Abstract
Carbapenem-resistant Enterobacterales are a growing problem in healthcare systems worldwide. While whole-genome sequencing (WGS) has become a powerful tool for analyzing transmission and possible outbreaks, it remains laborious, and the limitations in diagnostic workflows are not well studied. The aim of this study was to compare the performance of WGS and real-time multiplex PCR (RT-qPCR) for diagnosing carbapenem-resistant Enterobacterales. In this study, we analyzed 92 phenotypically carbapenem-resistant Enterobacterales, sent to the University Hospital Heidelberg in 2019, by the carbapenem inactivation method (CIM) and compared WGS and RT-qPCR as genotypic carbapenemase detection methods. In total, 80.4% of the collected isolates were identified as carbapenemase producers. For six isolates, discordant results were recorded for WGS, PCR and CIM, as the carbapenemase genes were initially not detected by WGS. A reanalysis using raw reads, rather than assembly, highlighted a coverage issue with failure to detect carbapenemases located in contigs with a coverage lower than 10×, which were then discarded. Our study shows that multiplex RT-qPCR and CIM can be a simple alternative to WGS for basic surveillance of carbapenemase-producing Enterobacterales. Using WGS in clinical workflow has some limitations, especially regarding coverage and sensitivity. We demonstrate that antimicrobial resistance gene detection should be performed on the raw reads or non-curated draft genome to increase sensitivity.
1. Introduction
Enterobacterales, including bacterial species such as Citrobacter freundii, Escherichia coli, Klebsiella pneumoniae and the Enterobacter cloacae complex, belong to the most common human pathogens and are able to cause a variety of infections [1,2].
In particular, infections with multidrug resistant Enterobacterales lead to high mortality since there are limited treatment options [3]. Carbapenemases are of great concern, as they are able to inactivate the last-resort drug carbapenems in addition to other beta-lactam antibiotics [3,4]. They are mostly plasmid encoded, which facilitates an easy transmission and dissemination through horizontal gene transfer [5]. Worldwide, the most common carbapenemases in Enterobacterales are KPC, NDM, VIM, IMP and OXA-48-like carbapenemases [2,6]. Another less frequent route of carbapenem resistance acquisition is via overexpression of the outer membrane efflux pumps or porin loss combined with the expression of extended-spectrum beta-lactamases or AmpC resistance genes [7,8].
Phenotypic screening for carbapenem resistance by Carba-NP test [9], the modified Hodge test [10] or the disc diffusion assay [11] is common in microbiology diagnostics, yet for epidemiological surveillance, high-resolution typing is useful and essential. A few real-time PCR (RT-qPCR)-based assays have been developed to detect carbapenem-resistance genes in Gram-negative bacteria [12,13,14]. However, these methods are technically limited to a certain number of targets. By contrast, whole-genome sequencing (WGS) provides more comprehensive information and thus has become a powerful tool for surveillance and outbreak investigation [15]. Although there are several studies comparing the performance of phenotypic and commercially available tests for carbapenemase detection [16,17,18], comparative studies on WGS and RT-qPCR remain scarce. Currently, the application of WGS in the clinical microbiological setting is limited to molecular typing. However, there is still an untapped potential for integrating WGS-based technologies into microbiological diagnostics. Although preparation and turnover time remains a major disadvantage for WGS, the performance and accuracy of WGS compared to those of faster nucleic acid amplification-based and simple phenotypic methods should be investigated.
Our study aimed to retrospectively evaluate the performance of WGS compared to that of RT-qPCR and phenotypic carbapenem-resistant Enterobacterales, identified by antimicrobial susceptibility testing and the carbapenem inactivation method (CIM).
2. Results
A total of 92 phenotypic carbapenem-resistant Enterobacterales were collected in 2019. Carbapenem-hydrolyzing activity could be detected in 74 isolates (80.4%) by CIM. These results were validated by WGS and RT-qPCR. For six isolates, different results occurred between the three methods, as carbapenemases were initially detected by CIM and PCR but not by WGS (Table 1 and Table 2). By reanalyzing the raw sequencing data and removing the coverage threshold blaNDM-1, blaKPC-2 (2x), blaVIM-1 (2x) and blaOXA-48 were identified (Table A1). For 18 isolates, all three methods revealed no carbapenemase.

Table 1.
Comparison of phenotypic and genotypic carbapenemase detection in Enterobacterales by CIM, RT-qPCR and WGS.

Table 2.
Comparison of genotypic carbapenemase detection in Enterobacterales by WGS and RT-qPCR.
The predominant species of the carbapenemase producers was E. cloacae (n = 30) followed by K. pneumoniae (n = 17) and E. coli (n = 15). C. freundii (n = 7), Klebsiella oxytoca (n = 3) and Serratia marcescens (n = 2) appeared less frequently (Figure 1). OXA-48 (40.5%) was the most prevalent carbapenemase and was detected in all species in this collection. VIM-1 (21.6%) was the second most common enzyme in our study, followed by KPC-2 (12.2%) and NDM-5 (9.5%). Other carbapenemase variants, such as NDM-1, OXA-244, KPC-3 and OXA-232, were less abundant (<3.0%), and isolates harboring two carbapenemases (8.1%) occurred sporadically (Figure 1, Table A1).
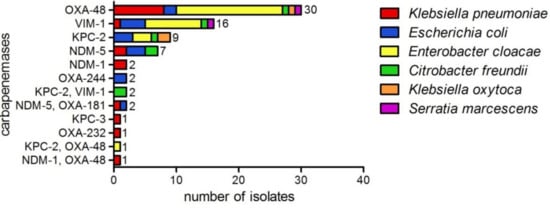
Figure 1.
Carbapenemases detected in Enterobacterales by WGS (n = 74), showing phenotypic resistance to carbapenem antibiotics. E. cloacae (n=30), K. pneumoniae (n = 17), E. coli (n = 15), C. freundii (n = 7), K. oxytoca (n = 3) and S. marcescens (n = 2).
3. Discussion
Rapid spreading of carbapenemase-producing Enterobacterales as well as outbreaks of different multidrug resistant bacteria is reported worldwide in clinical settings. For infection control and prevention of further dissemination, monitoring is necessary. Thus, we analyzed 92 phenotypically carbapenem-resistant Enterobacterales by CIM to confirm carbapenem-hydrolyzing activity. We then compared WGS and RT-qPCR to validate performance in detecting carbapenemase genes.
In total, 74 isolates were found to be carbapenemase producers (Figure 1). In six cases, discordant results occurred between WGS and the other two methods, since the carbapenemase was initially not detected by sequencing (Table 1 and Table 2 and Table A1). For analyzing WGS data, quality control is crucial, including coverage of the assembly, quality of de novo assembly and detection of potential DNA contamination. The read coverage is of particular importance, as it influences the sensitivity of sequencing [19]. In the initial assembly, we set up a limit of 25× coverage for the full genome, and contigs with a coverage <10× or smaller than 1000 bp were removed because they are potential contaminants or misassemblies. However, our study showed that true signals might be lost during the cleaning of the assembly, since the quality control parameters N50 and the coverage were in the desired range (Table A1). Low-copy number plasmids or plasmid loss during DNA extraction might have led to a low abundance of carbapenemase genes, and, thus, the antimicrobial resistance genes were not detected. While the establishment of such thresholds is crucial for genomic comparison and annotation of a draft genome, our data suggest that antimicrobial resistance gene detection should be performed on the non-curated draft genome to increase sensitivity.
Our findings on carbapenemase variants are in line with the data of the German national reference laboratory (NRL) in the years 2017–2019. In particular, blaOXA-48 was detected in all years, followed by blaVIM-1, blaKPC-2, blaNDM-1, blaKPC-3, blaOXA-181 and blaNDM-5 [20,21,22], which are detectable with our assay. However, depending on the geographic region, less frequent carbapenemase types, such as GES, GIM and IMI, can occur in Enterobacterales [20,21,22]. These genes are not included in our assay and, therefore, can lead to false-negative results. In 2019, these carbapenemases were not detected by WGS (Figure 1, Table A1). However, if the epidemiology changes, the PCR should be adapted to the new resistance situation.
The RT-qPCR provides a fast and inexpensive alternative for diagnostic labs without NGS facilities, although the PCR-based assay is limited to known targets. Compared to the RT-qPCR, WGS is an unbiased method that provides more information, such as genetic relationships and the full resistome. Besides the presence or absence of known resistance genes, novel resistance genes can be identified in phenotypic resistant isolates by WGS [23]. However, the analysis is more complex, and, therefore, bioinformatics expertise is needed.
4. Materials and Methods
4.1. Bacterial Isolates
Clinical samples and rectal swabs were screened for carbapenem-resistant Enterobacterales at the Department of Infectious Diseases, Medical Microbiology, University Hospital Heidelberg in 2019. During routine diagnostics, 92 Enterobacterales showing phenotypic resistance to meropenem and imipenem were collected. Non-duplicate strains were obtained from 79 patients. Multiple isolates (n = 13) from the same patient were included in the study due to different bacterial species as determined by MALDI TOF MS (Bruker Daltonics GmbH & Co. KG, Bremen, Germany). The antibiotic susceptibility was tested by the VITEK-2 system (bioMérieux Deutschland GmbH, Nürtingen, Germany) and evaluated according to the valid EUCAST guidelines in the respective year (v 9.0). The isolates were stored at −20 °C until usage.
4.2. Carbapenem Inactivation Method
CIM was performed, as described elsewhere [24], to examine whether the carbapenem-resistant isolates, identified by antimicrobial susceptibility testing, are able to hydrolyze carbapenem antibiotics.
4.3. DNA Extraction
The isolates were regrown on BD™ Columbia Agar with 5% Sheep Blood (Becton Dickinson GmbH, Heidelberg, Germany) at 37 °C. DNA for WGS and RT-qPCR was extracted using the DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s protocol.
4.4. Multiplex Real-Time PCR
The assay based on hydrolysis probes consists of two multiplex PCRs for the detection of blaNDM, blaKPC, blaVIM and blaIMP, and blaOXA-23-like, blaOXA-40/24-like, blaOXA-58-like and blaOXA-48-like, respectively. Amplification and detection were performed on the BD MAX™ system, using the protocol for the PCR-only mode, as described elsewhere [25].
4.5. Whole-Genome Sequencing
WGS was performed on the MIseq instrument (2 × 300 bp), using the Nextera DNA Flex Library Prep Kit (Illumina) for preparing sequencing libraries. Quality control of the raw sequences, assembly and curation (contigs >1000bp and >10× coverage) were performed as described elsewhere [26]. The databases ResFinder 3.0, ARG-ANNOT and CARD-NCBI-BARRGD using ABRIcate (https://git.lumc.nl/bvhhornung/antibiotic-resistancepipeline/tree/master/tools/abricate, accessed on 10 June 2020) were used to determine the resistance genes as previously described [27].
5. Conclusions
Whole-genome sequencing is a powerful tool with high molecular resolution, giving information about bacterial species, plasmid replicon types and the whole resistance pattern, which is needed for surveillance of transmission and outbreak investigation. Real-time PCR is faster but provides less information and cannot detect new carbapenemases that are not included in the panel, which is a general drawback of PCR-based assays. Nevertheless, the additional use of PCR and/or CIM for carbapenemase detection in Enterobacterales was beneficial in our study to ensure high sensitivity, as some carbapenemases would have remained undetected by WGS due to coverage issues.
6. Patents
K.P., K.H. and A.H.D. have a patent (No. 20203612.5) pending.
Author Contributions
Conceptualization, D.N., K.H., A.H.D. and S.B.; methodology, K.P. and A.-M.F.; writing—original draft preparation, K.P.; writing—review and editing, S.B., D.N., A.-M.F., K.H., A.H.D. and K.P.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Bio project PRJNA634442.
Acknowledgments
We would like to acknowledge the excellent technical support from Delal Sahin, Nicole Henny, Selina Hassel and Suzan Leccese.
Conflicts of Interest
K.P., K.H. and A.H.D. have a patent (No. 20203612.5) pending. The other authors declare no conflicts of interest.
Appendix A

Table A1.
Phenotypic carbapenem-resistant Enterobacterales collected in 2019, analyzed by CIM, RT-qPCR and WGS. Quality control parameters for WGS: coverage and N50.
Table A1.
Phenotypic carbapenem-resistant Enterobacterales collected in 2019, analyzed by CIM, RT-qPCR and WGS. Quality control parameters for WGS: coverage and N50.
| Sample ID | Species | CIM | RT-qPCR | WGS | WGS Reanalyzed | Coverage | N50 |
|---|---|---|---|---|---|---|---|
| KE9539 | E. coli | positive | blaKPC | blaKPC-2 | 48 | 535,993 | |
| KE9246 | E. coli | positive | blaKPC | blaKPC-2 | 53 | 135,761 | |
| KE9526 | E. cloacae | positive | blaKPC | blaKPC-2 | 52 | 363,822 | |
| KE9478 | E. cloacae | positive | blaKPC | blaKPC-2 | 96 | 363,822 | |
| BK31926 | E. coli | positive | blaKPC | blaKPC-2 | 29 | 120,862 | |
| KE9621 | K. pneumoniae | positive | blaKPC | blaKPC-3 | 35 | 386,401 | |
| KE9498 | C. freundii | positive | blaKPC | blaKPC-2 | 31 | 200,582 | |
| KE9038 | K. oxytoca | positive | blaKPC | blaKPC-2 | 50 | 285,607 | |
| KE9326 | K. oxytoca | positive | blaKPC | negative | blaKPC-2 | 42 | 109,274 |
| KE9511 | C. freundii | positive | blaKPC, blaVIM | blaKPC-2, blaVIM-1 | 30 | 198,406 | |
| KE9378 | C. freundii | positive | blaKPC, blaVIM | blaKPC-2 | blaKPC-2, blaVIM-1 | 39 | 201,178 |
| KE9132 | E. cloacae | positive | blaKPC | blaKPC-2 | 49 | 363,822 | |
| KE9520 | K. pneumoniae | positive | blaNDM | blaNDM-5 | 53 | 186,575 | |
| KE9434 | K. pneumoniae | positive | blaNDM | blaNDM-5 | 34 | 292,061 | |
| KE9521 | E. coli | positive | blaNDM, blaOXA-48-like | blaNDM-5, blaOXA-181 | 61 | 106,471 | |
| KE9395 | E. coli | positive | blaNDM | blaNDM-5 | 54 | 94,083 | |
| KE9433 | E. coli | positive | blaNDM | blaNDM-5 | 36 | 214,212 | |
| KE9636 | K. pneumoniae | positive | blaNDM, blaOXA-48-like | blaNDM-1, blaOXA-48 | 27 | 383,090 | |
| KE9616 | C. freundii | positive | blaNDM | blaNDM-5 | 50 | 186,958 | |
| KE9522 | E. coli | positive | blaNDM | blaNDM-5 | 103 | 269,697 | |
| KE9593 | K. pneumoniae | positive | blaNDM, blaOXA-48-like | blaNDM-5, blaOXA-181 | 38 | 296,725 | |
| D3014 | C. freundii | positive | blaNDM | blaNDM-5 | 36 | 186,959 | |
| KE9449 | K. pneumoniae | positive | blaNDM | negative | blaNDM-1 | 25 | 220,843 |
| KE9500 | K. pneumoniae | positive | blaNDM | blaNDM-1 | 33 | 536,321 | |
| KE9382 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 27 | 374,725 | |
| KE9492 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-232 | 30 | 242,997 | |
| KE9629 | E. coli | positive | blaOXA-48-like | blaOXA-244 | 33 | 238,467 | |
| KE9025 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 49 | 272,750 | |
| KE9469 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 76 | 374,315 | |
| KE9472 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 98 | 382,653 | |
| KE9424 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 36 | 184,292 | |
| KE9499 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 66 | 486,681 | |
| KE9400 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 45 | 208,351 | |
| KE9468 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 80 | 383,026 | |
| KE9638 | E. coli | positive | blaOXA-48-like | blaOXA-244 | 37 | 156,925 | |
| KE9493 | E. cloacae | positive | blaOXA-48-like, blaKPC | blaOXA-48 | blaKPC-2, blaOXA-48 | 44 | 530,933 |
| KE9456 | K. oxytoca | positive | blaOXA-48-like | blaOXA-48 | 28 | 223,596 | |
| KE9443 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 27 | 225,118 | |
| KE9354 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 66 | 486,663 | |
| BK32270 | E. coli | positive | blaOXA-48-like | blaOXA-48 | 35 | 117,967 | |
| KE9626 | E. coli | positive | blaOXA-48-like | blaOXA-48 | 53 | 196,578 | |
| KE9208 | S. marcescens | positive | blaOXA-48-like | blaOXA-48 | 58 | 2,797,497 | |
| D2902 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 64 | 302,960 | |
| KE9541 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 47 | 427,613 | |
| KE9554 | C. freundii | positive | blaOXA-48-like | blaOXA-48 | 39 | 165,554 | |
| KE9338 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 47 | 374,725 | |
| KE9355 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 109 | 486,681 | |
| KE9328 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 43 | 274,145 | |
| KE9510 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 31 | 491,022 | |
| D3070 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 44 | 372,768 | |
| KE9428 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 36 | 486,663 | |
| KE9527 | E. cloacae | positive | blaOXA-48-like | negative | blaOXA-48 | 27 | 339,153 |
| D3018 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 25 | 473,650 | |
| D3082 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 62 | 486,663 | |
| KE9637 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 36 | 876,600 | |
| D3081 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 85 | 383,026 | |
| EX1012 | K. pneumoniae | positive | blaOXA-48-like | blaOXA-48 | 39 | 223,327 | |
| D3078 | E. cloacae | positive | blaOXA-48-like | blaOXA-48 | 54 | 486,828 | |
| KE9366 | E. coli | positive | blaVIM | blaVIM-1 | 38 | 215,473 | |
| KE9563 | E. cloacae | positive | blaVIM | blaVIM-1 | 35 | 377,920 | |
| KE9409 | E. cloacae | positive | blaVIM | blaVIM-1 | 46 | 486,118 | |
| KE9414 | E. cloacae | positive | blaVIM | blaVIM-1 | 38 | 161,463 | |
| KE9365 | K. pneumoniae | positive | blaVIM | blaVIM-1 | 32 | 232,474 | |
| KE9538 | S. marcescens | positive | blaVIM | blaVIM-1 | 40 | 1,130,420 | |
| KE9585 | E. cloacae | positive | blaVIM | blaVIM-1 | 25 | 287,090 | |
| KE9559 | C. freundii | positive | blaVIM | blaVIM-1 | 41 | 163,976 | |
| KE9549 | E. coli | positive | blaVIM | blaVIM-1 | 47 | 279,067 | |
| KE9548 | E. cloacae | positive | blaVIM | blaVIM-1 | 46 | 230,814 | |
| KE9579 | E. coli | positive | blaVIM | blaVIM-1 | 39 | 112,495 | |
| KE9474 | E. cloacae | positive | blaVIM | blaVIM-1 | 38 | 290,132 | |
| KE9462 | E. cloacae | positive | blaVIM | blaVIM-1 | 33 | 502,528 | |
| KE9560 | E. cloacae | positive | blaVIM | blaVIM-1 | 40 | 290,117 | |
| KE9575 | E. cloacae | positive | blaVIM | blaVIM-1 | 38 | 389,538 | |
| KE9536 | E. coli | positive | blaVIM | negative | blaVIM-1 | 44 | 377,920 |
| D2923 | E. cloacae | negative | negative | negative | 58 | 203,439 | |
| KE9347 | E. cloacae | negative | negative | negative | 79 | 439,426 | |
| KE9591 | E. cloacae | negative | negative | negative | 47 | 279,225 | |
| KE9576 | E. coli | negative | negative | negative | 27 | 228,481 | |
| KE9599 | E. coli | negative | negative | negative | 37 | 281,932 | |
| KE9623 | E. coli | negative | negative | negative | 50 | 93,960 | |
| KE9633 | K. aerogenes | negative | negative | negative | 40 | 495,847 | |
| KE9068 | C. freundii | negative | negative | negative | 46 | 176,242 | |
| KE8986 | E. cloacae | negative | negative | negative | 47 | 230,847 | |
| KE9344 | E. cloacae | negative | negative | negative | 48 | 235,301 | |
| KE9475 | E. cloacae | negative | negative | negative | 40 | 208,042 | |
| KE9083 | E. coli | negative | negative | negative | 57 | 208,544 | |
| KE9425 | K. aerogenes | negative | negative | negative | 48 | 902,223 | |
| KE9614 | K. aerogenes | negative | negative | negative | 62 | 429,809 | |
| D3017 | K. pneumoniae | negative | negative | negative | 27 | 232,937 | |
| KE9095 | K. pneumoniae | negative | negative | negative | 66 | 237,389 | |
| KE9171 | K. pneumoniae | negative | negative | negative | 54 | 481,561 | |
| KE9039 | S. marcescens | negative | negative | negative | 40 | 1,228,444 |
References
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215 (Suppl. 1), S28–S36. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Delgado-Valverde, M.; Sojo-Dorado, J.; Pascual, Á.; Rodríguez-Baño, J. Clinical management of infections caused by multidrug-resistant Enterobacteriaceae. Ther. Adv. Infect. Dis. 2013, 1, 49–69. [Google Scholar] [CrossRef]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 2017, 12, 3. [Google Scholar]
- Paterson, D.; Doi, Y. Carbapenemase-Producing Enterobacteriaceae. Semin. Respir. Crit. Care Med. 2015, 36, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Cornaglia, G. Carbapenemase-producing Enterobacteriaceae: A call for action! Clin. Microbiol. Infect. 2012, 18, 411–412. [Google Scholar] [CrossRef]
- Tijet, N.; Boyd, D.; Patel, S.N.; Mulvey, M.R.; Melano, R.G. Evaluation of the Carba NP Test for Rapid Detection of Carbapenemase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4578–4580. [Google Scholar] [CrossRef]
- Amjad, A.; Mirza, I.; Abbasi, S.; Farwa, U.; Malik, N.; Zia, F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran. J. Microbiol. 2011, 3, 189–193. [Google Scholar]
- Van Dijk, K.; Voets, G.M.; Scharringa, J.; Voskuil, S.; Fluit, A.C.; Rottier, W.C.; Hall, M.A.L.; Stuart, J.W.T.C. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin. Microbiol. Infect. 2014, 20, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, A.; Arena, F.; Giani, T.; Colavecchio, O.L.; Valeva, S.V.; Paule, S.; Boleij, P.; Rossolini, G.M. Performance of the BD MAX™ instrument with Check-Direct CPE real-time PCR for the detection of carbapenemase genes from rectal swabs, in a setting with endemic dissemination of carbapenemase-producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2016, 86, 30–34. [Google Scholar] [CrossRef]
- Hofko, M.; Mischnik, A.; Kaase, M.; Zimmermann, S.; Dalpke, A.H. Detection of Carbapenemases by Real-Time PCR and Melt Curve Analysis on the BD Max System. J. Clin. Microbiol. 2014, 52, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Schurch, A.C.; van Schaik, W. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann. N. Y. Acad. Sci. 2017, 1388, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Baeza, L.L.; Pfennigwerth, N.; Greissl, C.; Göttig, S.; Saleh, A.; Stelzer, Y.; Gatermann, S.; Hamprecht, A. Comparison of five methods for detection of carbapenemases in Enterobacterales with proposal of a new algorithm. Clin. Microbiol. Infect. 2019, 25, 1286.e9–1286.e15. [Google Scholar] [CrossRef]
- Baeza, L.L.; Pfennigwerth, N.; Hamprecht, A. Rapid and Easy Detection of Carbapenemases in Enterobacterales in the Routine Laboratory Using the New Gene POC Carba/Revogene Carba C Assay. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Han, R.; Guo, Y.; Peng, M.; Shi, Q.; Wu, S.; Yang, Y.; Zheng, Y.; Yin, D.; Hu, F. Evaluation of the Immunochromatographic NG-Test Carba 5, RESIST-5 O.O.K.N.V., and IMP K-SeT for Rapid Detection of KPC-, NDM-, IMP-, VIM-type, and OXA-48-like Carbapenemase Among Enterobacterales. Front. Microbiol. 2020, 11, 609856. [Google Scholar] [CrossRef] [PubMed]
- Ellington, M.; Ekelund, O.; Aarestrup, F.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.; Hopkins, K.; et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef]
- Pfennigwerth, N. Bericht des Nationalen Referenzzentrums (NRZ) für gramnegative Krankenhauserreger – Zeitraum 1. Januar 2017 – 31. Dezember 2017. J. Epidemiol. Bull. 2018, 28, 263–267. [Google Scholar]
- Pfennigwerth, N. Bericht des Nationalen Referenzzentrums (NRZ) für gramnegative Krankenhauserreger, 2018. J. Epidemiol. Bull. 2019, 31, 289–294. [Google Scholar]
- Pfennigwerth, N. Bericht des Nationalen Referenzzentrums (NRZ) für gramnegative Krankenhauserreger, 2019. J. Epidemiol. Bull. 2020, 26, 3–10. [Google Scholar]
- Nurjadi, D.; Zizmann, E.; Chanthalangsy, Q.; Heeg, K.; Boutin, S. Integrative Analysis of Whole Genome Sequencing and Phenotypic Resistance Toward Prediction of Trimethoprim-Sulfamethoxazole Resistance in Staphylococcus aureus. Front. Microbiol. 2020, 11, 607842. [Google Scholar] [CrossRef] [PubMed]
- Van der Zwaluw, K.; de Haan, A.; Pluister, G.N.; Bootsma, H.J.; de Neeling, A.J.; Schouls, L.M. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef] [PubMed]
- Probst, K.; Boutin, S.; Bandilla, M.; Heeg, K.; Dalpke, A.H. Fast and automated detection of common carbapenemase genes using multiplex real-time PCR on the BD MAX™ system. J. Microbiol. Methods 2021, 185, 106224. [Google Scholar] [CrossRef] [PubMed]
- Nurjadi, D.; Boutin, S.; Dalpke, A.; Heeg, K.; Zanger, P. Draft Genome Sequence of Staphylococcus aureus Strain HD1410, Isolated from a Persistent Nasal Carrier. Genome Announc. 2018, 6, e00411–e00418. [Google Scholar] [CrossRef] [PubMed]
- Eichel, P.C.V.; Boutin, S.; Poschl, J.; Heeg, K.; Nurjadi, D. Altering antibiotic regimen as additional control measure in suspected multi-drug-resistant Enterobacter cloacae outbrake in neonatal intensive care unit. J. Hosp. Infect. 2019, 104, 144–149. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).