Abstract
Shared bikes as a public transport provide convenience for short-distance travel. Whilst they also act as a potential vector for antimicrobial resistant (AR) bacteria and antimicrobial resistance genes (ARGs). However, the understanding of the whole genome sequence of AR strains and ARGs-carrying plasmids collected from shared bikes is still lacking. Here, we used the HiSeq platform to sequence and analyze 24 Escherichia coli isolated from shared bikes around Metro Stations in Beijing. The isolates from shared bikes showed 14 STs and various genotypes. Two blaNDM-5 and blaCTX-M-199-producing ST167 E. coli have 16 resistance genes, four plasmid types and show >95% of similarities in core genomes compared with the ST167 E. coli strains from different origins. The blaNDM-5- or blaCTX-M-199-carrying plasmids sequencing by Nanopore were compared to plasmids with blaNDM-5- or blaCTX-M-199 originated from humans and animals. These two ST167 E. coli show high similarities in core genomes and the plasmid profiles with strains from hospital inpatients and farm animals. Our study indicated that ST167 E. coli is retained in diverse environments and carried with various plasmids. The analysis of strains such as ST167 can provide useful information for preventing or controlling the spread of AR bacteria between animals, humans and environments.
1. Introduction
Shared bikes as a public transport provide more choices and convenience for people’s travel. They also act as the last-mile connection between means of transport such as light rail stations or bus stops and people’s destinations such as home or the office. Some studies suggest that public transportation such as buses, subways, and taxis can act as a transmission media for bacteria or viruses [,], which could cause public health emergencies. Meanwhile, microorganisms on the surface of public transport arouses concern due to the severity of antimicrobial resistance worldwide [,,]. Previous studies indicated that antimicrobial resistant (AR) Enterobacteriaceae, Staphylococcus spp. and Enterococcus spp. were already isolated from shared bikes [,,,]. Additionally, various bacteria with antimicrobial resistance genes (ARGs) were found in buses, subways, and aircrafts [,,,].
Several studies showed that both Gram-positive bacteria and Gram-negative bacteria could be isolated from shared bikes. Among them, Staphylococci and Enterococci were widely distributed in shared bikes around schools, hospitals, metro stations, and from riders, with detection rates of 2.3–12.9% and 0.08–5.5%, respectively []. The multiple resistant Staphylococci showed diversity in SCCmec and sequence type (ST) []. Meanwhile, the prevalence of Enterobacteriaceae in shared bikes was 19.7%, which suggested that hospitals might increase the risk of AR Enterobacteriaceae based on the distance from the hospital to the subway station []. Wu et al. reported that Bacillus was the most abundant bacteria in the shared bicycle bacteria community, and the drug-resistant bacteria in the shared bicycle bacterial community of metro stations, shopping malls, and hospitals showed no significant differences [].
In recent years, the increasing reports of carbapenem resistance genes have increased the pressure on effective bacterial treatment. ST167 E. coli was often reported to carry carbapenem resistance genes such as blaKPC-3, blaNDM-5, and blaNDM-1 and was found in various species such as ducks, cattle, and mussels [,,,,,]. A study on hospitalized neonatal sepsis showed that E. coli (34.01%) was one of the main pathogens of neonatal bacteremia, and ST167 was the most prevalent ST []. More importantly, ST167 has been reported to spread between companion animals and their owners []. The spread of ST167 clones between countries has also been reported []. Although characterization of bacteria from shared bikes has attracted widespread attention in recent years, current studies have mainly focused on the prevalence and the phenotypes of strain descriptions in public transportation or the features of isolates themselves. To the best of our knowledge, the whole genome analysis with strains from different locations or biological sources and comparisons of their plasmid profiles are still lacking. E. coli is an important representative of Enterobacteriaceae, which can carry a variety of ARGs and has significance for public health safety. Herein, we used the E. coli isolates from the shared bikes to investigate the similarities and differences between strains from the shared bikes and other sources to find the relationship of the whole genome sequencing between the E. coli isolates from environmental and clinical samples.
2. Results and Discussion
2.1. E. coli Isolates from Shared Bikes
We identified 14 STs among all 24 E. coli isolates from shared bikes (14 from Metro Station nearing secondary/tertiary hospitals and ten from non-hospital stations, Supplementary material Table S1), and the ST10 clonal complex (n = 7) were the dominant clonal complex (Figure 1). There is no dominant ST or clonal complex related to hospitals, although ST10, ST48, and ST167 found in this study were the most prevalent STs in hospitals [,,].
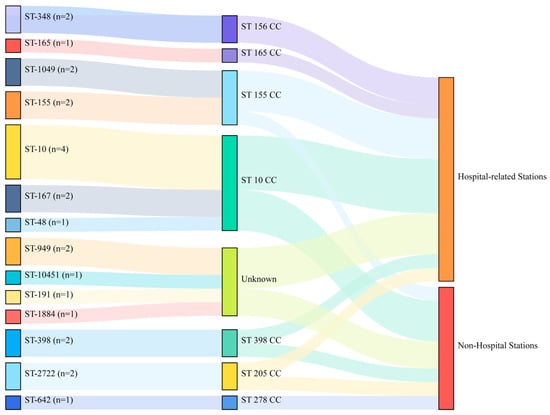
Figure 1.
The distribution of STs from 24 E. coli in shared bikes. (CC: clonal complex, hospital-related stations represent Metro Stations nearing secondary/tertiary hospitals).
The phylogenetic tree analysis showed that the 24 E. coli strains from shared bikes had different profiles. The number of ARGs in each of the strains ranged from one to sixteen, and plasmid types ranged from zero to five (Table S1). The resistance phenotypes showed that some strains (such as 770, 776) which have a higher number of resistance genes exhibited more resistance to antimicrobial agents than other strains. However, the number of strains exhibiting resistance phenotype mismatch the number of ARGs. Some strains showed high similarity in one small clade, for instance, 26, 25, 31 and 769, 780. Almost all AR strains have resistance genes of aminoglycosides, quinolones, sulfonamides, tetracyclines and beta-lactams. Despite most strains (66.7%) from hospital-related stations have resistance gene to different kinds of antimicrobial agents, there is no significant difference between multidrug resistance (MDR) E. coli from hospital-related stations and non-hospital stations (p > 0.05). The two strains (770 and 776) collected, respectively, from hospital-related stations and non-hospital stations carried the maximum number of resistance genes and plasmid types of all strains and showed >95% similarities in core genomes with the same sequence type ST167 (Figure 2).
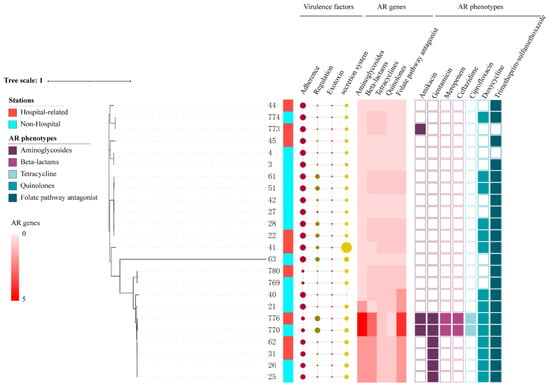
Figure 2.
The phylogenetic tree of 24 E. coli from shared bikes. (The size of circles represents the number of virulence factors. The color of the heatmap indicated the number of resistance genes found in different antibiotic classes. Different colors were used to distinguish AR phenotypes of each antibiotic class).
Furthermore, these two strains carried blaNDM-5 and blaCTX-M-199, and another 14 resistance genes including aminoglycoside resistance genes aadA2, aadA5, aph(3″)-Ib, aph(6)-Id, rmtB beta-lactam resistance genes blaEC-15 and blaTEM-1, phenicol resistance gene floR, macrolide resistance gene mph(A), tetracycline resistance gene tet(A), sulfonamide resistance genes sulI, sulII, trimethoprim resistant genes dfrA12 and dfrA17. In addition, the comparison of virulence factors between hospital-related and non-hospital stations showed no significant difference (p > 0.05). ST167 is one of the epidemic STs in E. coli that carried ARGs, especially β-lactamase genes []. Previous studies indicated that ARGs-carrying ST167 E. coli were isolated from humans, food animals, companion animals and environments [,,,]. Until now, ST167 E. coli were found in countries and districts across five continents, such as China, Tunisia, Switzerland, Italy, Finland, Canada, Brazil, and Tanzania [,,,,,,]. The ST167 E. coli carrying the blaNDM gene were previously identified in hospitals, livestock farms, poultry farms, and the environment [,,,,]. Growing evidence indicated that the public environment is of increasing concern as a reservoir for the transmission of MDR bacteria and genes. However, unlike strains from farm environments, the AR bacteria strains from public transportation mean that they can be transferred between individual populations due to personnel movement.
2.2. Comparison of Core Genome with ST167 E. coli from Different Origins
Due to the high prevalence of ST167 E. coli in the world, we would like to compare the profiles of ST167 E. coli from shared bikes and from other origins (Supplementary material Table S2). A total of 404 ST167 E. coli from the NCBI database were selected for comparative analysis with two E. coli from shared bikes. These strains were collected from human (n = 370), food animals (n = 11), companion animals (n = 15), environment (n = 8) samples (Figure 3) from 35 countries or districts (Supplementary material Figure S1). More than half of strains carried blaNDM (n = 288) and blaCTX-M gene (n = 272). blaNDM-5 were the most prevalent NDM type (n = 254) but blaCTX-M-199 were found on only one strain. The phylogenetic tree indicated that all ST167 E. coli strains exhibited various characterizations in the core genome and have 21~11,206 single nucleotide polymorphisms (SNPs) compared to strains from shared bikes. The two ST167 E. coli from shared bikes show high similarity (SNPs < 50) with 33 strains (Pink color range in the Figure 3) from samples of human (n = 18), dogs (n = 12), cats (n = 1), chicken (n = 1) as well as environment (n = 1). The human samples were identified from Bangladesh (n = 1), the United Kingdom (n = 7), China (n = 6), Switzerland (n = 1), Italy (n = 2) and the United States (n = 1). The dog strains were originated from Switzerland (n = 2) and the United States (n = 10). Other strains were collected from a cat in Italy, a chicken in China and an environmental source from the United States. All strains were collected from 2015 to 2021, while 32 of these strains carried blaNDM-5.
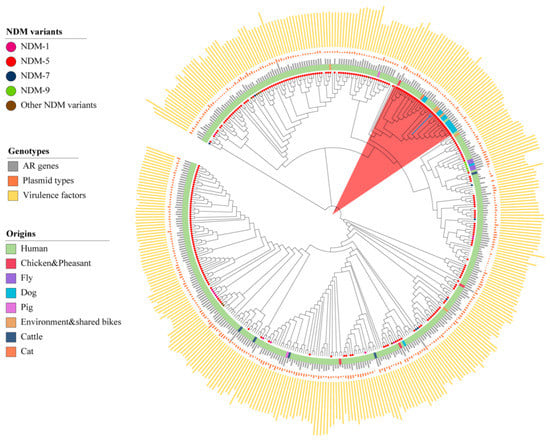
Figure 3.
The core genome phylogenetic tree of ST167 E. coli from humans, animals and the environment. (The blue branches are the strains from shared bikes, the length of the bar represents the number of ARGs/Plasmid types/Virulence factors. The circles attached to the leaves represent the NDM variants).
2.3. Comparison of Plasmid Profiles with ST167 E. coli from Different Origins
Illumina and Nanopore sequencing of blaNDM-5 or blaCTX-M-199-carrying isolates indicated that blaNDM-5 and blaCTX-M-199 were located on a ~98.5 kb IncFII plasmid and a ~113 kb IncFII plasmid, respectively. From the NCBI database, we downloaded nine plasmids that have the highest coverage and identities in sequences with blaNDM-5- or blaCTX-M-199-carrying plasmid of shared bikes (Supplementary material Table S3). Nine blaNDM-5-carrying plasmids belong to strains from patients in China (n = 3), Japan (n = 1), Tanzania (n = 1), Myanmar (n = 2) and Switzerland (n = 1), and from chicken meat in Laos (n = 1). Plasmid pNDM-EC16-50 in one E. coli strain from China showed >90% coverage and highest identifies with the blaNDM-5-carrying plasmid of shared bikes (Figure 4a). Nine blaCTX-M-199-carrying plasmids belong to strains from human (n = 3), chicken (n = 1), goose (n = 1) in China, humans in the United States (n = 1), Japan (n = 1), and Lebanon (n = 1), and water samples from India (n = 1). The nucleotide sequence of the blaCTX-M-199-carrying plasmid of shared bikes displayed the highest similarity with E. coli strain L100 plasmid pL100-3 and E. coli plasmid J-8 plasmid pCTX from goose and chicken in China (Figure 4b). According to the information of NCBI, we download the isolates that carried these similar plasmids (blaNDM-5 or blaCTX-M-199) and identified the ST of these isolates (except nine plasmids without the whole genome of isolates upload). The results showed that three blaNDM-5 plasmids were from ST167 E. coli of human origin, the other three plasmids were from non-ST167 E. coli, and three blaCTX-M-199 plasmids were from ST10 E. coli of human origin, ST148, ST156 E. coli of food animal origin. The results of the plasmid profiles comparison indicated that maybe some bacteria carrying plasmids with ARGs from patients and farm animals are possible to persist in the environment and further plasmid conjugative transfer to the bacteria of environments. Moreover, ST167 can acquire plasmids easily from other STs, which makes plasmids with ARGs commonly available.
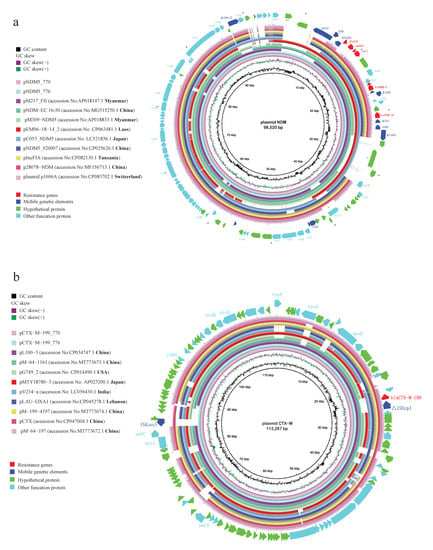
Figure 4.
The plasmid profiles of (a) blaNDM-5 and (b) blaCTX-M-199. (The reference sequences were blaNDM-5- or blaCTX-M-199-carrying plasmid of shared bikes. The shade of circles represents the number of identities, the blank means sequences were not consistent with the reference).
The plasmid analysis of ST167 E. coli from shared bikes showed that ST167 might be an important strain for plasmid-borne ARGs across different origins. Furthermore, combined with the results of the phylogenetic tree, the strains which have good environmental adaptability can increase the possibility of plasmid transfer between different bacteria which enhances the dissemination of ARGs among animals, humans and the environment, and threaten public health. Researchers are also concerned about the AR bacteria and gene transmission via the environment [,,]. Furthermore, these strains increased the difficulty of AR control. However, not only can ST167 act as the vector but also some other prevalent strains can play the same role as ST167, so the control of prevalent strains requires substantial concern.
We found a high similarity of strains from the shared bikes and other origins, which means that some well-adapted isolates can persist in different environments. Many studies proved that AR bacteria isolated from the same environment are convergence in molecular profile because the environment has prevalent AR bacteria and genes []. However, now some prevalent STs such as ST131, ST167 and ST 10 E. coli which have good environmental adaptability with ARGs can be a potential reservoir of ARGs in the environment []. Moreover, ST 167 E. coli carried important resistant genes, such as blaNDM, mcr-1, blaCTX-M [], which are stable in the environment and pose a threat to public health. The flow of population further accelerates ARGs spread to diverse environments or different species, which adds pressure to control antimicrobial resistance. Therefore, the dominant host of ARGs like ST 167 in the environment should be concerned and focused on.
3. Materials and Methods
3.1. Bacterial Isolates, Whole Genome Sequencing
A total of 444 Enterobacteriaceae were isolated from shared bikes in the previous study and E. coli was the species that exhibited more drug resistance than others []. Therefore, E. coli was chosen for further analysis. A total of 28 E. coli strains were isolated from samples in the previous study, excluding 4 from stations outside the fifth Ring Road of Beijing; finally, 24 E. coli isolates were collected. Genomic DNA was extracted using a HiPure Bacterial DNA Kit. DNA libraries were prepared and sequenced with HiSeq PE150. Two blaNDM-5 and blaCTX-M-199-producing E. coli were sequenced with Nanopore to obtain the complete plasmids. The sequences were assembled by SPAdes and Unicycler.
3.2. Assembled Data of ST167 E. coli from Different Sources
We searched all E. coli available in the NCBI database which were collected from January 2014 to December 2021 and downloaded those. We only selected assembled data of whole genome sequencing. Furthermore, we reorganized the detailed information related to the assembled data we downloaded and excluded strains without information on the host. All genomes were confirmed the ST using MLST. Additionally, only E. coli with ST167 were chosen for the following analysis.
3.3. Genomic Analysis of Sequenced and Collected E. coli Strains
ARGs and plasmid incompatibility groups were determined using the database (resfinder, plasmidfinder) from the Center for Epidemiology (http://www.genomicepidemiology.org/, accessed on 12 February 2022). According to the mechanism of resistance classified ARGs in different antibiotic classes. MLST was confirmed using MLST in the Center for Epidemiology and database from public databases for molecular typing and microbial genome diversity (https://pubmlst.org/, accessed on 23 February 2022 ). The Sankey diagram of the ST clonal complex was performed using plug-in components of Excel named EasyShu. Virulence genes were identified using the VFDB database and virulencefinder of Center for Epidemiology. The criteria of different groups of virulence genes in accordance with VFDB. The tests were used for the comparison of the number of virulence factors from hospital-related Stations and non-hospital Stations. Core genomes were extracted using Snippy []. Core genome phylogenetic trees were constructed using Snippy and Fasttree []. Phylogenetic tree of the core genomes with ARGs, plasmid types, stations and phenotypes displayed with iTOL []. Genes in the plasmids were annotated using PATRIC and NCBI. The comparison of plasmid profiles was performed using BLAST and BRIG. The reference plasmid was annotated using the DNAplotter.
4. Conclusions
ST167 E. coli found on shared bikes showed high similarities with strains from patients and food-producing animals, and the plasmids also showed high identities with those from humans and animals in this study. These AR bacteria may originate from hospitals or farms. Vectors such as shared bikes may contribute to the dissemination of these AR bacteria in the environment. Furthermore, the persistence of these AR bacteria in the environment challenges the control of AR bacteria and ARGs. In the future, we need to take measures to assess the risk of AR bacteria in the environment and cut off transmission.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11081030/s1, Figure S1. Number of ST 167 E. coli from different countries or districts. Table S1. The information of E. coli collected from shared bikes. Table S2. The information of ST167 E. coli of NCBI database. Table S3. The information of plasmids compared with blaNDM-5 and blaCTX-M-199-carrying plasmids from shared bikes.
Author Contributions
Conceptualization, B.S., Y.W. and L.L.; Methodology, Q.C., Z.Z. and L.L.; Validation, Q.C., Z.Z. and L.L.; Formal Analysis, L.L. and Q.C.; Data Curation, Q.C., H.L. and L.L.; Writing—Original Draft Preparation, Q.C., Z.Z. and L.L.; Writing—Review & Editing, Y.W., B.S. and C.C.; Visualization, Q.C., Z.Z. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (81861138051, 81991535, 32141002) and Beijing Natural Science Foundation (6222017).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data and material information used and analyzed in the current study are available from the corresponding author upon reasonable requests.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shen, C.; Feng, S.; Chen, H.; Dai, M.; Paterson, D.L.; Zheng, X.; Wu, X.; Zhong, L.L.; Liu, Y.; Xia, Y.; et al. Transmission of mcr-1-producing multidrug-resistant Enterobacteriaceae in public transportation in Guangzhou, China. Clin. Infect. Dis. 2018, 67 (Suppl. 2), S217–S224. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Ou, Q.; Lin, D.; Xu, P.; Li, Y.; Ye, X.; Zhou, J.; Yao, Z. Metro system in Guangzhou as a hazardous reservoir of methicillin-resistant Staphylococci: Findings from a point-prevalence molecular epidemiologic study. Sci. Rep. 2015, 5, 16087. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mendes, Â.; Martins da Costa, P.; Rego, D.; Beça, N.; Alves, C.; Moreira, T.; Conceição, T.; Aires-de-Sousa, M. Contamination of public transports by Staphylococcus aureus and its carriage by biomedical students: Point-prevalence, related risk factors and molecular characterization of methicillin-resistant strains. Public Health 2015, 129, 1125–1131. [Google Scholar] [CrossRef]
- Lutz, J.K.; van Balen, J.; Crawford, J.M.; Wilkins, J.R., 3rd; Lee, J.; Nava-Hoet, R.C.; Hoet, A.E. Methicillin-resistant Staphylococcus aureus in public transportation vehicles (buses): Another piece to the epidemiologic puzzle. Am. J. Infect. Control 2014, 42, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; Diamantino, F.; Coelho, C.; de Lencastre, H.; Aires-de-Sousa, M. Contamination of public buses with MRSA in Lisbon, Portugal: A possible transmission route of major MRSA clones within the community. PLoS ONE 2013, 8, e77812. [Google Scholar] [CrossRef]
- Wu, Y.; Xie, J.; Li, J.; Zhao, J.; Qiao, S.; Li, Y.; Zeng, J. Shared bicycle microbial community: A potential antibiotic-resistant bacteria warehouse. Folia Microbiol. 2021, 66, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xie, X.J.; Liu, J.X.; Shui, J.R.; Zhang, H.Y.; Feng, G.Y.; Liu, X.Y.; Li, L.C.; Lan, Q.W.; Jin, Q.H.; et al. Prevalence and transmission of antimicrobial-resistant Staphylococci and Enterococci from shared bicycles in Chengdu, China. Sci. Total Environ. 2020, 738, 139735. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, S.; Chen, L.; Liu, Y.; Tan, L.; Shen, J.; Zhang, W. Antimicrobial resistance and molecular characterization of methicillin-resistant coagulase-negative Staphylococci from public shared bicycles in Tianjin, China. J. Glob. Antimicrob. Resist. 2019, 19, 231–235. [Google Scholar] [CrossRef]
- Zou, Z.Y.; Lei, L.; Chen, Q.Y.; Wang, Y.Q.; Cai, C.; Li, W.Q.; Zhang, Z.; Shao, B.; Wang, Y. Prevalence and dissemination risk of antimicrobial-resistant Enterobacteriaceae from shared bikes in Beijing, China. Environ. Int. 2019, 132, 105119. [Google Scholar] [CrossRef] [PubMed]
- Mani, Y.; Mansour, W.; Mammeri, H.; Denamur, E.; Saras, E.; Boujâafar, N.; Bouallègue, O.; Madec, J.Y.; Haenni, M. KPC-3-producing ST167 Escherichia coli from mussels bought at a retail market in Tunisia. J. Antimicrob. Chemother. 2017, 72, 2403–2404. [Google Scholar] [CrossRef]
- Huang, J.L.; Jianjun Zhu, J.J.; Gong, D.J.; Wu, L.; Zhu, Y.Z.; Hu, L.Q. Whole genome sequence of EC16, a blaNDM-5-, blaCTX-M-55-, and fosA3-coproducing Escherichia coli ST167 clinical isolate from China. J. Glob. Antimicrob. Resist. 2022, 29, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Grönthal, T.; Österblad, M.; Eklund, M.; Jalava, J.; Nykäsenoja, S.; Pekkanen, K.; Rantala, M. Sharing more than friendship—transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Eurosurveillance 2018, 23, 1700497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lou, D.; Xu, Y.; Shang, Y.; Li, D.; Huang, X.; Li, Y.; Hu, L.; Wang, L.; Yu, F. First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol. 2013, 13, 282. [Google Scholar] [CrossRef]
- Wang, M.G.; Zhang, R.M.; Wang, L.L.; Sun, R.Y.; Bai, S.C.; Han, L.; Fang, L.X.; Sun, J.; Liu, Y.H.; Liao, X.P. Molecular epidemiology of carbapenemase-producing Escherichia coli from duck farms in south-east coastal China. J. Antimicrob. Chemother. 2021, 76, 322–329. [Google Scholar] [CrossRef] [PubMed]
- He, W.Y.; Zhang, X.X.; Gao, G.L.; Gao, M.Y.; Zhong, F.G.; Lv, L.C.; Cai, Z.P.; Si, X.F.; Yang, J.; Liu, J.H. Clonal spread of Escherichia coli O101: H9-ST10 and O101: H9-ST167 strains carrying fosA3 and blaCTX-M-14 among diarrheal calves in a Chinese farm, with Australian Chroicocephalus as the possible origin of E. coli O101: H9-ST10. Zool. Res. 2021, 42, 461–468. [Google Scholar] [CrossRef]
- Zou, H.; Jia, X.; He, X.; Su, Y.; Zhou, L.; Shen, Y.; Sheng, C.; Liao, A.; Li, C.; Li, Q. Emerging treat of multidrug resistant pathogens from neonatal sepsis. Front. Cell. Infect. Microbiol. 2021, 11, 694093. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, T.; Sadek, M.; Yao, Y.; Imirzalioglu, C.; Stephan, R.; Poirel, L.; Nordmann, P. Cross-border emergence of Escherichia coli producing the carbapenemase NDM-5 in Switzerland and Germany. J. Clin. Microbiol. 2021, 59, e02238-20. [Google Scholar] [CrossRef]
- Cantón, R.; Gijón, D.; Ruiz-Garbajosa, P. Antimicrobial resistance in ICUs: An update in the light of the COVID-19 pandemic. Curr. Opin. Crit. Care 2020, 26, 433–441. [Google Scholar] [CrossRef]
- Xu, L.; Wang, P.; Cheng, J.; Qin, S.; Xie, W. Characterization of a novel blaNDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 2019, 12, 511–519. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Zheng, S.; Yu, F.; Kong, H.; Yang, Q.; Cui, D.; Chen, N.; Lou, B.; Li, X.; et al. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China. Clin. Microbiol. Infect. 2014, 20, 52–58. [Google Scholar] [CrossRef]
- Garcia-Fernandez, A.; Villa, L.; Bibbolino, G.; Bressan, A.; Trancassini, M.; Pietropaolo, V.; Venditti, M.; Antonelli, G.; Carattoli, A. Novel insights and features of the NDM-5-producing Escherichia coli sequence type 167 high-risk clone. mSphere 2020, 5, e00269-20. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Taddei, R.; Cordaro, G.; Fontana, M.C.; Toschi, E.; Gaibani, P.; Marani, I.; Giacomi, A.; Diaconu, E.L.; Iurescia, M.; et al. Carbapenemase IncF-borne blaNDM-5 gene in the E. coli ST167 high-risk clone from canine clinical infection, Italy. Vet. Microbiol. 2021, 256, 109045. [Google Scholar] [CrossRef] [PubMed]
- Dziri, R.; Klibi, N.; Alonso, C.A.; Jouini, A.; Ben Said, L.; Chairat, S.; Bellaaj, R.; Boudabous, A.; Ben Slama, K.; Torres, C. Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST131-B2 and ST167-A in environmental samples of a Tunisian hospital. Microb. Drug Resist. 2016, 22, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Santos Tufic-Garutti, S.D.; de Araújo Longo, L.G.; Fontana, H.; Garutti, L.H.G.; de Carvalho Girão, V.B.; Fuga, B.; Lincopan, N.; de Pinho Rodrigues, K.M.; Moreira, B.M. OXA-181 carbapenemase carried on an IncX3 plasmid in high-risk Escherichia coli ST167 isolated from a traveler returning from Sub-Saharan Africa to Brazil. Diagn. Microbiol. Infect. Dis. 2022, 102, 115570. [Google Scholar] [CrossRef]
- Baloch, Z.; Lv, L.; Yi, L.; Wan, M.; Aslam, B.; Yang, J.; Liu, J.H. Emergence of almost identical F36: A-:B32 plasmids carrying blaNDM-5 and qepA in Escherichia coli from both Pakistan and Canada. Infect. Drug Resist. 2019, 12, 3981–3985. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yu, X.; Xie, M.; Wang, X.; Liao, K.; Xue, W.; Chan, E.W.; Zhang, R.; Chen, S. Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob. Agents Chemother. 2016, 60, 4364–4368. [Google Scholar] [CrossRef] [PubMed]
- Nukui, Y.; Ayibieke, A.; Taniguchi, M.; Aiso, Y.; Shibuya, Y.; Sonobe, K.; Nakajima, J.; Kanehira, S.; Hadano, Y.; Tohda, S.; et al. Whole-genome analysis of EC129, an NDM-5-, CTX-M-14-, OXA-10- and MCR-1-co-producing Escherichia coli ST167 strain isolated from Japan. J. Glob. Antimicrob. Resist. 2019, 18, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Peterhans, S.; Stevens, M.J.A.; Nüesch-Inderbinen, M.; Schmitt, S.; Stephan, R.; Zurfluh, K. First report of a blaNDM-5-harbouring Escherichia coli ST167 isolated from a wound infection in a dog in Switzerland. J. Glob. Antimicrob. Resist. 2018, 15, 226–227. [Google Scholar] [CrossRef]
- Sánchez-Benito, R.; Iglesias, M.R.; Quijada, N.M.; Campos, M.J.; Ugarte-Ruiz, M.; Hernández, M.; Pazos, C.; Rodríguez-Lázaro, D.; Garduño, E.; Domínguez, L.; et al. Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and blaCTX-M-15 resistance determinants isolated from a human respiratory infection. Int. J. Antimicrob. Agents 2017, 50, 285–286. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Z.; Xing, S.; Liao, X. The correlation between antibiotic resistance gene abundance and microbial community resistance in pig farm wastewater and surrounding rivers. Ecotoxicol. Environ. Saf. 2019, 182, 109452. [Google Scholar] [CrossRef]
- Jamborova, I.; Johnston, B.D.; Papousek, I.; Kachlikova, K.; Micenkova, L.; Clabots, C.; Skalova, A.; Chudejova, K.; Dolejska, M.; Literak, I.; et al. Extensive genetic commonality among wildlife, wastewater, community, and nosocomial isolates of Escherichia coli sequence type 131 (H30R1 and H30Rx subclones) that carry blaCTX-M-27 or blaCTX-M-15. Antimicrob. Agents Chemother. 2018, 62, e00519-18. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xu, C.; Sun, Q.; Schwarz, S.; Ou, Y.; Yang, L.; Huang, Z.; Eichhorn, I.; Walsh, T.R.; Wang, Y.; et al. Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 2018, 62, e00404-18. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bi, Z.; Ma, S.; Chen, B.; Cai, C.; He, J.; Schwarz, S.; Sun, C.; Zhou, Y.; Yin, J.; et al. Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ. Health Perspect. 2019, 127, 107009. [Google Scholar] [CrossRef]
- Seemann T Snippy: Fast Bacterial Variant Calling from NGS Reads. Available online: https://github.com/tseemann/snippy (accessed on 30 December 2018).
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).