Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and blaNDM-5 on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne blaCMY-2 from Egypt
Abstract
1. Introduction
2. Results and Discussion
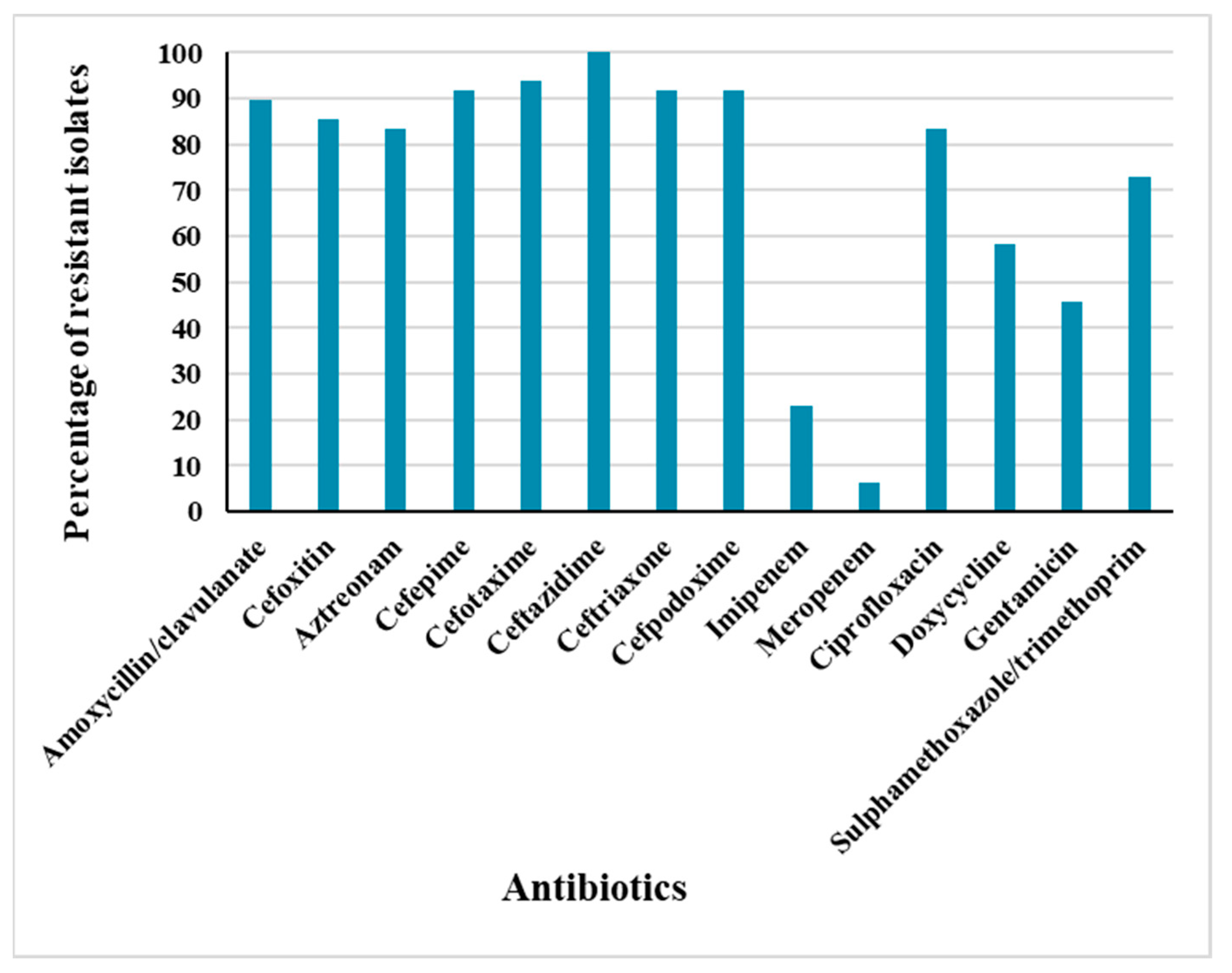
2.1. Antimicrobial Resistance Profiles
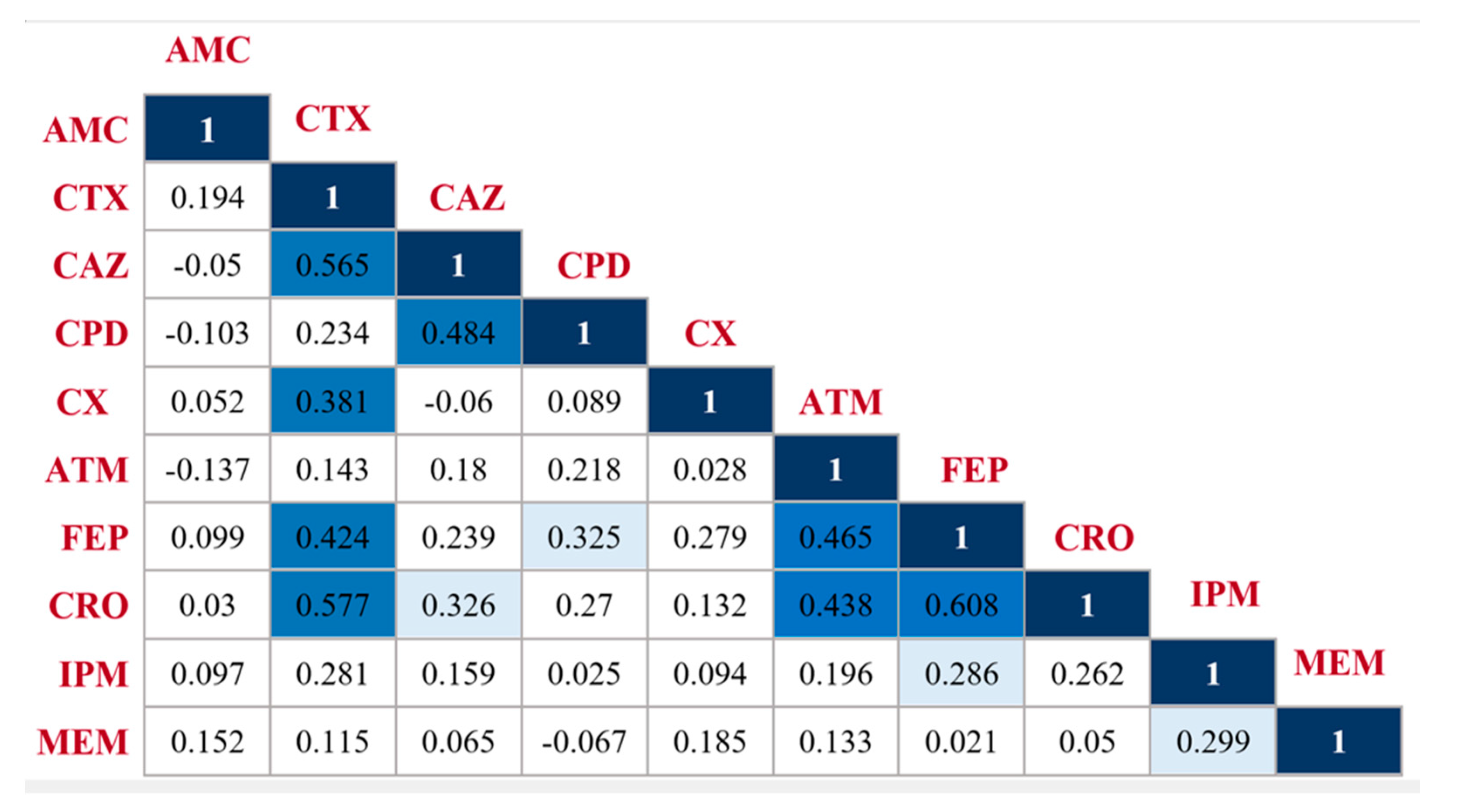
2.2. Pairwise β-Lactam-β-Lactam Correlation
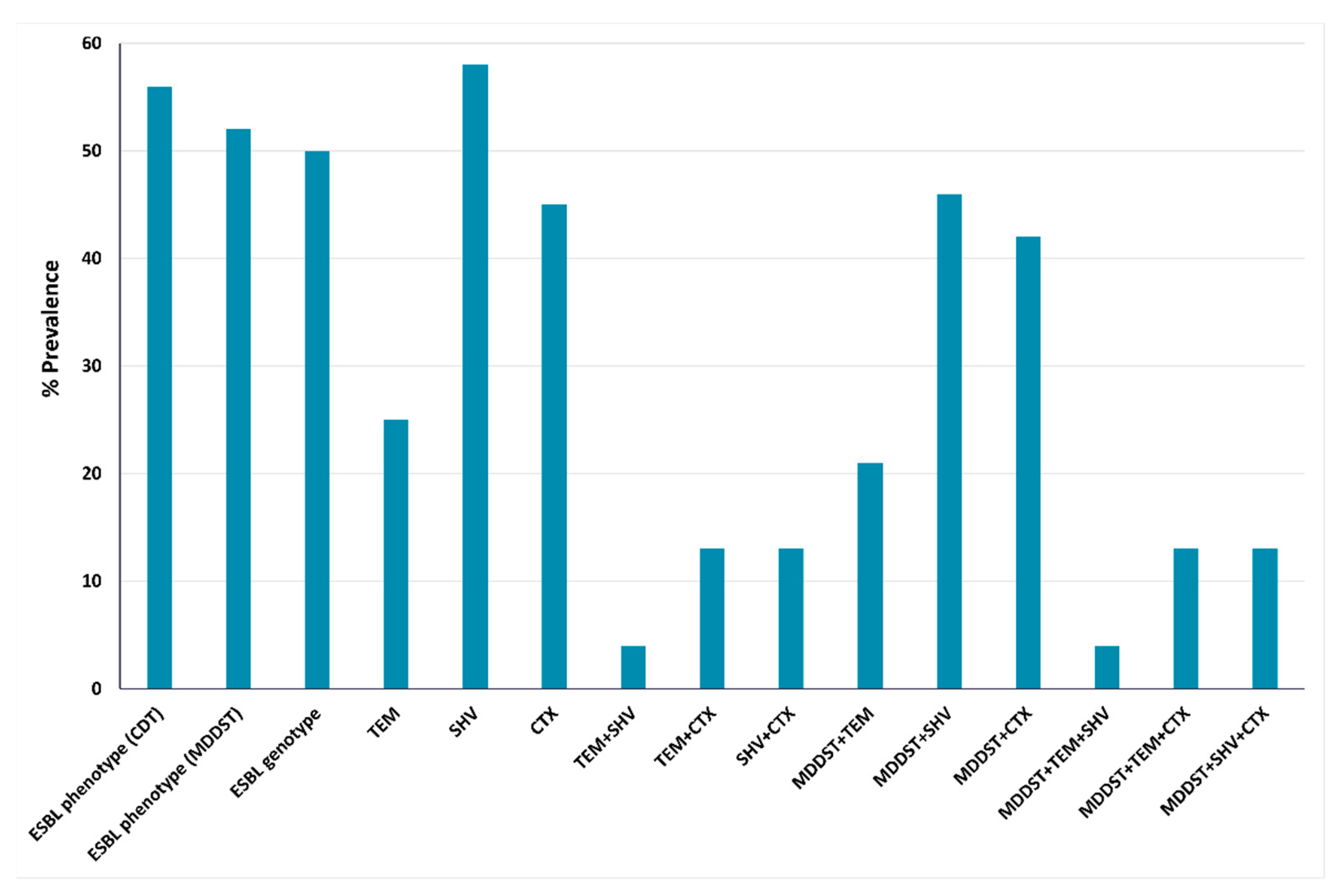
2.3. Phenotypic and Genotypic Detection of ESBLs and AmpC β-Lactamases
2.4. Conjugative Transfer of ESBL- and AmpC-Encoding Genes
2.5. Genomic Analysis of E. coli Strain EC13655
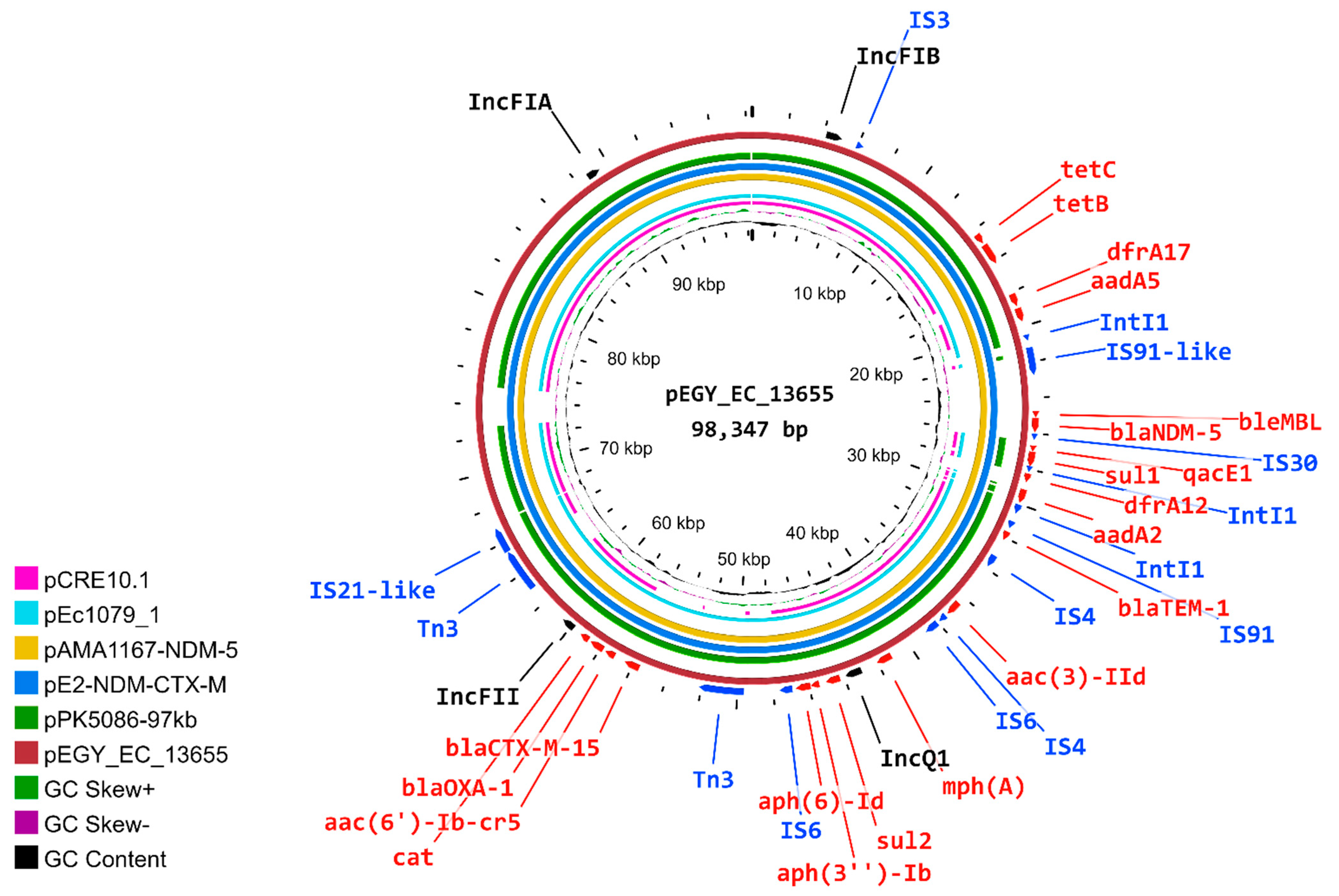
2.6. Characterization of pEGY_EC_13655 Plasmid
2.7. Similarity of pEGY_EC_13655 to Published Plasmids
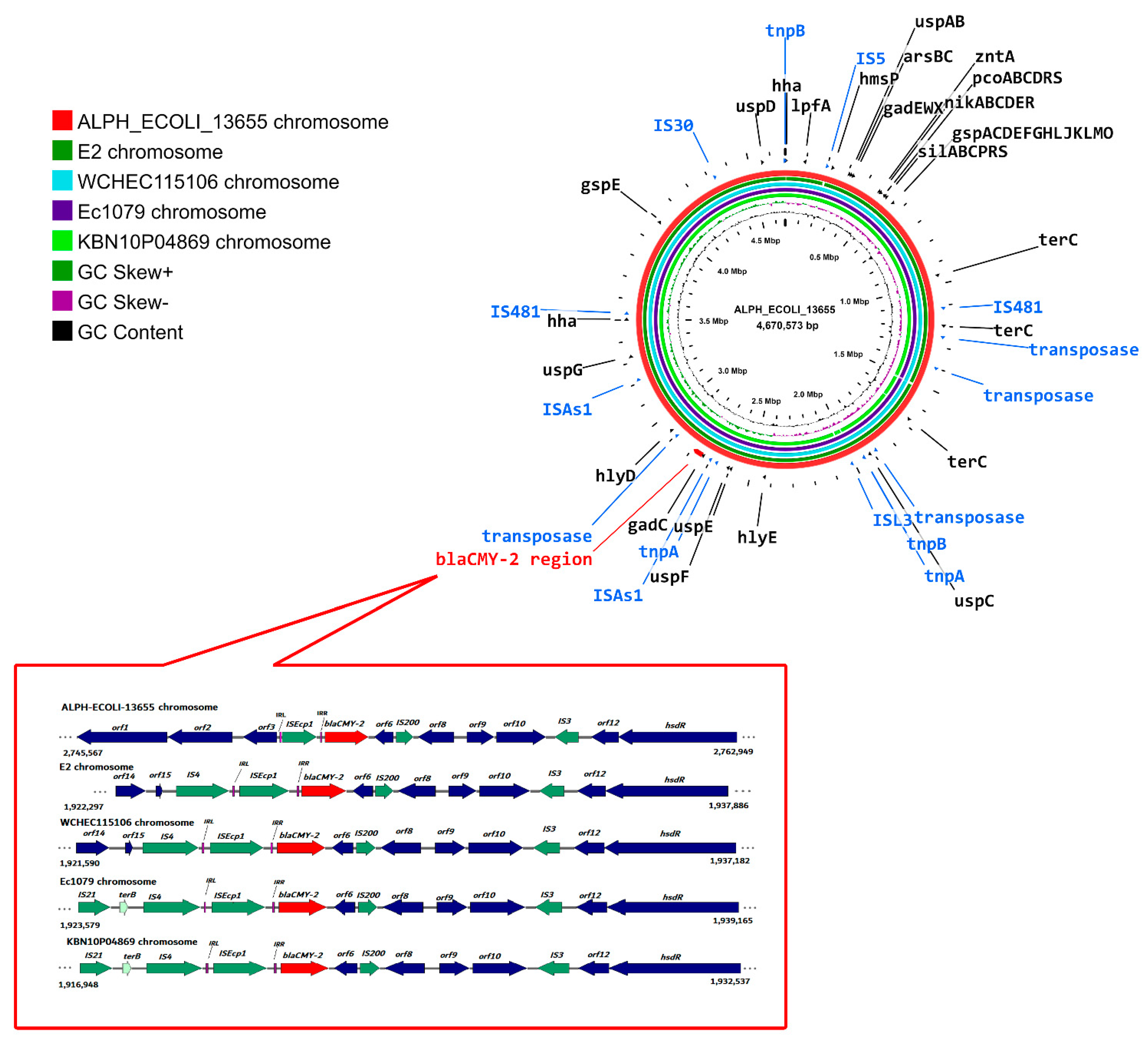
2.8. Location of blaCMY-2 on the ALPH_ECOLI_13655 Chromosome and Its Similarity with Closely Related Chromosomes
2.9. Genetic Context of blaCMY-2
3. Materials and Methods
3.1. Bacterial Isolates Collection and Identification
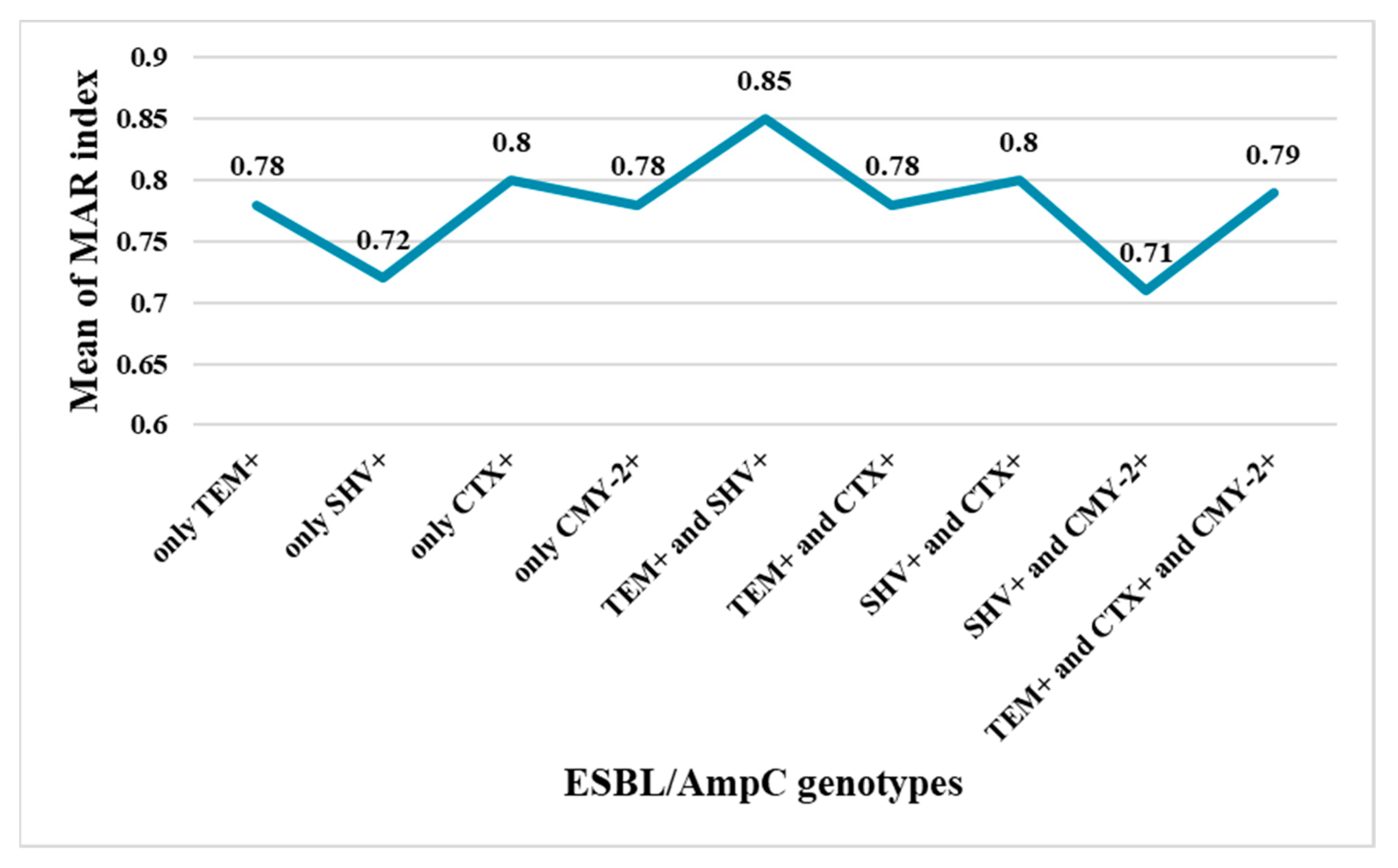
3.2. Antimicrobial Susceptibility Testing and Calculation of the Multiple Antibiotic Resistance Index (MARI)
3.3. Phenotypic Detection of ESBLs and AmpC β-Lactamases
3.3.1. Screening of ESBLs Using the Combined Disk Test (CDT)
3.3.2. Confirmation of ESBL Production
3.3.3. AmpC Detection
3.4. Detection of Genes Encoding ESBLs and Plasmid-Mediated AmpC β-Lactamase
3.5. Conjugation Experiments
3.6. Whole Genome Sequencing, Genome Assembly, and Genome Characterization
3.7. Plasmid Reconstruction and Detection of Chromosomal Location of blaCMY-2
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hassuna, N.A.; Khairalla, A.S.; Farahat, E.M.; Hammad, A.M.; Abdel-Fattah, M. Molecular characterization of Extended-spectrum β lactamase- producing E. coli recovered from community-acquired urinary tract infections in Upper Egypt. Sci. Rep. 2020, 10, 2772. [Google Scholar] [CrossRef] [PubMed]
- Pana, Z.D.; Zaoutis, T. Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections: What have we learned until now? F1000Research 2018, 7, 1347. [Google Scholar] [CrossRef] [PubMed]
- Masoud, S.; El-Baky, R.A.; Aly, S.; Ibrahem, R. Co-Existence of Certain ESBLs, MBLs and Plasmid Mediated Quinolone Resistance Genes among MDR E. coli Isolated from Different Clinical Specimens in Egypt. Antibiotics 2021, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Lob, S.; Biedenbach, D.; Badal, R.; Kazmierczak, K.; Sahm, D. Antimicrobial resistance and resistance mechanisms of Enterobacteriaceae in ICU and non-ICU wards in Europe and North America: SMART 2011–2013. J. Glob. Antimicrob. Resist. 2015, 3, 190–197. [Google Scholar] [CrossRef] [PubMed]
- El-Baky, R.M.A.; Ibrahim, R.A.; Mohamed, D.S.; Ahmed, E.F.; Hashem, Z.S. Prevalence of Virulence Genes and Their Association with Antimicrobial Resistance Among Pathogenic E. coli Isolated from Egyptian Patients with Different Clinical Infections. Infect. Drug Resist. 2020, 13, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Baño, J.; Gutiérrez-Gutiérrez, B.; Machuca, I.; Pascual, A. Treatment of Infections Caused by Extended-Spectrum-Beta-Lactamase-, AmpC-, and Carbapenemase-Producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018, 31, e00079-17. [Google Scholar] [CrossRef]
- Boyd, D.; Tyler, S.; Christianson, S.; McGeer, A.; Muller, M.P.; Willey, B.M.; Bryce, E.; Gardam, M.; Nordmann, P.; Mulvey, M.R. Complete Nucleotide Sequence of a 92-Kilobase Plasmid Harboring the CTX-M-15 Extended-Spectrum Beta-Lactamase Involved in an Outbreak in Long-Term-Care Facilities in Toronto, Canada. Antimicrob. Agents Chemother. 2004, 48, 3758–3764. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.A.; Abdelaziz, N.A.; Amin, M.A.; Aziz, R.K. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci. Rep. 2019, 9, 4224. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.-X.; Li, X.-P.; Li, L.; Chen, M.-Y.; Wu, C.-Y.; Li, L.-L.; Liao, X.-P.; Liu, Y.-H.; Sun, J. ISEcp1-mediated transposition of chromosome-borne blaCMY-2 into an endogenous ColE1-like plasmid in Escherichia coli. Infect. Drug Resist. 2018, 11, 995–1005. [Google Scholar] [CrossRef]
- Fang, L.-X.; Sun, J.; Li, L.; Deng, H.; Huang, T.; Yang, Q.-E.; Li, X.; Chen, M.-Y.; Liao, X.-P.; Liu, Y.-H. Dissemination of the chromosomally encoded CMY-2 cephalosporinase gene in Escherichia coli isolated from animals. Int. J. Antimicrob. Agents 2015, 46, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, M.; RESET Study Group; Irrgang, A.; Roschanski, N.; Michael, G.B.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef]
- Abdelaziz, S.; Aboshanab, K.; Yahia, I.; Yassien, M.; Hassouna, N. Correlation between the Antibiotic Resistance Genes and Susceptibility to Antibiotics among the Carbapenem-Resistant Gram-Negative Pathogens. Antibiotics 2021, 10, 255. [Google Scholar] [CrossRef]
- Abdelkhalik, A.M.; Agha, M.M.; Zaki, A.M.; Tahoun, A.T. Clinical and Lab-Assessed Antibiotic Resistance Pattern of Uropathogens among Women with Acute Uncomplicated Cystitis. Egypt. J. Hosp. Med. 2018, 73, 7860–7868. [Google Scholar] [CrossRef]
- El-Sokkary, R.H.; Ramadan, R.A.; El-Shabrawy, M.; El-Korashi, L.A.; Elhawary, A.; Embarak, S.; Tash, R.M.E.; Elantouny, N.G. Community acquired pneumonia among adult patients at an Egyptian university hospital: Bacterial etiology, susceptibility profile and evaluation of the response to initial empiric antibiotic therapy. Infect. Drug Resist. 2018, 11, 2141–2150. [Google Scholar] [CrossRef]
- Tadesse, B.T.; Ashley, E.A.; Ongarello, S.; Havumaki, J.; Wijegoonewardena, M.; González, I.J.; Dittrich, S. Antimicrobial resistance in Africa: A systematic review. BMC Infect. Dis. 2017, 17, 616. [Google Scholar] [CrossRef]
- Abdel-Moaty, M.; Mohamed, W.; Abdel-All, S.; El-Hendawy, H. Prevalence and molecular epidemiology of extended spectrum beta-lactamase producing Escherichia coli from hospital and community settings in Egypt. J. Appl. Pharm. Sci. 2016, 6, 042–047. [Google Scholar] [CrossRef][Green Version]
- Mohamed, E.S.; Khairy, R.M.M.; Abdelrahim, S.S. Prevalence and molecular characteristics of ESBL and AmpC β -lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrob. Resist. Infect. Control 2020, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Fam, N.; Gamal, D.; Said, E.M.; Aboul-Fadl, E.L.; Dabei, E.E.; Attar, S.; Sorur, A.; Fouad, S.; Klena, J. Detection of plas-mid-mediated AmpC beta-lactamases in clinically significant bacterial isolates in a research institute hospital in Egypt. Life Sci. J. 2013, 10, 2294–2304. [Google Scholar]
- Yilmaz, N.; Agus, N.; Bozcal, E.; Oner, O.; Uzel, A. Detection of plasmid-mediated AmpC β-lactamase in Escherichia coli and Klebsiella pneumoniae. Indian J. Med. Microbiol. 2013, 31, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Singhai, M.; Kumar, A.; Jha, P.K.; Goyal, R.; Rawat, V. Bacteriological and resistance profile in isolates from diabetic patients. N. Am. J. Med. Sci. 2012, 4, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Pages, J.-M.; Lavigne, J.-P.; Leflon-Guibout, V.; Marcon, E.; Bert, F.; Noussair, L.; Nicolas-Chanoine, M.-H. Efflux Pump, the Masked Side of ß-Lactam Resistance in Klebsiella pneumoniae Clinical Isolates. PLoS ONE 2009, 4, e4817. [Google Scholar] [CrossRef]
- Helmy, M.M.; Wasfi, R. Phenotypic and Molecular Characterization of Plasmid Mediated AmpCβ-Lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis Isolated from Urinary Tract Infections in Egyptian Hospitals. BioMed Res. Int. 2014, 2014, 171548. [Google Scholar] [CrossRef]
- Krishnamurthy, V. Phenotypic and Genotypic Methods for Detection of Extended Spectrum β Lactamase Producing Escherichia coli and Klebsiella pneumoniae Isolated from Ventilator Associated Pneumonia. J. Clin. Diagn. Res. 2013, 7, 1975–1978. [Google Scholar] [CrossRef]
- Dirar, M.H.; Bilal, N.E.; Ibrahim, M.E.; Hamid, M.E. Prevalence of extended-spectrum β-lactamase (ESBL) and molecular detection of bla TEM, bla SHV and bla CTX-M genotypes among Enterobacteriaceae isolates from patients in Khartoum, Sudan. Pan Afr. Med. J. 2020, 37, 213. [Google Scholar] [CrossRef]
- Bindayna, K.M.; Joji, R.M.; Ezzat, H.; Jahrami, H.A. Antibiotic-resistance genes in E. coli strains in GCC countries: A meta-analysis. Saudi J. Med. Med. Sci. 2022, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zorgani, A.; Almagatef, A.; Sufya, N.; Bashein, A.; Tubbal, A. Detection of CTX-M-15 Among Uropathogenic Escherichia coli Isolated from Five Major Hospitals in Tripoli, Libya. Oman Med. J. 2017, 32, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.; El-Halaby, H.; Elmansoury, E.; Zeid, M.; Khaled, K.; Nomir, M. Genetic Study of Extended Spectrum Beta-Lactamase and Carbapenemase Producing Escherichia Coli Causing Sepsis among Egyptian Children. Open Microbiol. J. 2019, 13, 128–137. [Google Scholar] [CrossRef]
- Meini, S.; Tascini, C.; Cei, M.; Sozio, E.; Rossolini, G.M. AmpC β-lactamase-producing Enterobacterales: What a clinician should know. Infection 2019, 47, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Mendes, R.E.; Jones, R.N.; Sader, H. Changes in the Frequencies of β-Lactamase Genes among Enterobacteriaceae Isolates in U.S. Hospitals, 2012 to 2014: Activity of Ceftazidime-Avibactam Tested against β-Lactamase-Producing Isolates. Antimicrob. Agents Chemother. 2016, 60, 4770–4777. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.-S.; Hsueh, P.-R.; SMART Asia-Pacific Group. Distribution of ESBLs, AmpC? lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 2017, 72, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.; Ali, S.; Hossain, M.; Uddin, S.Z.; Moniruzzaman, M.; Islam, M.R.; Shohael, A.M.; Islam, S.; Ananya, T.H.; Rahman, M.; et al. ESBL Producing Escherichia coli in Faecal Sludge Treatment Plants: An Invisible Threat to Public Health in Rohingya Camps, Cox’s Bazar, Bangladesh. Front. Public Health 2021, 9, 783019. [Google Scholar] [CrossRef] [PubMed]
- Roer, L.; Overballe-Petersen, S.; Hansen, F.; Schønning, K.; Wang, M.; Røder, B.L.; Hansen, D.S.; Justesen, U.S.; Andersen, L.P.; Fulgsang-Damgaard, D.; et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere 2018, 3, e00337-18. [Google Scholar] [CrossRef]
- Gamal, D.; Fernandez-Martinez, M.; El-Defrawy, I.; Ocampo-Sosa, A.A.; Martinez-Martinez, L. First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emerg. Microbes Infect. 2016, 5, e30. [Google Scholar] [CrossRef]
- Soliman, A.M.; Zarad, H.O.; Nariya, H.; Shimamoto, T.; Shimamoto, T. Genetic analysis of carbapenemase-producing Gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect. Genet. Evol. 2019, 77, 104065. [Google Scholar] [CrossRef]
- Mushtaq, S.; Irfan, S.; Sarma, J.B.; Doumith, M.; Pike, R.; Pitout, J.; Livermore, D.M.; Woodford, N. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 2011, 66, 2002–2005. [Google Scholar] [CrossRef] [PubMed]
- Temmerman, R.; Garmyn, A.; Antonissen, G.; Vanantwerpen, G.; Vanrobaeys, M.; Haesebrouck, F.; Devreese, M. Evaluation of Fluoroquinolone Resistance in Clinical Avian Pathogenic Escherichia coli Isolates from Flanders (Belgium). Antibiotics 2020, 9, 800. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Day, M.; Cleary, P.; Hopkins, K.L.; Toleman, M.A.; Wareham, D.W.; Wiuff, C.; Doumith, M.; Woodford, N. OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J. Antimicrob. Chemother. 2019, 74, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Fiett, J.; Baraniak, A.; Izdebski, R.; Sitkiewicz, I.; Żabicka, D.; Meler, A.; Filczak, K.; Hryniewicz, W.; Gniadkowski, M. The First NDM Metallo-β-Lactamase-Producing Enterobacteriaceae Isolate in Poland: Evolution of IncFII-Type Plasmids Carrying the bla NDM-1 Gene. Antimicrob. Agents Chemother. 2014, 58, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Akeda, Y.; Sakamoto, N.; Takeuchi, D.; Motooka, D.; Nakamura, S.; Hagiya, H.; Yamamoto, N.; Nishi, I.; Yoshida, H.; et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS ONE 2017, 12, e0184720. [Google Scholar] [CrossRef] [PubMed]
- Kucken, D.; Feucht, H.-H.; Kaulfers, P.-M. Association of qacE and qacEDelta1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol. Lett. 2000, 183, 95–98. [Google Scholar] [CrossRef]
- Dortet, L.; Girlich, D.; Virlouvet, A.-L.; Poirel, L.; Nordmann, P.; Iorga, B.I.; Naas, T. Characterization of BRPMBL, the Bleomycin Resistance Protein Associated with the Carbapenemase NDM. Antimicrob. Agents Chemother. 2017, 61, e02413-16. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Mohsin, M.; Lu, X.; Abdullah, S.; Munir, A.; Wang, Z. Emergence of Plasmid-Mediated Resistance Genes tet (X) and mcr-1 in Escherichia coli Clinical Isolates from Pakistan. mSphere 2021, 6, e0069521. [Google Scholar] [CrossRef] [PubMed]
- Mahazu, S.; Prah, I.; Ayibieke, A.; Sato, W.; Hayashi, T.; Suzuki, T.; Iwanaga, S.; Ablordey, A.; Saito, R. Possible Dissemination of Escherichia coli Sequence Type 410 Closely Related to B4/H24RxC in Ghana. Front. Microbiol. 2021, 12, 770130. [Google Scholar] [CrossRef]
- Hirabayashi, A.; Yanagisawa, H.; Takahashi, H.; Yahara, K.; Boeing, P.; Wolfenden, B.; Nov, V.; Lorn, V.; Veng, M.; Ann, V.; et al. On-Site Genomic Epidemiological Analysis of Antimicrobial-Resistant Bacteria in Cambodia with Portable Laboratory Equipment. Front. Microbiol. 2021, 12, 675463. [Google Scholar] [CrossRef]
- Weber, A.; Jung, K. Biochemical Properties of UspG, a Universal Stress Protein of Escherichia coli. Biochemistry 2006, 45, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.; von Rhein, C.; Bauer, S.; Hüttinger, C.; Goebel, W. Molecular Analysis of Cytolysin A (ClyA) in Pathogenic Escherichia coli Strains. J. Bacteriol. 2004, 186, 5311–5320. [Google Scholar] [CrossRef]
- Paytubi, S.; Dietrich, M.; Queiroz, M.H.; Juárez, A. Role of plasmid- and chromosomally encoded Hha proteins in modulation of gene expression in E. coli O157:H7. Plasmid 2013, 70, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Lallemand, M.; Login, F.H.; Guschinskaya, N.; Pineau, C.; Effantin, G.; Robert, X.; Shevchik, V.E. Dynamic Interplay between the Periplasmic and Transmembrane Domains of GspL and GspM in the Type II Secretion System. PLoS ONE 2013, 8, e79562. [Google Scholar] [CrossRef] [PubMed]
- Shankar, C.; Vasudevan, K.; Jacob, J.J.; Baker, S.; Isaac, B.J.; Neeravi, A.R.; Sethuvel, D.P.M.; George, B.; Veeraraghavan, B. Mosaic antimicrobial resistance/virulence plasmid in hypervirulent ST2096 Klebsiella pneumoniae in India: The rise of a new superbug? bioRxiv 2020. [Google Scholar] [CrossRef]
- Zakaria, A.; Edward, E.; Mohamed, N. Genomic Insights into a Colistin-Resistant Uropathogenic Escherichia coli Strain of O23:H4-ST641 Lineage Harboring mcr-1.1 on a Conjugative IncHI2 Plasmid from Egypt. Microorganisms 2021, 9, 799. [Google Scholar] [CrossRef] [PubMed]
- Tille, P. Bailey & Scott’s Diagnostic Microbiology-E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Supplement M100; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Kaur, J. Modified Double Disc Synergy Test to Detect ESBL Production in Urinary Isolates of Escherichia coli and Klebsiella pneumoniae. J. Clin. Diagn. Res. 2013, 7, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Polsfuss, S.; Bloemberg, G.V.; Giger, J.; Meyer, V.; Böttger, E.C.; Hombach, M. Practical Approach for Reliable Detection of AmpC Beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 2798–2803. [Google Scholar] [CrossRef] [PubMed]
- Perween, N. Phenotypic Methods of Detection of Beta-Lactamases. In Beta-Lactam Resistance in Gram-Negative Bacteria; Shahid, M., Singh, A., Sami, H., Eds.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Skulj, M.; Okrslar, V.; Jalen, S.; Jevsevar, S.; Slanc, P.; Strukelj, B.; Menart, V. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb. Cell Factories 2008, 7, 6–12. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.H.; Alqahtani, F.Y.; Aleanizy, F.S.; Alfaraj, R.; Mahmoud, A.Z.; Zakaria, A.S. Frequency of plasmid-mediated quinolone resistance determinants Qnr and QepA among clinical isolates of Escherichia coli and Klebsiella pneumoniae producing extended- spectrum βlactamases from Saudi Arabia intensive care units. Int. J. Microbiol. Res. 2017, 9, 924–929. [Google Scholar]
- Peymani, A.; Naserpour-Farivar, T.; Yeylagh-Beygi, M.; Bolori, S. Emergence of CMY-2- and DHA-1-type AmpC be-ta-lactamases in Enterobacter cloacae isolated from several hospitals of Qazvin and Tehran, Iran. Iran J. Microbiol. 2016, 8, 168–174. [Google Scholar] [PubMed]
| E. coli Isolates | Conjugation Frequency a (CFU/Donor Cell) | Resistance Genes | Resistance Profile |
|---|---|---|---|
| EC13655 | blaTEM, blaCTX, blaCMY-2 | CTX, CAZ, CPD, ATM, CIP, CRO, DO, FEP, CX, IPM, SXT | |
| Transconjugant of EC13655 | 1.02 × 10−6 | blaTEM, blaCTX | CTX, CAZ, CPD, ATM, CIP, CRO, DO, FEP, CX, IPM, SXT, RD |
| EC14149 | blaSHV, blaCMY-2 | CTX, CAZ, CPD, ATM, CIP, CN, CRO, FEP, CX, IPM, SXT | |
| Transconjugant of EC14149 | 2.27 × 10−5 | blaSHV, blaCMY-2 | CTX, CAZ, CPD, ATM, CIP, CRO, FEP, CX, IPM, RD |
| EC14437 | blaCTX, blaCMY-2 | AMC, CTX, CAZ, CPD, ATM, CIP, CN, CRO, DO, FEP, CX, SXT | |
| Transconjugant of EC14437 | 3.53 × 10−4 | blaCTX, blaCMY-2 | AMC, CTX, CAZ, CPD, CIP, CRO, FEP, SXT, RD |
| EC11994 | blaSHV, blaCMY-2 | AMC, CTX, CAZ, CPD, ATM, CRO, FEP, CX, SXT | |
| Transconjugant of EC11994 | 6.53 × 10−6 | blaSHV, blaCMY-2 | CTX, CAZ, CPD, ATM, CRO, FEP, CX, RD |
| Serotype a | Phylogroup b | Sequence Type (ST) c | Clonotype d | Resistance Profile e | Virulence f | Plasmid Replicon Type h | ||
|---|---|---|---|---|---|---|---|---|
| Antimicrobial Class | ARGs and Point Mutations | Virulence-Associated Genes g | Heavy Metal Resistance Genes | |||||
| O8:H9 | Group A | 410 | CH4-24 | β-lactams | blaTEM-1B, blaOXA-1, blaCTX-M-15, blaNDM-5, blaCMY-2 | gadECWX lpfA uspABCDEF hlyD, hlyE hha hmsP gspAC-H, gspJKLMO | terC silABCPRS pcoABCDRS nikABCDER arsBC zntA | Col(BS512) IncFIA IncFIB IncFII IncQ1 p0111 |
| Aminoglycosides | aadA2, aadA5, aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, aac(3)-IId | |||||||
| Amphenicol | catB3 | |||||||
| Quinolones | aac(6′)-Ib-cr, gyrAS83L, gyrA D87N, parC S80I, parE S458A | |||||||
| Macrolide | mph(A) | |||||||
| Folate pathway antagonist | sul1, sul2, dfrA12, dfrA17 | |||||||
| Tetracycline | tetB, tetC | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, N.M.; Zakaria, A.S.; Edward, E.A. Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and blaNDM-5 on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne blaCMY-2 from Egypt. Antibiotics 2022, 11, 1031. https://doi.org/10.3390/antibiotics11081031
Mohamed NM, Zakaria AS, Edward EA. Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and blaNDM-5 on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne blaCMY-2 from Egypt. Antibiotics. 2022; 11(8):1031. https://doi.org/10.3390/antibiotics11081031
Chicago/Turabian StyleMohamed, Nelly M., Azza S. Zakaria, and Eva A. Edward. 2022. "Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and blaNDM-5 on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne blaCMY-2 from Egypt" Antibiotics 11, no. 8: 1031. https://doi.org/10.3390/antibiotics11081031
APA StyleMohamed, N. M., Zakaria, A. S., & Edward, E. A. (2022). Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and blaNDM-5 on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne blaCMY-2 from Egypt. Antibiotics, 11(8), 1031. https://doi.org/10.3390/antibiotics11081031






