. Infantis and S. Kentucky stains.
2.5. Assays for Tolerance to Simulated Gastrointestinal Conditions of Chickens
The eight chosen lactobacilli were further evaluated for their survival capacity in a medium simulating the GIT conditions of chickens (Figure 4). All strains were able to tolerate acidity and 0.1% (w/v) bile salts. However, at 0.3% bile salts, the survival rate was reduced for Ligilactobacillus salivarius 16/i4 and A33/i26 to 0% and 37%, respectively.
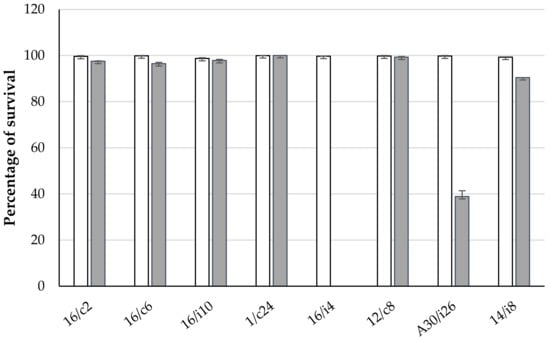
Figure 4.
Effects of simulated gastrointestinal conditions on lactobacilli viability. White and grey columns correspond to lactobacilli subjected to 0.15% and 0.3% bile salts, respectively. L. crispatus 16/c2, Ligilactobacillus salivarius (16/c6, 16/i4, A30/i26, 14/i8), Lactobacillus sp. 16/i10 and Limosilactobacillus reuteri (1/c24 and 12/c8).
2.6. Adhesion Assays
The attachment of the lactobacilli isolates varied from 0.53 to 10.78% (Figure 5). Ligilactobacillus salivarius (A30/i26, 16/c6 and 16/i4) and Limosilactobacillus reuteri 1/c24 showed the highest adhesion abilities (p < 0.05) of 10.78% ± 4.2, 6.5% ± 1.82, 5% ± 0.99 and 6.43% ± 2.26, respectively. The remaining Lactobacillus spp. 16/i10, Ligilactobacillus salivarius 14/i8, Limosilactobacillus reuteri 12/c8 and L. crispatus 16/c2 strains showed no significant differences, with low adhesion capacities of 3.61% ± 1.14, 2.35% ± 0.86, 1.99% ± 0.66 and 0.53% ± 0.21, respectively.
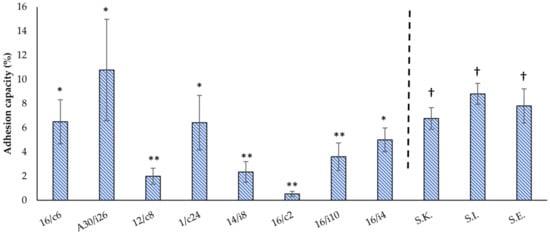
Figure 5.
Adhesion capacities of the eight-native poultry-derived lactobacilli strains and the three Salmonella serotypes (S. Kentucky ST 198 (S.K.), S. Infantis (S.I.) and S. Enteritidis (S.E.)) to Caco-2 monolayers. The means and standard deviations of the two independent experiments are shown, each with three replicates. The differences between the levels of strain adhesion were evaluated separately for the lactobacilli strains and Salmonella serotypes. Ligilactobacillus salivarius 16/c6, 16/i4 and A30/i26 and Limosilactobacillus reuteri 1/c24 revealed no significant differences (*) in their adhesion capacity, a finding which was dissimilar from the four remaining tested strains (**). The differences in the adhesion capacities of S. Enteritidis, S. Infantis and S. Kentucky ST198 were also not significant among the three serotypes (†).
S. Infantis, S. Enteritidis and S. Kentucky ST198 attached to the Caco-2 cells at a percentage of 8.81% ± 0.87, 7.81% ± 1.41 and 6.77% ± 0.89, respectively. No significant difference was found between the different serotypes (Figure 5).
2.7. Competition/Exclusion Assays
Three lactobacilli strains that showed the highest adhesion capacity, namely, Ligilactobacillus salivarius A30/i26 and 16/c6 and Limosilactobacillus reuteri 1/c24, were assessed for their ability to compete with the pathogen for the adhesion site on the Caco-2 monolayers (Figure 6). The results showed that none of these strains displayed an effect on the pathogen adhesion to the Caco-2 cells. In the exclusion assays, Ligilactobacillus salivarius 16/c6 excluded the pathogens to a better degree than Ligilactobacillus salivarius A30/i26 and Limosilactobacillus reuteri 1/c24. The percentages of anti-adhesion to the Caco-2 cells of S. Enteritidis, S. Infantis and S. Kentucky ST198 were 70.30% ± 6.22, 86.57% ± 9.22 and 79.54% ± 9.26, respectively (p < 0.05).
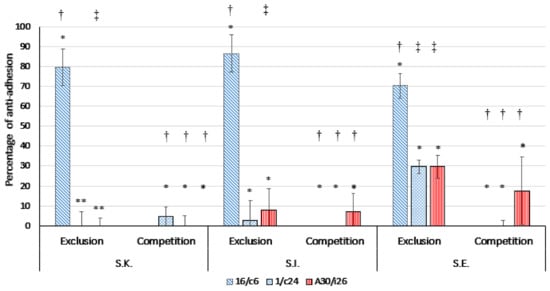
Figure 6.
Inhibition of the adherence of S. Kentucky ST 198 (S.K.), S. Infantis (S.I.) and S. Enteritidis (S.E.) to Caco-2 cells by Ligilactobacillus salivarius 16/c6 and A30/i26 and Limosilactobacillus reuteri 1/c24 in competition and exclusion assays. The means and standard deviations of three independent experiments are shown, each with three replicates. (*) Lactobacillus strains were fixed and the differences in inhibition were calculated between the three serotypes in the same assay. (*) p > 0.05, (**) p ≤ 0.05. (†) Salmonella serotypes were fixed, and the differences in inhibition were calculated between the three lactobacilli strains in the same assay. (†) p > 0.05, (‡) p ≤ 0.05.
2.8. Co-Culture Growth Kinetics
Since Ligilactobacillus salivarius 16/c6 was able to inhibit the adhesion of Salmonella to the Caco-2 monolayers, its ability to inhibit the growth of Salmonella serotypes was assessed in a broth co-culture assay. Pure cultures of the lactobacilli and Salmonella serotypes (S.E., S.K., and S.I.) grew very well in the chosen Laptg medium (Figure 7).
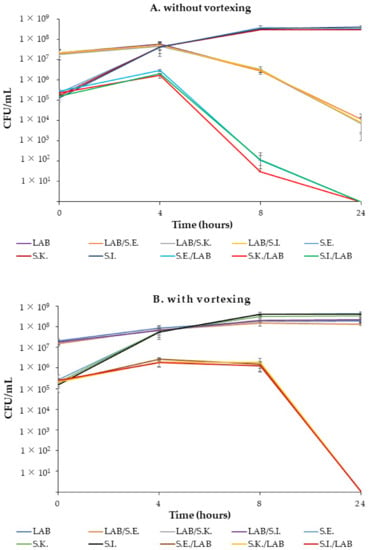
Figure 7.
Kinetic growths of the pure cultures and co-cultures of Ligilactobacillus salivarius 16/c6 and S. Enteritidis, S. Infantis and S. Kentucky ST198 without (A) or with (B) vortexing. The means and standard deviations of the three independent experiments are shown, each with three replicates.
In both experiments, without (Figure 7 A) or with (Figure 7 B) vortexing, differences in the CFUs between the control cultures of Salmonella (S.E., S.K., and S.I.) and co-cultures (S.E./LAB, S.K./LAB and S.I./LAB) were observed from the early incubation hours. However, the numbers of CFUs estimated from the co-cultures without vortexing were lower than those determined from the vortexed co-cultures and those of the control cultures at 8 h. In fact, the Salmonella in the co-cultures increased from 105 to 106 CFU /mL in the first 4 h and then sharply decreased to 102 and 101 CFU/mL, until a negligible cell viability was obtained between 8 h and 24 h. Simultaneously, the Ligilactobacillus salivarius count decreased from 107 to 106 at 8 h, then was reduced to almost 104 at 24 h in the monoculture (16/c6) and co-cultures (LAB/S.E., LAB/S.K and LAB/S.I.).
In the second set of experiments, Salmonella counts from the co-cultures (S.E./LAB, S.K./LAB and S.I./LAB) slightly increased in 4 h and remained constant until 8 h, then decreased to an undetectable level (˂10 CFU/mL) at 24 h. However, the pure cultures of Salmonella slightly increased at 8 h and remained constant until the end of the experiments. The lactobacilli counts in the LAB–Salmonella mix (LAB/S.E., LAB/S.K and LAB/S.I.) were similar to those of the control monoculture. The pH value in both the mono- and co-cultures dropped from approximately 6.97 to 3.9 at 24 h.
3. Discussion
LAB are considered the principal residents of the GIT, where they provide the host with protection against enteric pathogen colonization (competition for nutrients and secretion of inhibitory substances). These LAB were the focus of many works, substituting the use of probiotics as growth promotors and/or subtherapeutic additives in animal feeds [22]. Numerous factors affect the microbial biodiversity of the poultry GIT, such as the GIT section (ileum or caeca) and the breed, diet and age of the chicken. The microbiota change significantly in the first 2–3 weeks until their stabilization at 5–6 weeks of age. It was found that, when broilers were fed with antibiotic and an additive-free corn-soy diet, 70% of their ileum population was comprised of Lactobacillus spp. The use of antibiotics in broilers was shown to induce changes in the composition of the intestinal bacterial community, namely Ligilactobacillus salivarius [23]. In this regard, our experiments did not detect an important species diversity among the lactobacilli isolates identified. Strains of Lactobacillus acidophilus, Ligilactobacillus salivarius and Limosilactobacillus fermentum were permanently found in all birds aged from two days old to market age. Babot and colleagues showed that the most common Lactobacillus species were L. crispatus, Limosilactobacillus reuteri and Ligilactobacillus salivarius, which was also the case in our findings [24].
In vitro tests have been used to assess the probiotic potential of lactobacilli. The production of hydrogen peroxide, organic acids and bacteriocins are the main strategies of Lactobacillus in inhibiting Salmonella growth [18]. However, in the present study, hydrogen peroxide production was unlikely to be the cause of this inhibition in the agar diffusion test due to the anaerobic growth conditions of the lactobacilli [25]. The well-diffusion antagonism method did not show any inhibition, thereby excluding the hypothesis of secreted bacteriocins or bacteriocin-like as Salmonella inhibitors. Decreasing the pH by organic acid production was likely to be the cause of such an effect [26]. Although the bacteriocin or bacteriocin-like activity produced by LAB is commonly more effective against Gram-positive bacteria, such as Listeria monocytogenes [27], the inhibition of the Gram-negative Salmonella has also been reported [28].
The adhesion behavior of bacteria is a complex multistep process which includes specific and non-specific ligand–receptor mechanisms [29]. The latter are controlled by physicochemical reactions of the cell wall, including electrostatic and Van der Waals interactions, as well as hydrophobic properties. These are the most reliable long-range non-covalent interactions (Lewis acid–base) due to the surface proteins and lipoteichoic acids covering the peptidoglycan, and conferring a net negative bacterial surface charge in physiological conditions [24]. According to the authors, this feature is strain-specific and may vary depending on the medium, age and surface structures of bacteria. Indeed, considerable variability in the hydrophobicity capacity has been observed in our study, with 65% of the isolates showing high hydrophobicity (70%).
Auto-aggregation properties, together with the co-aggregation ability, of a probiotic strain are necessary for adhering to the intestinal tract by forming a defensive barrier against the colonization of foodborne pathogens [30]. Moreover, the LAB co-aggregating ability might regulate the pathogen microenvironment and stimulate the excretion of antimicrobial substances [31]. Lactobacillus spp. also favors many aggregation-promoting factors (APFs) involved in auto-aggregation and/or adhesion in a strain-specific manner [32]. Furthermore, exopolysaccharides (EPS) are believed to play an essential role in cell aggregation, biofilm formation and adhesion. Polak-Berecka and colleagues concluded that the adherence and/or co-aggregation ability of Lactobacillus rhamnosus are strongly related to specific interactions based on surface proteins and specific fatty acids, whereas polysaccharides (hydrophilic nature) hinder the adhesion and aggregation by masking protein receptors [33].
Aggregation values have been shown to increase over time, typically at 20 h of incubation, in a strain-dependent manner [34], which is in accordance with our results. All isolates with the (Agg+) phenotype were identified as Ligilactobacillus salivarius, thus corroborating the findings of Ait Seddik and colleagues, who demonstrated the high auto-aggregation ability of this strain [35]. According to Solieri and colleagues, co-aggregation values below 20% are indicative of a weak co-aggregation ability [36]. Our isolates differed in their co-aggregation abilities (0 to 94.6%), highlighting once again these strain-specific characteristics.
Another probiotic protective mechanism involves the competition for adhesion sites [37]. Ligilactobacillus salivarius (16/c6, 16/i4, 14/i8 and A30/i26), Limosilactobacillus reuteri (1/c24), L. crispatus (16/c2) and Limosilactobacillus fermentum (12/c8 and 16/i10) were selected according to their cell hydrophobicity and auto/co-aggregation abilities. The adherence capacity differed significantly between the lactobacilli strains isolated, which is consistent with other studies, showing that this ability is species and strain-dependent [38]. The highest adhesion ability was shown in four isolates of lactobacilli: Ligilactobacillus salivarius A30/i26 and 16/i4, being highly auto-aggregative and hydrophobic, as well as in Ligilactobacillus salivarius 16/c6 and Limosilactobacillus reuteri 1/c24, showing great co-aggregation and hydrophobicity abilities. L. crispatus 16/c2, Limosilactobacillus reuteri 12/c8 and Ligilactobacillus salivarius 14/i8 had the lowest adhesion percentages, despite their high co-aggregation capacities. Interestingly, Limosilactobacillus sp.16/i10, a high hydrophobic strain, also exhibited a low adhesion percentage.
The studied parameters, i.e., hydrophobicity, aggregation / co-aggregation and adhesion, illustrated no interrelation. However, some studies mentioned that the cell surface hydrophobicity is related to the attachment to epithelial cells [39,40], while others have excluded this relationship in their analyses [27]. García-Cayuela and colleagues revealed a correlation between auto-aggregation and co-aggregation [29], which disagrees with our results. Del Re and colleagues proposed that auto-aggregation and hydrophobicity are independent characteristics, but both are necessary for adhesion [41]. Multitude interrelated surface factors (fatty acids, surface proteins, LPS and EPS) may have unpredictable effects on adherence, co-aggregation and cell-to-cell interactions [38].
Survival in the GIT is a critical probiotic property. Bile tolerance is strain-specific and related to the hydrolase activity [42]. By mimicking the GIT conditions, all the eight lactobacilli strains were capable of growing at 0.1% (w/v) bile salts, but two Ligilactobacillus salivarius strains, namely, A30/i26 and 16/i4, were affected by 0.3%. This concentration is considered critical for screening for resistant probiotic strains [27]. Genes involved in bile salt hydrolysis, bsh-1 and bsh-2, were found to be responsible for the acid and bile tolerance in Ligilactobacillus salivarius UCC118 [26]. In favor of our findings, a significantly decreasing cell count in most of the Ligilactobacillus salivarius isolates has been observed when the strains were incubated with a high concentration of bile salts (0.5%), whereas most of the Limosilactobacillus reuteri isolates showed a high tolerance [43].
Ligilactobacillussalivarius A30/i26 and 16/c6 and Limosilactobacillus reuteri 1/c24 were selected for their high adhesion abilities and were further evaluated for their potential to compete with the three Salmonella serotypes in, or exclude them from, epithelial adhesion using the Caco2 cells as an experimental model. The inhibition of the pathogen adhesion by the three probiotic strains indicated a high variability in a strain-dependent manner. Ligilactobacillus salivarius 16/c6 significantly inhibited the adhesion of the three Salmonella serotypes to the Caco-2 cell monolayers only by exclusion assays, which is in accordance with findings of Campana, Van Hemert and Baffone [38]. The authors suggested that Ligilactobacillus salivarius W24 could significantly inhibit the adhesion of pathogens to Caco-2 cells only by exclusion. Jankowska and colleagues showed that L. paracasei reduced Salmonella’s adhesion to Caco-2 cells by 4- and 7-fold in competition and exclusion experiments, respectively [44]. However, the inhibition of Salmonella’s attachment to Caco-2 cells by exclusion, as well as by competition, has been frequently reported [37,45,46].
The inhibition of the Salmonella serotypes by Ligilactobacillus salivarius 16/c6 was similarly demonstrated by a mixed co-culture assay. When the co-cultures were tested without vortexing, the kinetic growth results of the lactobacilli and the pathogens confirmed what was previously distinguished by the auto-aggregation and co-aggregation assays and emphasized the ability for these features over time. Indeed, both co-cultures and the Ligilactobacillus salivarius monoculture revealed a clear supernatant after 8 h of incubation. Additionally, it has been demonstrated that the efficient aggregation and proper settling of flocs are essential for the management of effluent in the activated sludge process [47]. In this regard, such a feature of our strain might be promising in regard to the purification and decontamination of wastewater of slaughterhouses, which is mainly polluted by pathogens and organic materials.
When Ligilactobacillus salivarius 16/c6 and the Salmonella serotypes were subjected to the same co-culture assay but with vortexing, the reduction in the Salmonella counts in the mixed cultures co-occurred with a decrease in the pH, which is in accordance with findings of other studies [43]. Some bacterial strains have acid-adaptation systems that enable them to survive at pH < 2 [2]. Other non-negligible antimicrobial factors are involved in the Salmonella inhibition, such as competition for nutrients [43] and the contact-dependent inhibition (CDI) mechanism [48]. The latter case, where cell-to-cell contact is required, could be explained by the exchange of and interactions between bacteria mediated by conjugation, secretion systems, contact-dependent inhibition, allolysis and nanotubes. In fact, in our study, the low count was observed at 4 h among the Salmonella monocultures and mixed co-cultures.
4. Materials and Methods
4.1. Isolation and Phenotypic Characterization of Lactobacillus spp.
The different lactobacilli were isolated from the digestive tracts (ileum and cecum) of 16 antibiotic-free healthy broiler groups of different ages (four levels), breeds (four species) and diet formulas (four levels), as well as from 10 antibiotic-treated commercial broilers (Table 3). Experiments coded from 1 to 16 corresponded to the antibiotic-free broiler group, the “A”-coded experiment represents the group of antibiotic-treated commercial broilers, and the sample origin was designated as “i” for ileum and “c” for cecum. Samples of the ileum or cecum content of each category were homogenized at a ratio of 1:10 (10 g of ileum or cecum content in 90 mL of buffered peptone water (Scharlau-Chemie, Barcelona, Spain)). The homogenate was diluted 107-fold and 0.1 mL was plated onto de Man, Rogosa and Sharpe (MRS) agar (Sigma-Aldrich, Burlington, MA, USA). The plates were incubated anaerobically for 3 to 4 days at 37 °C. In total, 210 randomly selected strains were first characterized by Gram staining, motility and the detection of catalase activity. Gram-positive, catalase-negative bacilli were presumptively considered as Lactobacillus for further identification. Isolates were preserved in MRS broth with 20% glycerol at −70 °C until further use. All strains were revivified by successive streaking on MRS agar prior to performing any assay.

Table 3.
List of coded experiments numbered according to age, breed and diet formula of the broilers and hens deprived of any antibiotics and feed additives. The A-coded sample refers to non-antibiotic-free commercial broilers.
4.2. Salmonella Isolates
The antagonistic activity and co-aggregation ability of the lactobacilli strains were tested on three native avian Salmonella strains isolated from our previous study [4,49]. S. Enteritidis is the most predominant avian pulsotype causing human illness, whereas S. Kentucky ST198 and S. Infantis were chosen for their MDR pattern and their high prevalence in Lebanese poultry production. Strains were inoculated into 15 mL tryptic soy broth (TSB) (Sigma-Aldrich) and incubated at 37 °C for 18 h for further analyses.
4.3. Assessment of the Lactobacilli Antagonism
The anti-Salmonella activities of 210 presumptive lactobacilli were preliminarily screened using the simple spot-on-lawn antimicrobial assay and the agar well-diffusion method [25], with minor modifications. In brief, 10 µL of overnight lactobacilli cultures were spotted onto the surfaces of MRS agar plates and then incubated anaerobically for 18 h at 37 °C. In parallel, an overnight culture of each selected Salmonella serotype was inoculated at 105 CFU/mL into 7 mL of TSB soft agar (0.7% agar) and then poured onto the agar plates previously cultured with a strain of lactobacilli. After solidification, the plates were incubated for an additional 18 h at 37 °C under anaerobic conditions. The anti-Salmonella activity was evaluated by observing the inhibition zones around lactobacilli spots.
The agar well-diffusion assay was performed to identify the inhibitory substances secreted in the culture supernatants. The lactobacilli isolates showing antagonism were grown overnight at 37 °C in 15 mL of MRS broth. The cell-free supernatants (CFSs) were obtained by centrifugation (4000× g, 20 min, 4 °C) and filtration using 0.22 μm-pore Hi-MED syringe filters. The pH of the CFSs was then adjusted to 6.5 by 1 N NaOH. The Salmonella isolates were added at 106 CFU/mL to 20 mL of TSB supplemented with 0.75% agar-agar (semi-solid) and then poured into an empty Petri dish. After solidification, 6 mm wells were punched and 50 µL of the CFS was added to each well. The plates were left to settle at 8 °C for 24 h to enable the diffusion of the secreted antimicrobial substances, then incubated at 37 °C for 24 h. The absence or presence of any inhibitory zones was recorded after 24 h of incubation at 37 °C. The two assays were performed in triplicate.
4.4. Selection of Strains Based on Their Phenotypic Aggregation
A preliminary visual aggregation screening was performed according to Del Re et al. [41], with minor modifications. In brief, the lactobacilli cultured in the MRS broth were classified into three categories: category I for strains with an aggregation phenotype (Agg+) showing visible aggregates even after vigorous vortexing, category II for strains with constant turbidity and without precipitate (Agg−), and category III for strains with a mixed phenotype forming a precipitate and a clear or small turbid supernatant (Agg+/Agg−).
4.5. Species Identification and Phylogenetic Relationships
The 48 selected lactobacilli isolates were identified by 16S rRNA gene sequence analysis. The DNA extraction was achieved with a Qiamp DNA Mini Kit (Qiagen, Hilden, Germany). The amplification of the 16S rRNA gene sequence was performed in a Veriti thermal cycler (Applied Biosystem, CA, USA) under the following cycling conditions:: denaturation at 95 °C for 15 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 52 °C for 1 min and extension at 72 °C for 2 min, followed by another extension at 72 °C for 7 min. Reaction mixtures (50 μL) were prepared as follows: reaction buffer 10 × (5 μL), 10 mM dNTPs mix (1 μL), 0.5 mM of primer (27F (5′-GTGCTGCAGAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-CACGGATCCTACGGGTACCTTGTTACGACTT-3′), bacterial DNA (5 μL) and 2.5 U of HotStarTaq DNA polymerase (Qiagen, Hilden, Germany). Th amplicon separation was completed by electrophoresis at 100 V on 1% (w/v) agarose, stained with ethidium bromide in 1 × TBE buffer and purified using the GenElute TM PCR Clean-Up Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The DNA sequencing was carried out on a SeqStudio Genetic Analyzer (Applied Biosystem, USA). The editing was performed with Bioedit (version 7.2.5, 2013), and the 16S rDNA sequences were compared with other sequences using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 25 April 2022). A phylogenetic tree was assembled using the neighboring methods [50], with the tree builder function of MEGA X [51].
4.6. Cell Surface Properties
4.6.1. Auto-Aggregation and Co-Aggregation Assays
Auto-aggregation and co-aggregation capacities of the selected lactobacilli strains, according to their auto-aggregation visual features, were further assessed by spectrophotometric analysis at 4 h and 24 h, as described by Collado and colleagues [34], with minor modifications. An overnight lactobacilli culture (108 CFU/mL) was centrifuged (4000× g, 20 min, 4 °C) and the pellet was washed with phosphate-buffered saline (PBS) at pH 7.1 and then resuspended in the same buffer. The cell suspension (4 mL) was placed into glass bijou bottles and incubated at room temperature for 24 h. The absorbances at 600 nm (A600 nm) were measured at different times of incubation (t0, t4 and t24).
The auto-aggregation percentage was calculated using the following formula: 1 − (At/A0) × 100, where At represents the absorbance at different times (4 and 24 h) and A0 the absorbance at time = 0 (t0). The aggregation ability was classified according to Del Re and colleagues [41], with minor modifications. Results with values ≥ 65% and ≤10% were considered as highly auto-aggregative and non-auto-aggregative, respectively.
For the co-aggregation assay, mixed cultures with equal volumes (2 mL) of lactobacilli and Salmonella strains, as well as monocultures (4 mL), were prepared and incubated at room temperature without agitation. A600 nm were measured at 24 h of incubation.
The percentage of co-aggregation was calculated according to Handley and colleagues [39], as follows: (1− Amix/ (ASal + ALac)/2) × 100, where ASal and ALac represent the absorbances of the monocultures, Salmonella and lactobacilli, respectively, while Amix represents the absorbance of the mixed culture at 24 h. Values below 20% are indicative of a weak co-aggregation capability [36].
4.6.2. Hydrophobicity Assays
The microbial adhesion to the hydrocarbons (MATH) test was evaluated as defined by Rosenberg and colleagues [52], with slight changes. Lactobacilli cultures were centrifuged, and the pellet was washed with PBS buffer pH = 7.1 and suspended in the same buffer to adjust the concentration to 108 CFU/mL. An equal volume of 2 mL cell culture and xylene (nonpolar solvent) were mixed and vigorously vortexed for 5 min before measuring the A600 nm (A0). After incubation at room temperature for 1 h, the aqueous phase was cautiously removed, and its A600 nm (A1) was measured.
The cell surface hydrophobicity (H) was calculated as follows: H% = (1 − A1/A0) × 100. Isolates with (H) values > 70%, between 50–70% and < 50% were classified as highly, moderately and low-hydrophobic, respectively [53].
4.7. In Vitro Cell Tolerance to Gastrointestinal Conditions
The tolerance to gastrointestinal conditions of the eight lactobacilli strains, which were chosen according to their hydrophobicity and auto/ co-aggregation capacities, was assessed according to Babot and colleagues [24], with minor modifications. Overnight lactobacilli cultures were centrifuged (4000× g, 4 °C, 20 min) and adjusted to approximately 108 CFU/mL in PBS buffer. A volume of 1.75 mL was inoculated in 2.25 mL of a simulated gastric juice (125 mM NaCl, 7 mM KCl, 45 mM NaHCO3, 3 g/L pepsin pH 2.0). After incubation at 41.5 °C (poultry corporal temperature) for 1 h (mean retention time in the proventriculus and gizzard), the suspension was centrifuged and washed twice with PBS buffer. The pellet was then suspended in 3 mL of simulated intestinal juice (NaCl 22 mM, KCl 3.2 mM, NaHCO3 7.6 mM, pancreatin 0.1% w/v, bile salts 0.15% or 0.3% w/v, pH = 8.00) and incubated at 41.5 °C for 2 h (mean retention time in the small intestine). The concentrations of bile salts were selected to simulate the 0.1 to 1% bile concentration range of the poultry GIT, with approximately 0.25% in the ileum and 0.1% in the cecum [12]. After the serial dilutions, 0.1 mL of the suspensions was plated onto the MRS agar and incubated anaerobically for three days at 37 °C.
Tolerance to the GIT conditions was evaluated as follows: % survival = (Log10 N1/Log10 N0) × 100, where Log10 N0 is the number of bacterial cells in the PBS, and Log10 N1 represents the number of viable cells after exposure to gastrointestinal conditions.
4.8. Cell Culture
4.8.1. Cell Line and Growth Conditions
The human colorectal adenocarcinoma Caco-2 cell line was used to perform the adhesion assays. Cells were grown in a 75 cm2 flask containing Dulbecco’s modified Eagle’s medium (DMEM) (1× DMEM, 1M-1Glutamax, Gibco), supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Eurobio), 1× non-essential amino acids (NEAA), 100 U/mL penicillin and 10 mg/mL streptomycin (Sigma-Aldrich). Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 until 80% confluence was reached. Prior to the adhesion assay, 5 × 104 cells were seeded in 24-well tissue culture plates and incubated in the same conditions as mentioned above for 16 days (full differentiation). At the end of the incubation time, the cell line monolayers were washed twice with Dubelcco’s PBS (Eurobio) to remove antibiotics before adding the bacterial suspension.
4.8.2. Adhesion to Caco-2 Cells
Overnight cultures of the selected lactobacilli strains (16/c6, 16/c2, 16/i10, 16/c4, 14/i8, 12/c8, 1/c24 and A30/i26) and Salmonella serotypes were centrifuged, washed twice in Dulbecco’s PBS (Eurobio) and suspended in an antibiotic-free DMEM medium at a concentration of 108 CFU/mL. Then, 1 mL of bacterial culture was added to each cell well and incubated for 1 h at 37 °C in a humidified atmosphere containing 5% CO2. After that, the supernatant was removed and the wells were gently washed three times with Dulbecco’s PBS buffer to eliminate non-adherent bacteria. Finally, the Caco-2 monolayers were trypsinized with 0.25% trypsin-EDTA solution (Eurobio), and the adherent bacteria were enumerated by plating serial dilutions onto MRS agar medium for Lactobacillus and TSA agar medium for Salmonella. The adhesion ability was calculated as (N1/N0) × 100, where N1 and N0 represent the CFUs of the total adhered and added bacteria, respectively. Two independent experiments were conducted in triplicate for each condition.
4.8.3. Inhibition of the Adhesion of Salmonella to Caco-2 Cells
Two different protocols were followed to evaluate the ability of the selected lactobacilli strains to inhibit Salmonella’s adhesion to the Caco-2 cells. The Ligilactobacillus salivarius (16/c6 and A30/i26) and Limosilactobacillus reuteri (1/c24) strains were selected based on their adhesion properties. The competition adhesion assay was performed by seeding Caco-2 cell monolayers with a mixed culture of each of the selected lactobacilli (108 CFU/mL) with each of the Salmonella strains (107 CFU/mL) in complete DMEM. The Salmonella monocultures were used as controls. After an incubation period of 2 h at 37 °C in a humidified atmosphere containing 5% CO2, the supernatants with the non-adherent bacteria were removed, and then the Caco-2 cells were trypsinized. The adherent bacterial cells were serially diluted and plated onto TSA agar medium and MRS agar medium to enumerate the Salmonella and Lactobacillus, respectively.
The ability of the Salmonella strains to adhere to the Caco-2 cells in the absence (NSal) and the presence (NMix) of lactobacilli was calculated as follows:
Anti-adhesion ability% = 1 − (NMix/NSal)% [54].
For the exclusion assays, the Caco-2 cell monolayers were pre-exposed to lactobacilli strains (108 CFU/mL) for 1 h [37]. Then, the Caco-2 cell monolayers were gently washed three times with Dulbecco’s PBS prior to the addition of the Salmonella strains (107 CFU/mL) and incubation for 2 h. At the end of incubation time, supernatants with the non-adherent bacteria were removed, and the Caco-2 cells were then trypsinized. The adherent bacterial cells were serially diluted and plated onto TSA and MRS agar media to enumerate the Salmonella and Lactobacillus, respectively. Two independent experiments for each strain were conducted in triplicate for each condition.
4.9. Co-Culture Growth Kinetic Study
Two series of experiments were carried out to evaluate the effects of Ligilactobacillus salivarius 16 / c6 on the growth of the Salmonella strains in a co-culture model. In the first co-culture experiment, an 18 h-old culture of the 16 /c6 strain (107 CFU/mL) was co-inoculated with each culture of the three Salmonella strains (approximately 105 CFU/mL) into 100 mL of Laptg medium (peptone 15 g/L, tryptone 10 g/L, yeast extract 10 g/L, glucose 10 g/L, tween 80 0.1%; all media/chemicals were purchased from Sigma-Aldrich) at pH 6.9 and then incubated in a shaker incubator at 100 rpm at 37 °C for 24 h. Pure cultures of each of the strains served as controls. Before enumeration, the culture was left for 10 min without vortexing to evaluate the auto/co-aggregation capacity of Ligilactobacillus salivarius. Then, 0.1 mL of supernatant was plated out at different times (0, 4, 8 and 24 h) in triplicate onto the selective media (XLD agar for Salmonella and MRS agar for Lactobacillus) for counting. The pH of the culture medium was checked regularly. In the second experiment, the bacterial cultures were prepared as described above. Before enumeration, the culture was vigorously vortexed.
4.10. Statistical Analyses
Statistical analyses were performed using XLSTAT version 2021.4 (Addinsoft Inc. Paris, France). The surface properties of the forty-eight lactobacilli (n = 3) strains were assessed by principal component analysis (PCA). The index of Pearson was used to evaluate the correlation between the six assays, hydrophobicity, auto-aggregation and co-aggregation between a given lactobacilli strain and each of the S. Enteritidis, S. Infantis and S. Kentucky strains. Differences between the results for the adhesion and inhibition by competitive/exclusion were performed by one-way ANOVA, and p-values ≤ 0.05 were considered statistically significant.
5. Conclusions
This work was the first national tentative attempt to isolate potential candidates from the lactobacilli of Lebanese poultry to act as probiotics. The Ligilactobacillus. salivarius 16/c6 isolate that we highlighted here could be used as a potent probiotic dietary supplement in order to reinforce the intestinal microbiota of newly hatched chickens due to its high viability and long persistence in the poultry intestinal tract, as well as its ability to block the adhesion sites against Salmonella spp. The adhesion of lactobacilli strains to epithelial cells should also be investigated using the chicken LMH cell line to evaluate its probiotic potential in poultry. The study of these parameters is a crucial step in disseminating such native probiotic strains. However, further in vivo studies are required in order to ultimately better our understanding of how lactobacilli strains interact with and affect the fitness of Salmonella in the GITs of chicken hosts.
Author Contributions
Conceptualization, R.E.H. and Y.E.R.; methodology, R.E.H., S.P.S., F.M. and Y.E.R.; software, R.E.H., Z.A.K. and Y.E.R.; validation, R.E.H., F.M., J.-M.S., Z.A.K. and Y.E.R.; formal analysis, R.E.H. and Z.A.K.; investigation, R.E.H., J.E.H., S.P.S., J.T. and R.R.; resources, R.E.H., J.E.H., F.M. and J.-M.S.; data curation, R.E.H. and Z.A.K.; writing—original draft preparation, R.E.H. and I.A.; writing—review and editing, S.P.S., F.M., J.-M.S., Z.A.K. and Y.E.R.; visualization, R.E.H., J.E.H., Z.A.K. and Y.E.R.; supervision, F.M. and Y.E.R.; project administration, F.M. and Y.E.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to thank the general director of the LARI, Michel Afram, and INP-ENSAT in Toulouse for their full support. The authors also thank Chantal Ghanem for her help in plotting the principal component analysis (PCA).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- European Food Safety Authority/European Centre for Disease Prevention and Control. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2016. EFSA J. 2017, 15, e04676. [Google Scholar] [CrossRef]
- Tan, S.M.; Lee, S.M.; Dykes, G.A. Buffering Effect of Chicken Skin and Meat Protects Salmonella Enterica Strains against Hydrochloric Acid but Not Organic Acid Treatment. Food Control 2014, 42, 329–334. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The Role of Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed]
- El Hage, R.; El Rayess, Y.; Bonifait, L.; El Hafi, B.; Baugé, L.; Viscogliosi, E.; Hamze, M.; Mathieu, F.; Matar, G.M.; Chemaly, M. A National Study through a ‘Farm-to-Fork’ Approach to Determine Salmonella Dissemination along with the Lebanese Poultry Production Chain. Zoonoses Public Health 2022, 69, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Yu, Z. Intestinal Microbiome of Poultry and Its Interaction with Host and Diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial Resistance: A Global Emerging Threat to Public Health Systems. Crit. Rev. Food Sci. Nutr. 2017, 53, 2857–2876. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e144802. [Google Scholar] [CrossRef]
- Regulation (EC) No 1831/2003. Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003 on Additives for Use in Animal Nutrition. Off. J. Eur. Communities 2003, L268, 15.
- Krysiak, K.; Konkol, D.; Korczyński, M. Review Overview of the Use of Probiotics in Poultry Production. Animals 2021, 11, 1620. [Google Scholar] [CrossRef]
- Foley, S.L.; Nayak, R.; Hanning, I.B.; Johnson, T.J.; Han, J.; Ricke, S.C. Population Dynamics of Salmonella Enterica Serotypes in Commercial Egg and Poultry Production. Appl. Environ. Microbiol. 2011, 77, 4273–4279. [Google Scholar] [CrossRef]
- FAO; WHO. Guidelines for the Evaluation of Probiotics in Food; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Spivey, M.A.; Dunn-Horrocks, S.L.; Duong, T. Epithelial Cell Adhesion and Gastrointestinal Colonization of Lactobacillus in Poultry. Poult. Sci. 2014, 93, 2910–2919. [Google Scholar] [CrossRef] [PubMed]
- Saint-Cyr, M.J.; Guyard-Nicodème, M.; Messaoudi, S.; Chemaly, M.; Cappelier, J.M.; Dousset, X.; Haddad, N. Recent Advances in Screening of Anti-Campylobacter Activity in Probiotics for Use in Poultry. Front. Microbiol. 2016, 7, 553. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Forssten, S.; Hibberd, A.A.; Lyra, A.; Stahl, B. Probiotic Approach to Prevent Antibiotic Resistance. Ann. Med. 2016, 48, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Quezada, S.; Gomez-Llorente, C.; Plaza-Diaz, J.; Chenoll, E.; Ramón, D.; Matencio, E.; Bermudez-Brito, M.; Genovés, S.; Romero, F.; Gil, A.; et al. Competitive Inhibition of Three Novel Bacteria Isolated from Faeces of Breast Milk-Fed Infants against Selected Enteropathogens. Br. J. Nutr. 2013, 109 (Suppl. 2), S63–S69. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, L.; Zhou, L.; Yang, X.; Zhao, X. Using in Vitro Immunomodulatory Properties of Lactic Acid Bacteria for Selection of Probiotics against Salmonella Infection in Broiler Chicks. PLoS ONE 2016, 11, e0147630. [Google Scholar] [CrossRef] [PubMed]
- Rantala, M.; Nurmi, E. Prevention of the Growth of Salmonella Infantis in Chicks by the Flora of the Alimentary Tract of Chickens. Br. Poult. Sci. 1973, 14, 627–630. [Google Scholar] [CrossRef]
- Ayeni, A.O.; Ruppitsch, W.; Ayeni, F.A. Characterization of Bacteria in Nigerian Yogurt as Promising Alternative to Antibiotics in Gastrointestinal Infections. J. Diet. Suppl. 2018, 211, 1–11. [Google Scholar] [CrossRef]
- Yadav, A.K.; Tyagi, A.; Kumar, A.; Panwar, S.; Grover, S.; Saklani, A.C.; Hemalatha, R.; Batish, V.K. Adhesion of Lactobacilli and Their Anti-Infectivity Potential. Crit. Rev. Food Sci. Nutr. 2017, 57, 2042–2056. [Google Scholar] [CrossRef]
- Vineetha, P.G.; Tomar, S.; Saxena, V.K.; Susan, C.; Sandeep, S.; Adil, K.; Mukesh, K. Screening of Lactobacillus Isolates from Gastrointestinal Tract of Guinea Fowl for Probiotic Qualities Using in vitro Tests to Select Species-Specific Probiotic Candidates. Br. Poult. Sci. 2016, 57, 474–482. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap; Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Chen, X.; Xu, J.; Shuai, J.; Chen, J.; Zhang, Z.; Fang, W. The S-Layer Proteins of Lactobacillus Crispatus Strain ZJ001 Is Responsible for Competitive Exclusion against Escherichia coli O157:H7 and Salmonella Typhimurium. Int. J. Food Microbiol. 2007, 115, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Albazaz, R.I.; Byukunal Bal, E.B. Microflora of Digestive Tract in Poultry. Kahramanmaraş Sütçü İmam Üniversitesi Doğa Bilim. Derg. 2014, 17, 39–42. [Google Scholar] [CrossRef]
- Babot, J.D.; Argañaraz-Martínez, E.; Saavedra, L.; Apella, M.C.; Perez Chaia, A. Selection of Indigenous Lactic Acid Bacteria to Reinforce the Intestinal Microbiota of Newly Hatched Chicken—Relevance of in Vitro and Ex Vivo Methods for Strains Characterization. Res. Vet. Sci. 2014, 97, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Schillinger, U.; Lucke, F.K. Antimicrobial Activity of Lactobacillus Sake Isolated from Meat. Appl. Environ. Microbiol. 1989, 55, 1901–1906. [Google Scholar] [CrossRef]
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and Anti-Salmonella Activities of Lactic Acid Bacteria Isolated from Cattle Faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-Specific Probiotics Properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus crevis Isolates from Brazilian Food Products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef]
- Gupta, A.; Tiwari, S.K. Plantaricin LD1: A Bacteriocin Produced by Food Isolate of Lactobacillus plantarum LD1. Appl. Biochem. Biotechnol. 2014, 172, 3354–3362. [Google Scholar] [CrossRef]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; Gómez de Cadiñanos, L.P.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion Abilities of Dairy Lactobacillus plantarum Strains Showing an Aggregation Phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Kos, B.; Šušković, J.; Vuković, S.; Šimpraga, M.; Frece, J.; Matošić, S. Adhesion and Aggregation Ability of Probiotic Strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef]
- Potočnjak, M.; Pušić, P.; Frece, J.; Abram, M.; Janković, T.; Gobin, I. Three New Lactobacillus plantarum Strains in the Probiotic Toolbox against Gut Pathogen Salmonella enterica Serotype Typhimurium. Food Technol. Biotechnol. 2017, 55, 48–54. [Google Scholar] [CrossRef]
- Nishiyama, K.; Sugiyama, M.; Mukai, T. Adhesion Properties of Lactic Acid Bacteria on Intestinal Mucin. Microorganisms 2016, 4, 34. [Google Scholar] [CrossRef]
- Polak-Berecka, M.; Waśko, A.; Paduch, R.; Skrzypek, T.; Sroka-Bartnicka, A. The Effect of Cell Surface Components on Adhesion Ability of Lactobacillus rhamnosus. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2014, 106, 751–762. [Google Scholar] [CrossRef]
- Collado, M.C.; Surono, I.; Meriluoto, J.; Salminen, S. Indigenous Dadih Lactic Acid Bacteria: Cell-Surface Properties and Interactions with Pathogens. J. Food Sci. 2007, 72, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Ait Seddik, H.; Bendali, F.; Cudennec, B.; Drider, D. Anti-Pathogenic and Probiotic Attributes of Lactobacillus salivarius and Lactobacillus plantarum Strains Isolated from Feces of Algerian Infants and Adults. Res. Microbiol. 2017, 168, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Solieri, L.; Bianchi, A.; Mottolese, G.; Lemmetti, F.; Giudici, P. Tailoring the Probiotic Potential of Non-Starter Lactobacillus Strains from Ripened Parmigiano Reggiano Cheese by in Vitro Screening and Principal Component Analysis. Food Microbiol. 2014, 38, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.P.; Kaur, G.; Kapila, S.; Malik, R.K. Antagonistic Activity of Lactobacillus reuteri Strains on the Adhesion Characteristics of Selected Pathogens. Front. Microbiol. 2017, 8, 486. [Google Scholar] [CrossRef]
- Campana, R.; Van Hemert, S.; Baffone, W. Strain-Specific Probiotic Properties of Lactic Acid Bacteria and Their Interference with Human Intestinal Pathogens Invasion. Gut Pathog. 2017, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Handley, P.S.; Harty, D.W.; Wyatt, J.E.; Brown, C.R.; Doran, J.P.; Gibbs, A.C. A Comparison of the Adhesion, Coaggregation and Cell-Surface Hydrophobicity Properties of Fibrillar and Fimbriate Strains of Streptococcus salivarius. J. Gen. Microbiol. 1987, 133, 3207–3217. [Google Scholar] [CrossRef]
- De Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Lactobacillus casei and Lactobacillus fermentum Strains Isolated from Mozzarella Cheese: Probiotic Potential, Safety, Acidifying Kinetic Parameters and Viability under Gastrointestinal Tract Conditions. Probiotics Antimicrob. Proteins 2018, 11, 382–396. [Google Scholar] [CrossRef]
- Del Re, B.; Sgorbati, B.; Miglioli, M.; Palenzona, D. Adhesion, Autoaggregation and Hydrophobicity of 13 Strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000, 31, 438–442. [Google Scholar] [CrossRef]
- Zommiti, M.; Connil, N.; Hamida, J.B.; Ferchichi, M. Probiotic Characteristics of Lactobacillus curvatus DN317, a Strain Isolated from Chicken Ceca. Probiotics Antimicrob. Proteins 2017, 9, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Abhisingha, M.; Dumnil, J.; Pitaksutheepong, C. Selection of Potential Probiotic Lactobacillus with Inhibitory Activity Against Salmonella and Fecal Coliform Bacteria. Probiotics Antimicrob. Proteins 2018, 10, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, A.; Laubitz, D.; Antushevich, H.; Zabielski, R.; Grzesiuk, E. Competition of Lactobacillus paracasei with Salmonella Enterica for Adhesion to Caco-2 Cells. J. Biomed. Biotechnol. 2008, 2008, 357964. [Google Scholar] [CrossRef] [PubMed]
- Jessie Lau, L.Y.; Chye, F.Y. Antagonistic Effects of Lactobacillus plantarum 0612 on the Adhesion of Selected Foodborne Enteropathogens in Various Colonic Environments. Food Control 2018, 91, 237–247. [Google Scholar] [CrossRef]
- Hai, D.; Lu, Z.; Huang, X.; Lv, F.; Bie, X. In Vitro Screening of Chicken-Derived Lactobacillus Strains That Effectively Inhibit Salmonella Colonization and Adhesion. Foods 2021, 10, 569. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Sakamoto, M.; Hanazaki, S.; Osawa, M.; Suzuki, T.; Tochigi, M.; Kakii, K. Coaggregation among Nonflocculating Bacteria Isolated from Activated Sludge. Appl. Environ. Microbiol. 2003, 69, 6056–6063. [Google Scholar] [CrossRef]
- Bian, X.; Evivie, S.E.; Muhammad, Z.; Luo, G.W.; Liang, H.Z.; Wang, N.N.; Huo, G.C. In Vitro Assessment of the Antimicrobial Potentials of Lactobacillus helveticus Strains Isolated from Traditional Cheese in Sinkiang China against Food-Borne Pathogens. Food Funct. 2016, 7, 789–797. [Google Scholar] [CrossRef]
- El Hage, R.; Losasso, C.; Longo, A.; Petrin, S.; Ricci, A.; Mathieu, F.; Abi Khattar, Z.; El Rayess, Y. Whole-Genome Characterisation of TEM-1 and CMY-2 β-Lactamase-Producing Salmonella Kentucky ST198 in Lebanese Broiler Chain. J. Glob. Antimicrob. Resist. 2020, 23, 408–416. [Google Scholar] [CrossRef]
- Saitou, N.N.M.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees’. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rosenberg, M.; Gutnick, D.; Rosenberg, E. Adherence of Bacteria to Hydrocarbons: A Simple Method for Measuring Cell-Surface Hydrophobicity. FEMS Microbiol. Lett. 1980, 9, 29–33. [Google Scholar] [CrossRef]
- Buahom, J.; Siripornadulsil, S.; Siripornadulsil, W. Feeding with Single Strains Versus Mixed Cultures of Lactic Acid Bacteria and Bacillus subtilis KKU213 Affects the Bacterial Community and Growth Performance of Broiler Chickens. Arab. J. Sci. Eng. 2018, 43, 3417–3427. [Google Scholar] [CrossRef]
- Son, S.H.; Jeon, H.L.; Yang, S.J.; Lee, N.K.; Paik, H.D. In Vitro Characterization of Lactobacillus brevis KU15006, an Isolate from Kimchi, Reveals Anti-Adhesion Activity against Foodborne Pathogens and Antidiabetic Properties. Microb. Pathog. 2017, 112, 135–141. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).