Counteraction of Biofilm Formation and Antimicrobial Potential of Terminalia catappa Functionalized Silver Nanoparticles against Candida albicans and Multidrug-Resistant Gram-Negative and Gram-Positive Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of T. catappa Leaf Extract (TCE)
2.2. Bio-Synthesis of TCE-Ag-NPs
2.3. Physico-Chemical Characterization of TCE-Ag-NPs
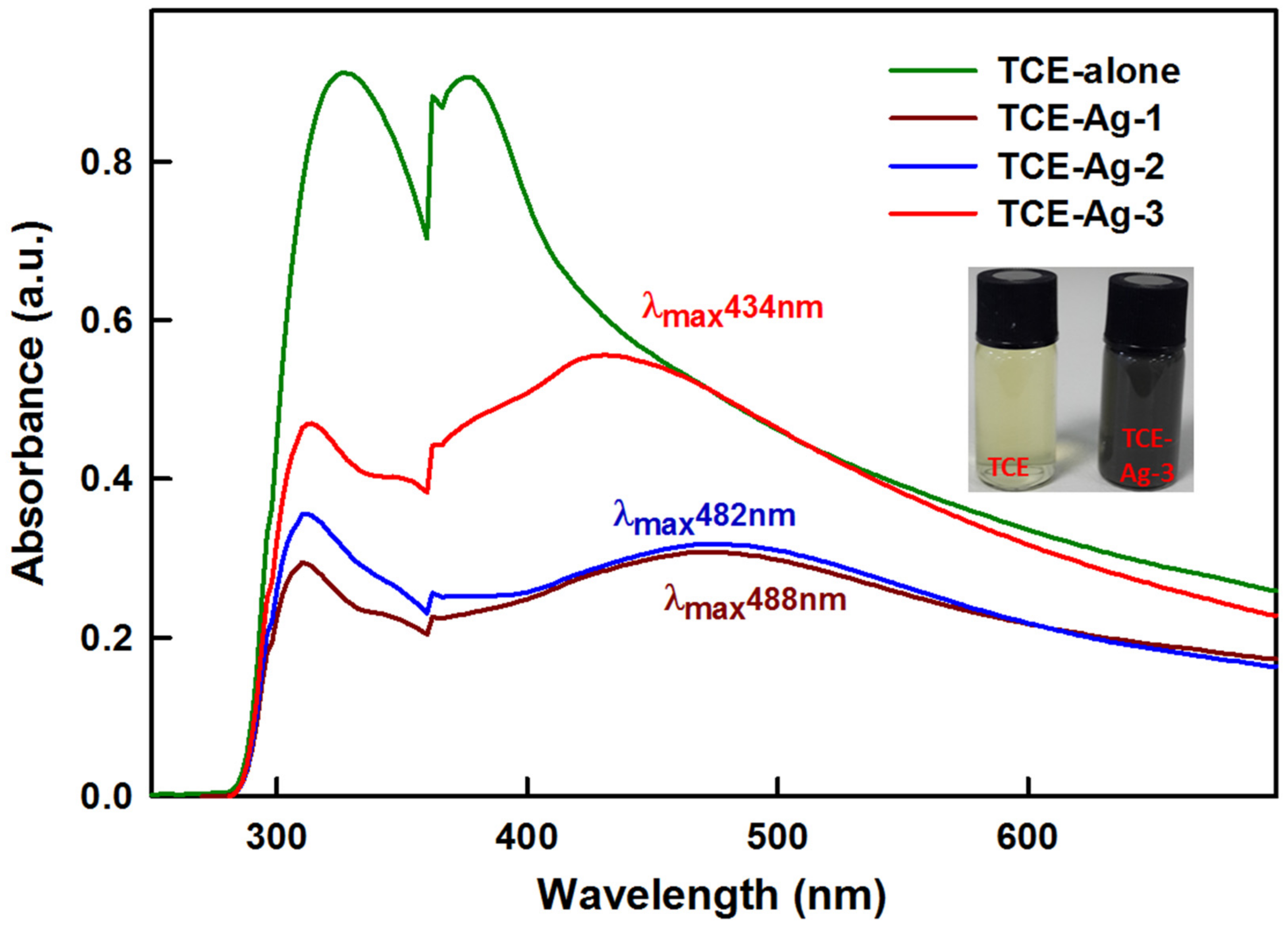
2.3.1. UV-Visible Spectroscopy
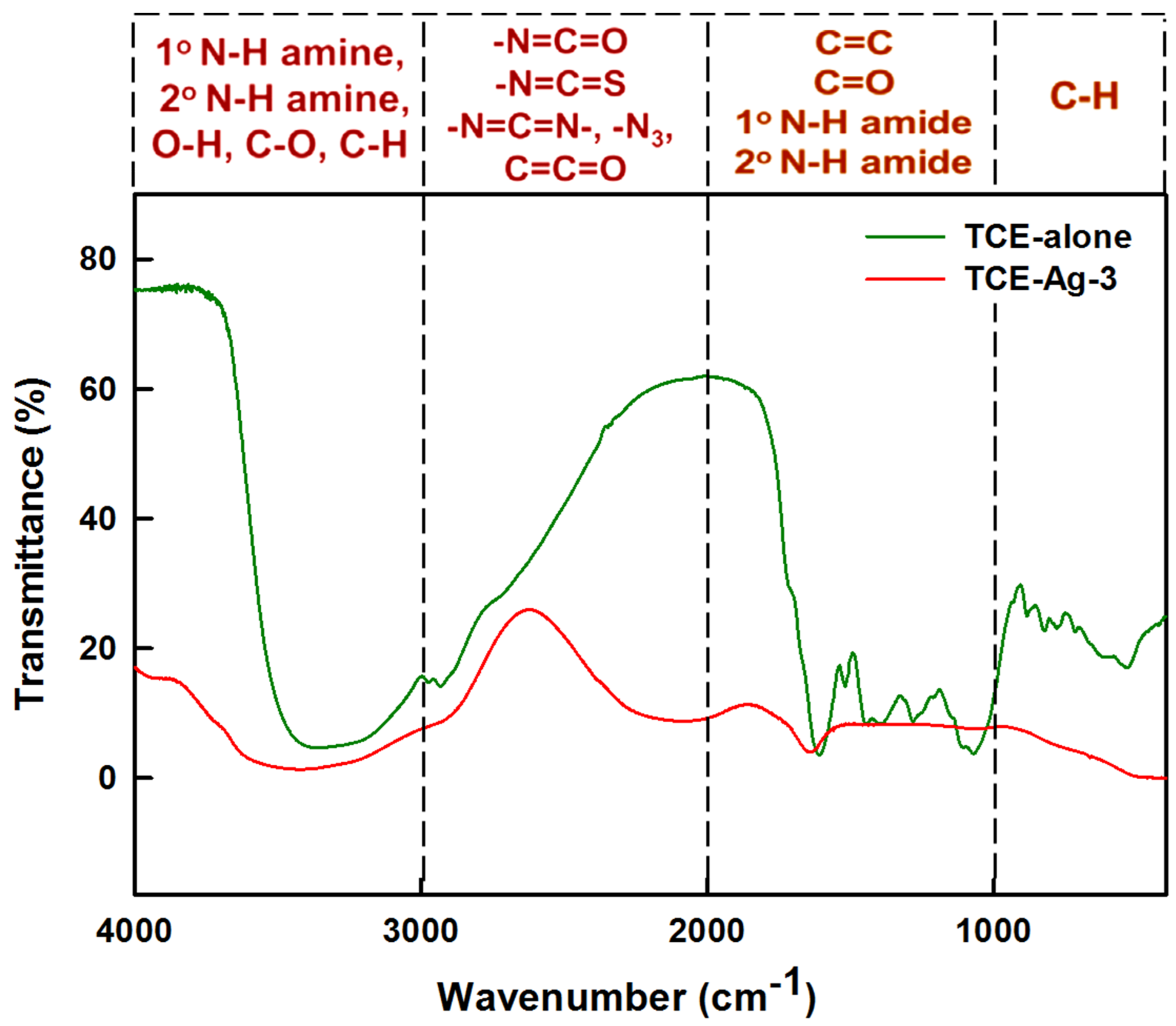
2.3.2. FTIR Spectroscopy of TCE-Ag-3
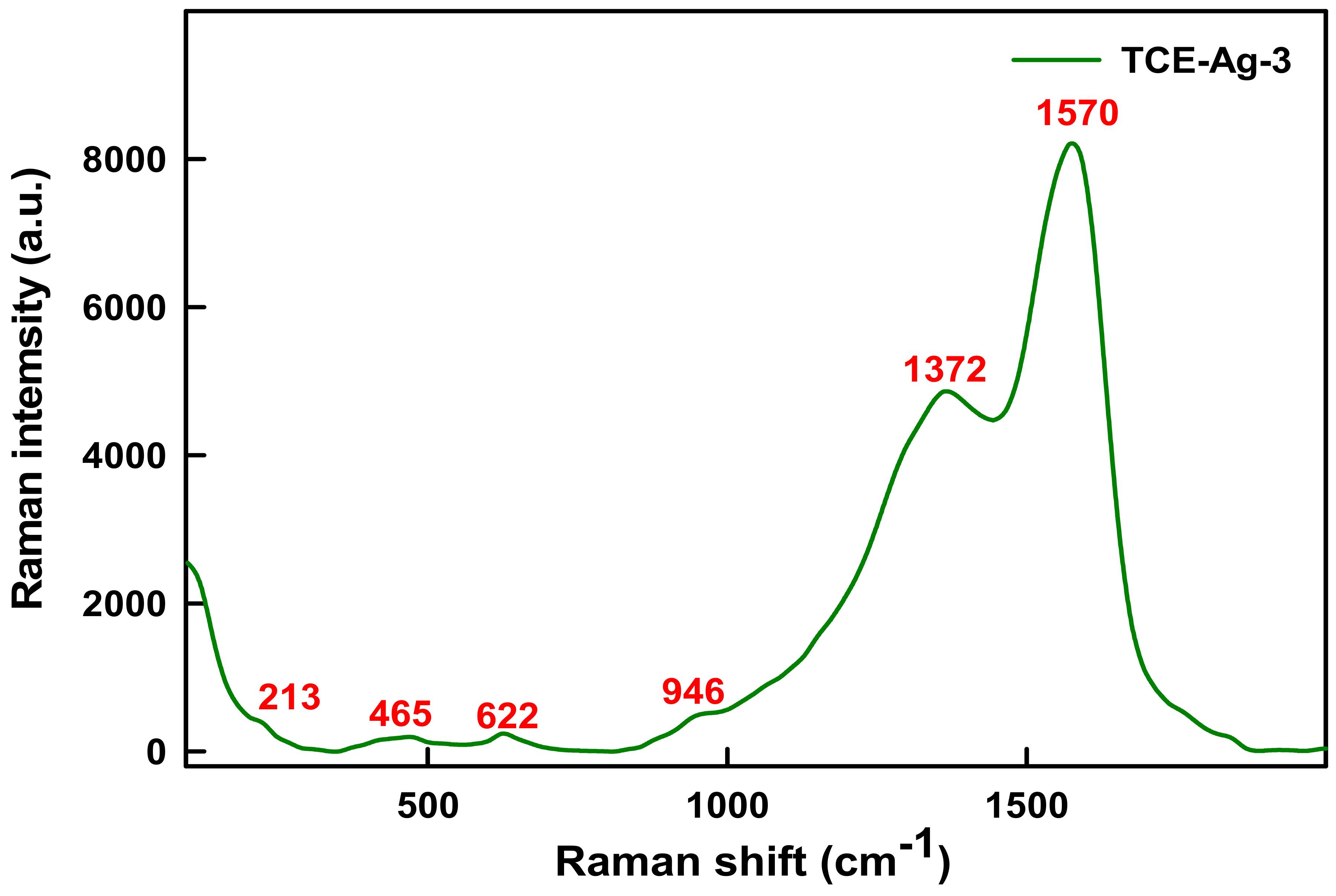
2.3.3. Raman Spectroscopy of TCE-Ag-3
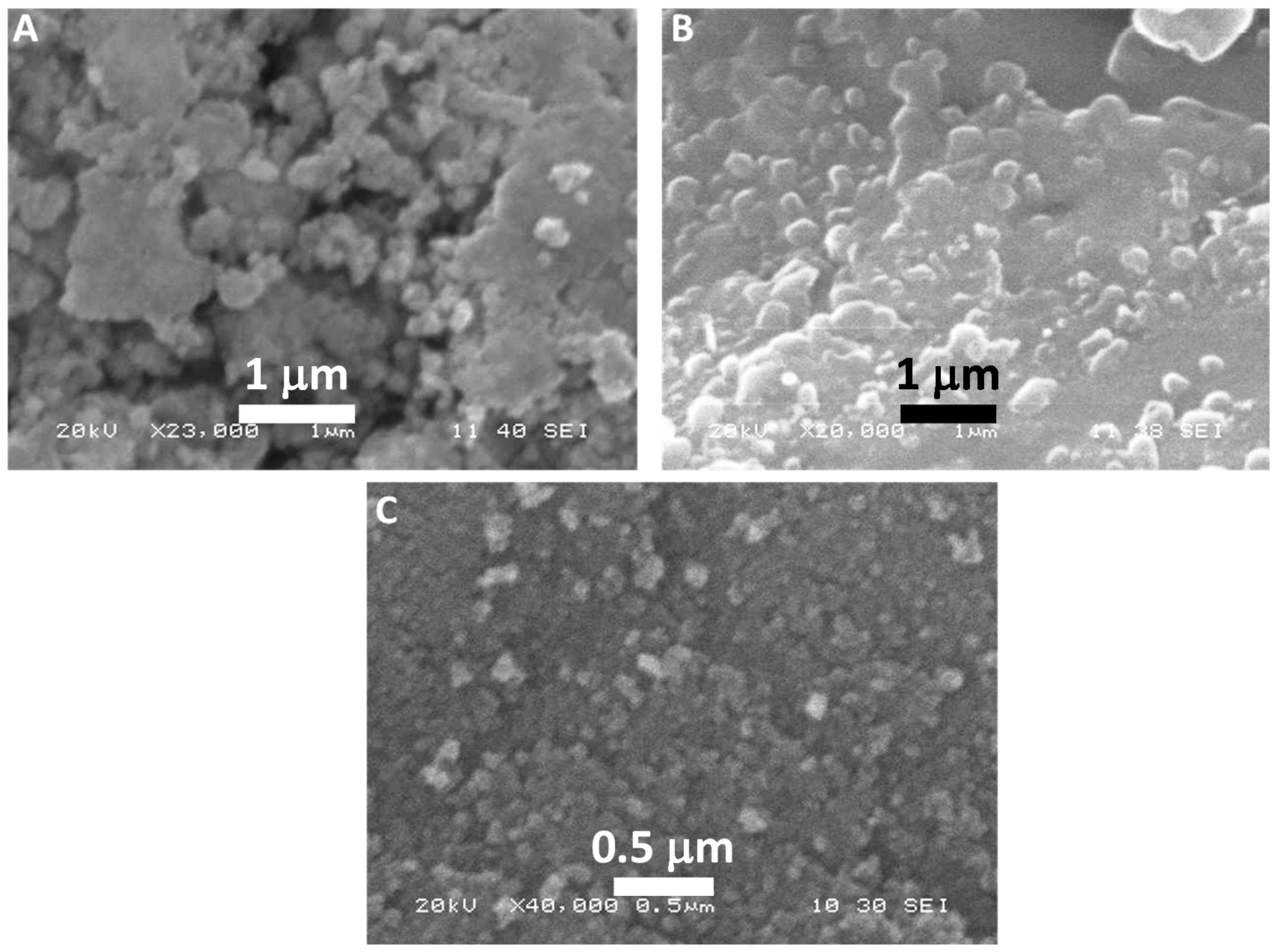
2.3.4. Electron Microscopy-Based Analyses of TCE-Ag-NPs
2.4. Antibacterial Activities of TCE-Ag-NPs
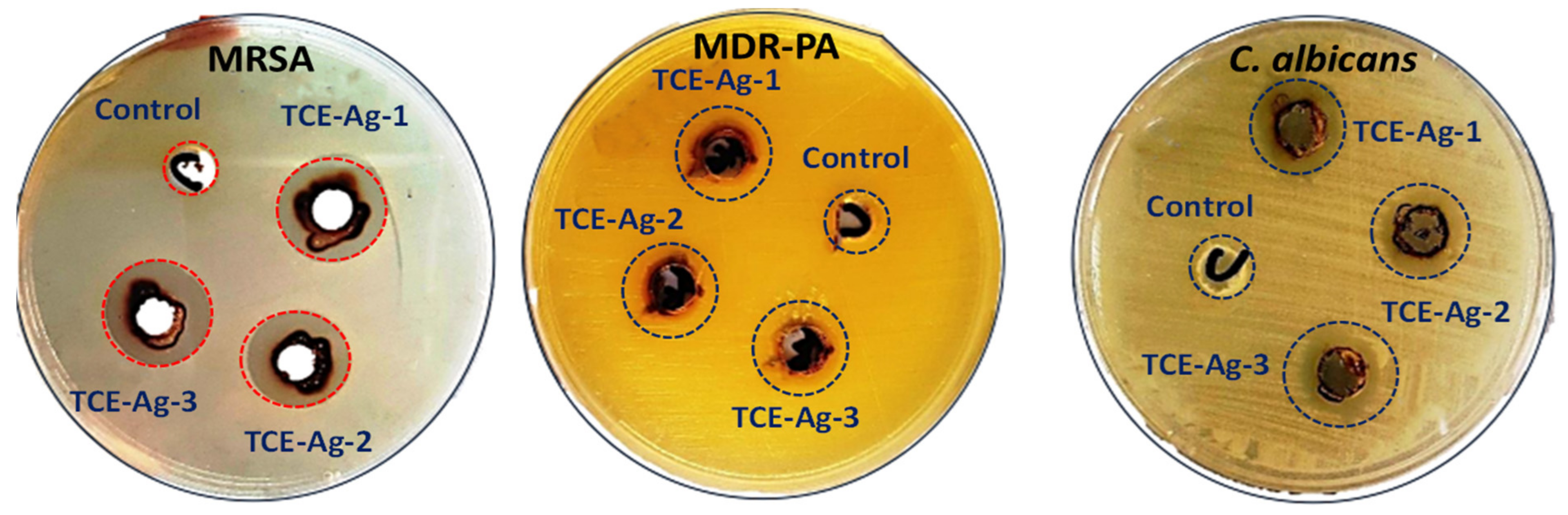
2.4.1. Assessment of Antibacterial Activities of TCE-Ag-NPs by Well Diffusion Assay
2.4.2. Determination of MIC, MBC and MFC Values of TCE-Ag-NPs against Test Bacterial and Fungal Strains
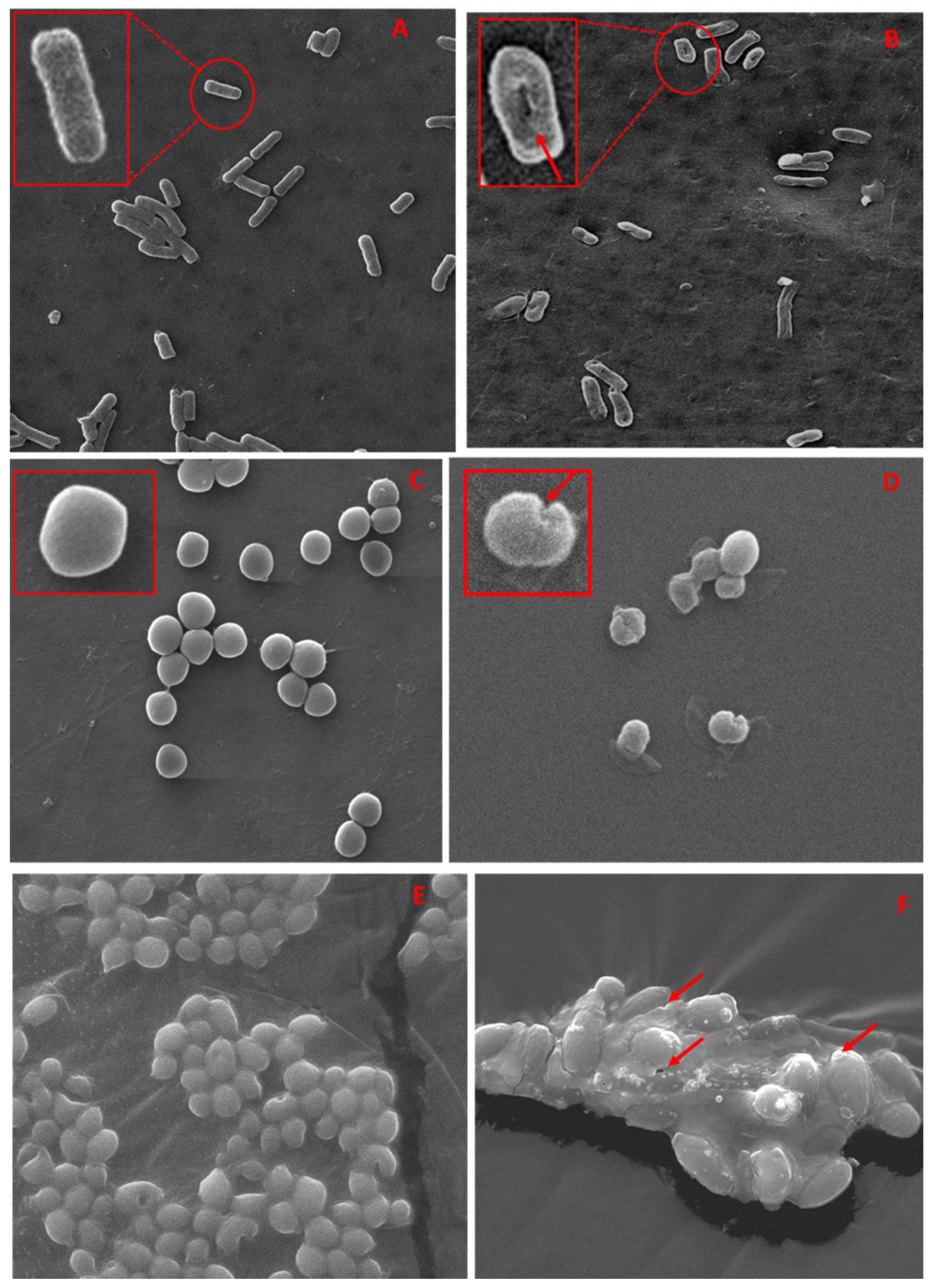
2.4.3. SEM Based Imaging of Test Strains and TCE-Ag-3
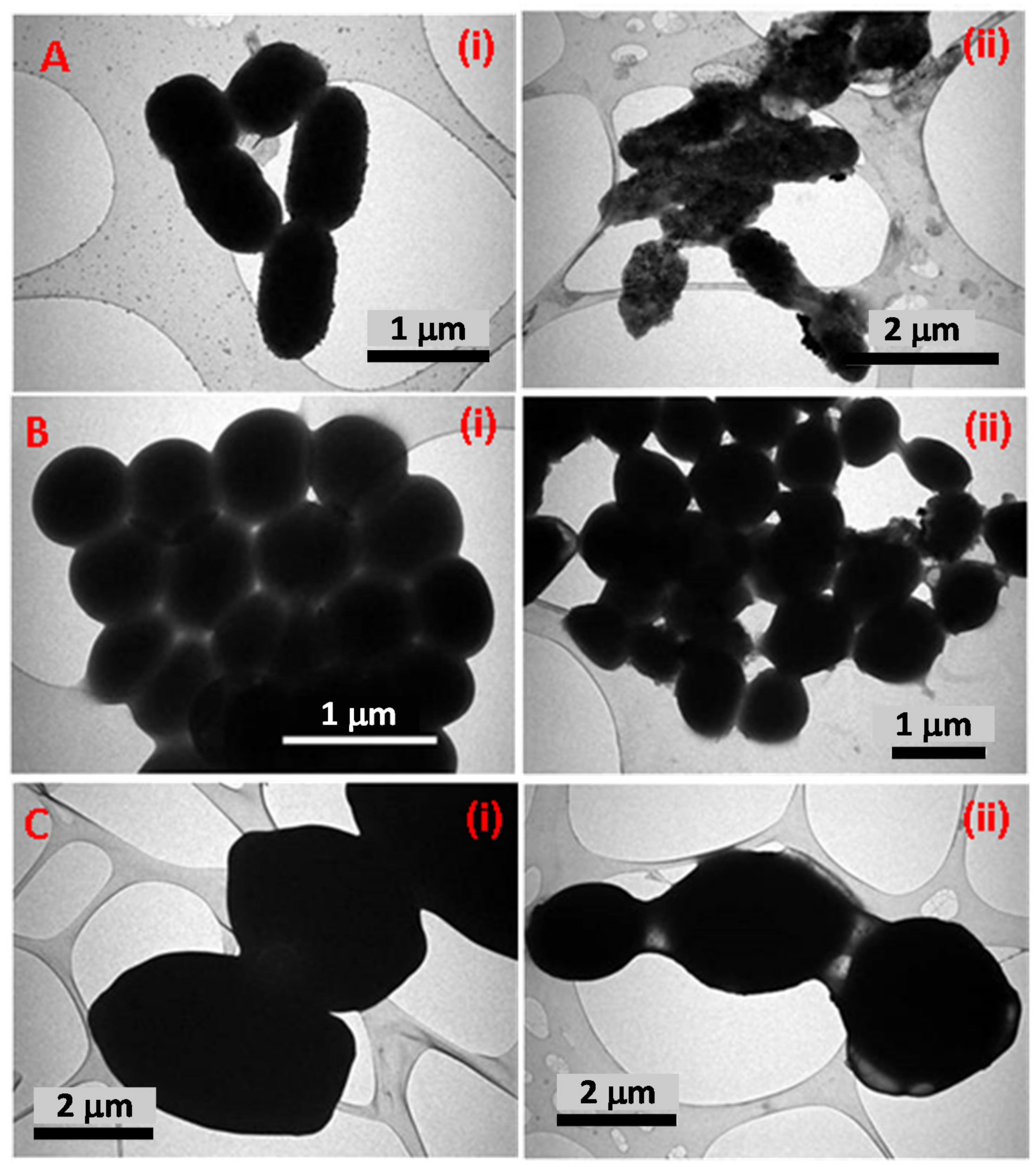
2.4.4. TEM Based Assessment of Antimicrobial Activity of TCE-Ag-3
2.4.5. Evaluation of Antibiofilm Activity of TCE-Ag-3 against Test Strains
2.4.6. Effect of TCE-Ag-3 on Biofilm Structure of Tested Strains
3. Results and Discussion
3.1. T. catappa Leaf Extract (TCE) Mediated Synthesis of Ag-NPs
3.2. FTIR Spectral Analysis of TCE-Ag-3
3.3. Raman Spectral Analysis of TCE-Ag-3
3.4. Morphological Characterization of TCE-Ag-NPs
3.5. Antimicrobial Activities
3.5.1. Assessment of Size-Specific Antimicrobial Activities by Well Diffusion Assay
3.5.2. Size-Specific MIC, MBC, and MFC Values Determination of TCE-Ag-NPs against Test Strains
3.5.3. SEM and TEM Based Analysis of the Interaction of TCE-Ag-3 with Planktonic Cells of Test Strains
3.5.4. Antibiofilm Activity of TCE-Ag-3 against Test Strains
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bühl, M.; Peter, S.; Willmann, M. Prevalence and risk factors associated with colonization and infection of extensively drug-resistant Pseudomonas aeruginosa: A systematic review. Expert Rev. Anti-Infect. Ther. 2015, 13, 1159–1170. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, C.; Li, C.; Lin, L. Inhibition mechanism of cardamom essential oil on methicillin-resistant Staphylococcus aureus biofilm. LWT 2020, 122, 109057. [Google Scholar] [CrossRef]
- Firacative, C. Invasive fungal disease in humans: Are we aware of the real impact? Memórias Inst. Oswaldo Cruz 2020, 115. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.; Lui, G.C.; Chen, N.; Li, A.; Lu, W.; Chan, F.K.; et al. Alterations in Fecal Fungal Microbiome of Patients With COVID-19 During Time of Hospitalization until Discharge. Gastroenterology 2020, 159, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Howell, P.L. Biofilm Exopolysaccharides of Pathogenic Fungi: Lessons from Bacteria. J. Biol. Chem. 2016, 291, 12529–12537. [Google Scholar] [CrossRef]
- Jiang, Y.; Geng, M.; Bai, L. Targeting Biofilms Therapy: Current Research Strategies and Development Hurdles. Microorganisms 2020, 8, 1222. [Google Scholar] [CrossRef] [PubMed]
- Limoli, D.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3, 223–247. [Google Scholar] [CrossRef]
- Ali, K.; Ahmed, B.; Dwivedi, S.; Saquib, Q.; Al-Khedhairy, A.; Musarrat, J. Microwave Accelerated Green Synthesis of Stable Silver Nanoparticles with Eucalyptus globulus Leaf Extract and Their Antibacterial and Antibiofilm Activity on Clinical Isolates. PLoS ONE 2015, 10, e0131178. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.G.; Ansari, M.A.; Jamal, Q.M.S.; Almatroudi, A.; Alzohairy, M.A.; Alomary, M.N.; Rehman, S.; Mahadevamurthy, M.; Jalal, M.; Khan, H.M.; et al. Butea monosperma seed extract mediated biosynthesis of ZnO NPs and their antibacte-rial, antibiofilm and anti-quorum sensing potentialities. Arab. J. Chem. 2021, 14, 103044. [Google Scholar] [CrossRef]
- Ansari, M.A.; Asiri, S.M.M.; Alzohairy, M.A.; Alomary, M.N.; Almatroudi, A.; Khan, F.A. Biofabricated Fatty Ac-ids-Capped Silver Nanoparticles as Potential Antibacterial, Antifungal, Antibiofilm and Anticancer Agents. Pharmaceuticals 2021, 14, 139. [Google Scholar] [CrossRef]
- Alomary, M.N.; Mohammad, A.A. Proanthocyanins-capped biogenic TiO2 nanoparticles with en-hanced penetration, antibacterial and ROS mediated inhibition of bacteria proliferation and biofilm formation: A comparative approach. Chem. Eur. J. 2021, 27, 5817–5829. [Google Scholar] [CrossRef]
- Murali, M.; Anandan, S.; Ansari, M.A.; Alzohairy, M.A.; Alomary, M.N.; Asiri, S.M.M.; Almatroudi, A.; Thriveni, M.C.; Singh, S.B.; Gowtham, H.G.; et al. Genotoxic and Cytotoxic Properties of Zinc Oxide Nanoparticles Phy-to-Fabricated from the Obscure Morning Glory Plant Ipomoea obscura (L.) Ker Gawl. Molecules 2021, 26, 891. [Google Scholar] [CrossRef]
- Lin, C.-C.; Yea-Ling, C.; Jer, M.L.; Takashi, U. Evaluation of the antioxidant and hepatoprotective activity of Terminalia catappa. Am. J. Chin. Med. 1997, 25, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, R.; Sayeed, S.A.; Naz, S.; Saeed, S.M.G. Antioxidant Activity of the Extracts Derived from Terminalia catappa. Biol. Sci. PJSIR 2011, 54, 93–98. [Google Scholar] [CrossRef]
- Khan, A.A.; Kumar, V.; Singh, B.K.; Singh, R. Evaluation of Wound Healing Property of Terminalia catappa on Excision Wound Models in Wistar Rats. Drug Res. 2013, 64, 225–228. [Google Scholar] [CrossRef]
- Ko, T.-F.; Weng, Y.-M.; Chiou, R.Y.-Y. Squalene Content and Antioxidant Activity of Terminalia catappa Leaves and Seeds. J. Agric. Food Chem. 2002, 50, 5343–5348. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Yu-Fang, H.; Ta-Chen, L. Antioxidant and free radical scavenging effects of the tannins of Terminalia catappa L. Anticancer Res. 2001, 21, 237–243. [Google Scholar]
- Chyau, C.-C.; Tsai, S.-Y.; Ko, P.-T.; Mau, J.-L. Antioxidant properties of solvent extracts from Terminalia catappa leaves. Food Chem. 2002, 78, 483–488. [Google Scholar] [CrossRef]
- Rajendhiran, R.; Deivasigamani, V.; Palanisamy, J.; Masan, S.; Pitchaiya, S. Terminalia catappa and Carissa carandas assisted synthesis of TiO2 nanoparticles—A green synthesis approach. In Proceedings of the Materials Today: Proceedings; Elsevier BV: Amsterdam, The Netherlands, 2021; Volume 45, pp. 2232–2238. [Google Scholar]
- Ankamwar, B. Biosynthesis of Gold Nanoparticles (Green-gold) Using Leaf Extract of Terminalia catappa. E-J. Chem. 2010, 7, 1334–1339. [Google Scholar] [CrossRef]
- Muthulakshmi, L.; Rajini, N.; Nellaiah, H.; Kathiresan, T.; Jawaid, M.; Rajulu, A. Preparation and properties of cellulose nanocomposite films with in situ generated copper nanoparticles using Terminalia catappa leaf extract. Int. J. Biol. Macromol. 2017, 95, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Lembang, M.S.; Yoki, Y.; Sudirman, S.; Apriandanu, D.O.B. A facile method for green synthesis of Nd2O3 nano-particles using aqueous extract of Terminalia catappa leaf. In Proceedings of the AIP Conference Proceedings, 2023: 020093; AIP Publishing LLC: Melville, NY, USA, 2018. [Google Scholar]
- Devadiga, A.; Shetty, K.V.; Saidutta, M. Highly stable silver nanoparticles synthesized using Terminalia catappa leaves as antibacterial agent and colorimetric mercury sensor. Mater. Lett. 2017, 207, 66–71. [Google Scholar] [CrossRef]
- Marambio-Jones, C.; Hoek, E.M.V. A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. J. Nanoparticle Res. 2010, 12, 1531–1551. [Google Scholar] [CrossRef]
- Chamakura, K.; Perez-Ballestero, R.; Luo, Z.; Bashir, S.; Liu, J. Comparison of bactericidal activities of silver nanoparticles with common chemical disinfectants. Colloids Surf. B Biointerfaces 2011, 84, 88–96. [Google Scholar] [CrossRef]
- Crisan, C.M.; Mocan, T.; Manolea, M.; Lasca, L.I.; Tăbăran, F.-A.; Mocan, L. Review on Silver Nanoparticles as a Novel Class of Antibacterial Solutions. Appl. Sci. 2021, 11, 1120. [Google Scholar] [CrossRef]
- Joshi, A.S.; Singh, P.; Mijakovic, I. Interactions of Gold and Silver Nanoparticles with Bacterial Biofilms: Molecular Interactions behind Inhibition and Resistance. Int. J. Mol. Sci. 2020, 21, 7658. [Google Scholar] [CrossRef] [PubMed]
- Padnya, P.; Gorbachuk, V.; Stoikov, I. The Role of Calix[n]arenes and Pillar[n]arenes in the Design of Silver Nanoparticles: Self-Assembly and Application. Int. J. Mol. Sci. 2020, 21, 1425. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-Negative Bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef]
- Shahverdi, A.R.; Fakhimi, A.; Shahverdi, H.R.; Minaian, S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 168–171. [Google Scholar] [CrossRef]
- Jalal, M.; Ansari, M.A.; Ali, S.G.; Khan, H.M.; Eldaif, W.A.H.; Alrumman, S.A. Green synthesis of silver nanoparticles using leaf extract of Cinnamomum tamala and its antimicrobial activity against clinical isolates of bacteria and fungi. Int. J. Adv. Res. 2016, 4, 428–440. [Google Scholar] [CrossRef]
- Cherian, T.; Ali, K.; Fatima, S.; Saquib, Q.; Ansari, S.M.; Alwathnani, H.A.; Al-Khedhairy, A.A.; Al-Shaeri, M.; Musarrat, J. Myristica fragrans bioactive ester functionalized ZnO nanoparticles exhibit antibacterial and antibiofilm activities in clinical isolates. J. Microbiol. Methods 2019, 166, 105716. [Google Scholar] [CrossRef]
- Ansari, M.A.; Khan, H.M.; Alzohairy, M.A.; Jalal, M.; Ali, S.G.; Pal, R.; Musarrat, J. Green synthesis of Al2O3 nanoparticles and their bactericidal potential against clinical isolates of multi-drug resistant Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2014, 31, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Klaus, T.; Joerger, R.; Olsson, E.; Granqvist, C.-G. Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. USA 1999, 96, 13611–13614. [Google Scholar] [CrossRef]
- Ahmad, A.; Mukherjee, P.; Senapati, S.; Mandal, D.; Khan, M.I.; Kumar, R.; Sastry, M. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids Surf. B Biointerfaces 2003, 28, 313–318. [Google Scholar] [CrossRef]
- Borse, S.; Murthy, Z.V.P.; Kailasa, S.K. Chicken egg white mediated synthesis of platinum nanoclusters for the selective detection of carbidopa. Opt. Mater. 2020, 107, 110085. [Google Scholar] [CrossRef]
- Thombre, R.; Parekh, F.; Lekshminarayanan, P.; Francis, G. Studies on antibacterial and antifungal activity of silver nanoparticles synthesized using Artocarpus heterophyllus leaf extract. Biotechnol. Bioinf. Bioeng. 2012, 2, 632–637. [Google Scholar]
- Černík, M.; Padil, V.V.T. Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application. Int. J. Nanomed. 2013, 8, 889–898. [Google Scholar] [CrossRef]
- Ali, K.; Dwivedi, S.; Azam, A.; Saquib, Q.; Al-Said, M.S.; Alkhedhairy, A.A.; Musarrat, J. Aloe vera extract functionalized zinc oxide nanoparticles as nanoantibiotics against multi-drug resistant clinical bacterial isolates. J. Colloid Interface Sci. 2016, 472, 145–156. [Google Scholar] [CrossRef]
- Bobby, M.N.; Wesely, E.G.; Johnson, M. FT-IR studies on the leaves of Albizia lebbeck benth. Int. J. Pharm. Pharm. Sci. 2012, 4, 293–296. [Google Scholar]
- Parlinska-Wojtan, M.; Kus-Liskiewicz, M.; Depciuch, J.; Sadik, O. Green synthesis and antibacterial effects of aqueous colloidal solutions of silver nanoparticles using camomile terpenoids as a combined reducing and capping agent. Bioprocess Biosyst. Eng. 2016, 39, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Moskovits, M. Surface-enhanced spectroscopy. Rev. Mod. Phys. 1985, 57, 783. [Google Scholar] [CrossRef]
- Jurasekova, Z.; Garcia-Ramos, J.V.; Domingo, C.; Sanchez-Cortes, S. Surface-enhanced Raman scattering of flavonoids. J. Raman Spectrosc. 2006, 37, 1239–1241. [Google Scholar] [CrossRef]
- Garcia-Bucio, M.A.; Maynez-Rojas, M.Á.; Casanova-González, E.; Cárcamo-Vega, J.J.; Ruvalcaba-Sil, J.L.; Mitrani, A. Raman and surface-enhanced Raman spectroscopy for the analysis of Mexican yellow dyestuff. J. Raman Spectrosc. 2019, 50, 1546–1554. [Google Scholar] [CrossRef]
- Oyeleye, S.I.; Adebayo, A.A.; Ogunsuyi, O.B.; Dada, F.A.; Oboh, G. Phenolic profile and Enzyme Inhibitory activities of Almond (Terminalia catappa) leaf and Stem bark. Int. J. Food Prop. 2017, 20, S2810–S2821. [Google Scholar] [CrossRef]
- Jones, R.S.; Parker, M.D.; Morris, M.E. Quercetin, Morin, Luteolin, and Phloretin Are Dietary Flavonoid Inhibitors of Monocarboxylate Transporter 6. Mol. Pharm. 2017, 14, 2930–2936. [Google Scholar] [CrossRef]
- Wijeratne, S.S.K.; Abou-Zaid, M.M.; Shahidi, F. Antioxidant Polyphenols in Almond and Its Coproducts. J. Agric. Food Chem. 2006, 54, 312–318. [Google Scholar] [CrossRef]
- Harnly, J.M.; Doherty, R.F.; Beecher, G.R.; Holden, J.M.; Haytowitz, D.B.; Bhagwat, A.S.; Gebhardt, S. Flavonoid Content of U.S. Fruits, Vegetables, and Nuts. J. Agric. Food Chem. 2006, 54, 9966–9977. [Google Scholar] [CrossRef] [PubMed]
- Siess, M.H.; Le Bon, A.M.; Canivenc-Lavier, M.C.; Amiot, M.J.; Sabatier, S.; Aubert, S.Y.; Suschetet, M. Flavonoids of Honey and Propolis: Characterization and Effects on Hepatic Drug-Metabolizing Enzymes and Benzo[a]pyrene—DNA Binding in Rats. J. Agric. Food Chem. 1996, 44, 2297–2301. [Google Scholar] [CrossRef]
- Biswas, N.; Kapoor, S.; Mahal, H.S.; Mukherjee, T. Adsorption of CGA on colloidal silver particles: DFT and SERS study. Chem. Phys. Lett. 2007, 444, 338–345. [Google Scholar] [CrossRef]
- Kora, A.J.; Arunachalam, J. Green fabrication of silver nanoparticles by gum Tragacanth (Astragalus gummifer): A dual func-tional reductant and stabilizer. J. Nanomater. 2012, 1, 69. [Google Scholar] [CrossRef]
- Ranoszek-Soliwoda, K.; Tomaszewska, E.; Małek, K.; Celichowski, G.; Orlowski, P.; Krzyzowska, M.; Grobelny, J. The synthesis of monodisperse silver nanoparticles with plant extracts. Colloids Surf. B Biointerfaces 2019, 177, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Korshed, P.; Li, L.; Liu, Z.; Mironov, A.; Wang, T. Size-dependent antibacterial activity for laser-generated silver nano-particles. J. Interdiscip. Nanomed. 2019, 4, 24–33. [Google Scholar] [CrossRef]
- Skomorokhova, E.A.; Sankova, T.P.; Orlov, I.A.; Savelev, A.N.; Magazenkova, D.N.; Pliss, M.G.; Skvortsov, A.N.; Sosnin, I.M.; Kirilenko, D.A.; Grishchuk, I.V.; et al. Size-Dependent Bioactivity of Silver Nanoparticles: Antibacterial Properties, Influence on Copper Status in Mice, and Whole-Body Turnover. Nanotechnol. Sci. Appl. 2020, 13, 137–157. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.Y.; Rukayadi, Y.; Nor-Khaizura, M.-A.-R.; Kuan, C.H.; Chieng, B.W.; Nishibuchi, M.; Radu, S. In Vitro Antimicrobial Activity of Green Synthesized Silver Nanoparticles against Selected Gram-negative Foodborne Pathogens. Front. Microbiol. 2018, 9, 1555. [Google Scholar] [CrossRef]
- Alqahtani, M.A.; Al Othman, M.R.; Mohammed, A.E. Bio fabrication of silver nanoparticles with antibacterial and cytotoxic abilities using lichens. Sci. Rep. 2020, 10, 1–17. [Google Scholar]
- Tolaymat, T.M.; El Badawy, A.M.; Genaidy, A.; Scheckel, K.; Luxton, T.P.; Suidan, M. An evidence-based environmental perspective of manufactured silver nanoparticle in syntheses and applications: A systematic review and critical appraisal of peer-reviewed scientific papers. Sci. Total. Environ. 2010, 408, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Piccapietra, F.; Wagner, B.; Marconi, F.; Kaegi, R.; Odzak, N.; Sigg, L.; Behra, R. Toxicity of Silver Nanoparticles to Chlamydomonas reinhardtii. Environ. Sci. Technol. 2010, 42, 8959–8964. [Google Scholar] [CrossRef] [PubMed]
- Miao, A.J.; Schwehr, K.A.; Xu, C.; Zhang, S.J.; Luo, Z.; Quigg, A.; Santschi, P.H. The algal toxicity of silver engineered nanopar-ticles and detoxification by exopolymeric substances. Environ. Pollut. 2008, 157, 3034–3041. [Google Scholar] [CrossRef] [PubMed]
- Fabrega, J.; Fawcett, S.R.; Renshaw, J.C.; Lead, J.R. Silver nanoparticle impact on bacterial growth: Effect of pH, concentration, and organic matter. Environ. Sci. Technol. 2009, 43, 7285–7290. [Google Scholar] [CrossRef] [PubMed]
- Kawata, K.; Osawa, M.; Okabe, S. In Vitro Toxicity of Silver Nanoparticles at Noncytotoxic Doses to HepG2 Human Hepatoma Cells. Environ. Sci. Technol. 2009, 43, 6046–6051. [Google Scholar] [CrossRef]
- Sotiriou, G.A.; Teleki, A.; Camenzind, A.; Krumeich, F.; Meyer, A.; Panke, S.; Pratsinis, S.E. Nanosilver on nanostructured sili-ca: Antibacterial activity and Ag surface area. Chem. Eng. J. 2011, 170, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-X.; Geng, L.; Ding, S.; Xu, S.-K. Facile Synthesis of Silver Nanoparticles and their Antimicrobial Activity against Several Representative Microbial Species. In Proceedings of the 2012 International Conference on Biomedical Engineering and Biotechnology, Macau, Macao, 28–30 May 2012; Institute of Electrical and Electronics Engineers (IEEE): Washington, DC, USA, 2012; pp. 287–290. [Google Scholar]
- Tormena, R.P.; Rosa, E.V.; Mota, B.D.; Chaker, J.A.; Fagg, C.W.; Freire, D.O.; Martins, P.M.; da Silva, I.C.; Sousa, M.H. Evaluation of the antimicrobial activity of silver nanoparticles obtained by microwave-assisted green synthesis using Handroanthus impe-tiginosus (Mart. ex DC.) Mattos underbark extract. RSC Adv. 2020, 10, 20676–20681. [Google Scholar] [CrossRef]
- Mandal, D.; Dash, S.K.; Das, B.; Chattopadhyay, S.; Ghosh, T.; Das, D.; Roy, S. Bio-fabricated silver nanoparticles preferentially targets Gram positive depending on cell surface charge. Biomed. Pharmacother. 2016, 83, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The effect of charge at the surface of silver nanoparticles on antimicrobial activity against gram-positive and gram-negative bacteria: A preliminary study. J. Nanomater. 2015, 16, 53. [Google Scholar] [CrossRef]
- Mohanta, Y.K.; Biswas, K.; Jena, S.K.; Hashem, A.; Allah, E.F.A.; Mohanta, T.K. Anti-biofilm and Antibacterial Activities of Silver Nanoparticles Synthesized by the Reducing Activity of Phytoconstituents Present in the Indian Medicinal Plants. Front. Microbiol. 2020, 11, 1143. [Google Scholar] [CrossRef] [PubMed]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; González Bardanca, M.; Ambroa, A.; Tomás, M. Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiotics 2020, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Cucarella, C.; Tormo-Mas, M.Á.; Úbeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Amorena, B.; Lasa, I.; Penadés, J.R. Role of Biofilm-Associated Protein Bap in the Pathogenesis of Bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Taganna, J.C.; Quanico, J.P.; Perono, R.M.G.; Amor, E.C.; Rivera, W.L. Tannin-rich fraction from Terminalia catappa inhibits quorum sensing (QS) in Chromobacterium violaceum and the QS-controlled biofilm maturation and LasA staphylolytic activity in Pseudomonas aeruginosa. J. Ethnopharmacol. 2011, 134, 865–871. [Google Scholar] [CrossRef]
- Machado-Gonçalves, L.; Tavares-Santos, A.; Santos-Costa, F.; Soares-Diniz, R.; Carvalho-Galvão, L.C.-D.; Sousa, E.M.-D.; Beninni-Paschoal, M.-A. Effects of Terminalia catappa Linn. Extract on Candida albicans biofilms developed on denture acrylic resin discs. J. Clin. Exp. Dent. 2018, 10, e642–e647. [Google Scholar] [CrossRef]




| Sample | Band Wave number/cm−1 | Plausible Bond Assignments | Plausible TCE Bio-Active Molecules on NPs Surface | References |
|---|---|---|---|---|
| TCE-Ag-3 | 1570 | C=C of ring B vibrations | Morin (C15H10O7) | [43,44,47] |
| 1372 | C=O of ring A vibrations | Morin (C15H10O7) | [43,44,47] | |
| 946 | cyclic ring bend, cyclic CH, COH bend | Luteolin (C15H10O6) | [44,45,48,50] | |
| 622 | δ (CCO) and δ (COC) vibrations | Chrysin (C15H10O4) | [44] | |
| 465 | Cyclic CH, COH bend, CO(H) Ag bend, and CO2 twist | Luteolin (C15H10O6) | [44,45,48] | |
| 213 | Ag–O vibrations | Unassigned | [50] |
| Sr. No. | Bacterial Strains | Zone of Inhibition (mm) | ||
|---|---|---|---|---|
| TCE-Ag-1 | TCE-Ag-2 | TCE-Ag-3 | ||
| 1 | MR-S. aureus | 11.07 ± 0.25 | 12.13 ± 0.35 | 14 ± 0.15 |
| 2 | MDR-P. aeruginosa | 19.03 ± 0.21 | 20.20 ± 0.20 | 21.13 ± 0.15 |
| 3 | C. albicans | 14.17 ± 0.06 | 15.03 ± 0.15 | 17.03 ± 0.21 |
| Strains | TCE-Ag-1 | TCE-Ag-2 | TCE-Ag-3 | |||
|---|---|---|---|---|---|---|
| MIC | MBC/MFC * | MIC | MBC/MFC * | MIC | MBC/MFC * | |
| MRSA | 31.08 ± 1.01 | 124.33 ± 4.04 | 7.77 ± 0.25 | 31.08 ± 1.01 | 7.77 ± 0.25 | 31.08 ± 1.01 |
| MDR-P. aeruginosa | 7.77 ± 0.25 | 15.54 ± 0.50 | 3.88 ± 0.13 | 7.77 ± 0.25 | 3.88 ± 0.13 | 7.77 ± 0.25 |
| C. albicans * | 124.33 ± 4.04 | 248.67 ± 8.08 * | 62.17 ± 2.02 | 248.67 ± 8.08 * | 62.17 ± 2.02 | 124.33 ± 4.04 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ansari, M.A.; Kalam, A.; Al-Sehemi, A.G.; Alomary, M.N.; AlYahya, S.; Aziz, M.K.; Srivastava, S.; Alghamdi, S.; Akhtar, S.; Almalki, H.D.; et al. Counteraction of Biofilm Formation and Antimicrobial Potential of Terminalia catappa Functionalized Silver Nanoparticles against Candida albicans and Multidrug-Resistant Gram-Negative and Gram-Positive Bacteria. Antibiotics 2021, 10, 725. https://doi.org/10.3390/antibiotics10060725
Ansari MA, Kalam A, Al-Sehemi AG, Alomary MN, AlYahya S, Aziz MK, Srivastava S, Alghamdi S, Akhtar S, Almalki HD, et al. Counteraction of Biofilm Formation and Antimicrobial Potential of Terminalia catappa Functionalized Silver Nanoparticles against Candida albicans and Multidrug-Resistant Gram-Negative and Gram-Positive Bacteria. Antibiotics. 2021; 10(6):725. https://doi.org/10.3390/antibiotics10060725
Chicago/Turabian StyleAnsari, Mohammad Azam, Abul Kalam, Abdullah G. Al-Sehemi, Mohammad N. Alomary, Sami AlYahya, Mohammad Kashif Aziz, Shekhar Srivastava, Saad Alghamdi, Sultan Akhtar, Hussain D. Almalki, and et al. 2021. "Counteraction of Biofilm Formation and Antimicrobial Potential of Terminalia catappa Functionalized Silver Nanoparticles against Candida albicans and Multidrug-Resistant Gram-Negative and Gram-Positive Bacteria" Antibiotics 10, no. 6: 725. https://doi.org/10.3390/antibiotics10060725
APA StyleAnsari, M. A., Kalam, A., Al-Sehemi, A. G., Alomary, M. N., AlYahya, S., Aziz, M. K., Srivastava, S., Alghamdi, S., Akhtar, S., Almalki, H. D., Adil, S. F., Khan, M., & Hatshan, M. R. (2021). Counteraction of Biofilm Formation and Antimicrobial Potential of Terminalia catappa Functionalized Silver Nanoparticles against Candida albicans and Multidrug-Resistant Gram-Negative and Gram-Positive Bacteria. Antibiotics, 10(6), 725. https://doi.org/10.3390/antibiotics10060725











