Abstract
Heterocapsa bohaiensis is an emerging harmful dinoflagellate increasingly reported from coastal regions of the Pacific. However, an available molecular assay offering rapid and sensitive detection is still lacking. This study developed a SYBR Green real-time quantitative PCR (qPCR) assay for the identification and quantification of H. bohaiensis. Species-specific primers (F: 5′-CCATCGAACCAGAACTCCGT-3′; R: 5′-AGTGTAGTGCACCGCATGTC-3′) were designed and the assay was optimized and evaluated using laboratory cultures for specificity, sensitivity, and quantitative performance. Primer screening and melt-curve analysis confirmed that the selected primer pair produced a single, specific amplification peak for H. bohaiensis, with no cross-reactivity observed in non-target species (Chlorella pyrenoidosa, Phaeocystis globosa, Skeletonema costatum, Alexandrium tamarense) or mixed algal communities. The standard curve displayed strong linearity (R2 = 0.9868) and a high amplification efficiency (102.5%). The limit of detection (LOD) was approximately 2–3 cells per reaction, as determined from 24 replicates of 5-cell equivalents and verified at ~2.7-cell equivalents. This sensitivity was comparable to or exceeded that reported for assays targeting other HABs forming dinoflagellates. Quantitative results derived from the qPCR assay closely matched microscopic cell counts, with a relative error of 10.79%, falling within the acceptable threshold for phytoplankton surveys. In summary, this study established and validates a species-specific qPCR assay for H. bohaiensis under controlled laboratory conditions. The method shows strong potential for incorporation into HAB monitoring programs, early-warning systems, and future ecological investigations of this emerging species.
1. Introduction
Harmful algal blooms (HABs) have become an increasingly serious environmental and socio-economic issue in marine coastal ecosystems worldwide [1]. Their rising frequency, intensity, and geographic expansion pose substantial threats to marine biodiversity, aquaculture production, public health, and coastal economies [2]. Bloom-forming microalgae can produce potent toxins or generate excessive biomass in a way that leads to hypoxia, fish mortality, and large-scale ecological disturbances [3,4]. In the Bohai Sea of China, dinoflagellates are one of the primary drivers of HABs [5], posing risks to seafood safety, degrading marine environments, and significantly threating public health. These events also threaten the sustainable development of coastal tourism and may interfere with the safe operation of major coastal infrastructures [6]. Given these far-reaching impacts, the development of rapid, sensitive, and accurate detection methods for HAB species is crucial for effective monitoring, risk assessment, and early warning systems.
Heterocapsa bohaiensis is a newly identified dinoflagellate species from the Bohai Sea [7], causing significant economic losses due to crab stress and mortality [8]. Since its discovery, it has been increasingly documented in several regions across the Pacific, including Malaysia [9], New Caledonia [9], and Mexico [10]. It is characterized by distinct cell morphology (14.4 ± 1.6 μm, ellipsoid shape, and a unique thecal plate arrangement) and a significant genetic divergence from other Heterocapsa species, as confirmed by ITS and LSU rDNA sequencing [7]. The species produces hemolytic toxins, which are involved in its toxicity to Brachionus plicatilis, suggesting a potential ecological impact on zooplankton communities [11]. In addition, H. bohaiensis exhibits a competitive advantage over co-occurring phytoplankton species when N:P ratios exceed the Redfield ratio, indicating that nutrient imbalance may promote its expansion [8]. Laboratory culture studies further revealed that temperature significantly affects its growth, with an optimal growth range between 25 and 30 °C [12]. These findings suggest that H. bohaiensis may be more widely distributed than previously recognized and may represent an emerging harmful algal species in warm and temperate waters. Despite its ecological relevance and growing geographic footprint, research on this species remains limited, and fundamental aspects of its biology, distribution, and bloom dynamics are still poorly understood, particularly the lack of rapid and sensitive methods for early detection of H. bohaiensis blooms.
Routine monitoring of phytoplankton communities and early detection of HAB-forming species have traditionally relied on light microscopy [13]. Although microscopy-based methods remain indispensable, they require considerable taxonomic expertise and are time-consuming, especially when distinguishing morphologically similar dinoflagellates [14]. So microscopy lacks the sensitivity needed to detect low cell densities during the early stages of bloom formation, which is a critical period for effective environmental management and mitigation [15]. Over the past two decades, molecular techniques, particularly real-time quantitative PCR (qPCR), have greatly advanced HAB monitoring [16,17]. qPCR offers several advantages over conventional microscopy, including higher sensitivity, better specificity, rapid processing speed, and the ability to provide accurate quantitative information. SYBR Green qPCR assays have been successfully developed for numerous harmful algal taxa, such as Alexandrium spp. [17], Prorocentrum donghaiense [18], and Karenia spp. [19]. In real-time PCR, the cycle threshold (Ct) value corresponds to the number of cycles required for the fluorescence signal to exceed a predetermined threshold. This value is inversely proportional to the logarithm of the initial template copy number, thereby providing a reliable indicator of the starting DNA concentration in the sample. By establishing a standard curve, the Ct value can be converted into the absolute initial copy number of the target DNA. Given the direct correlation between DNA copy number and algal cell density, this approach allows for accurate estimation of algal abundance in environmental samples [20]. These assays have proven effective in both laboratory cultures and environmental samples, enabling early detection and improving the temporal resolution of HAB monitoring programs. However, despite the growing importance of H. bohaiensis, no qPCR assay or species-specific primers have been reported to date. This gap severely limits the ability of monitoring agencies and researchers to detect, quantify, and track this species in natural waters.
To address this need, this study aimed to develop a rapid, sensitive, and specific SYBR Green qPCR assay for the detection and quantification of H. bohaiensis. The objectives of this work were to (1) design and screen species-specific primers targeting H. bohaiensis; (2) establish and optimize a qPCR amplification system; (3) evaluate primer specificity using both target and non-target algal species; (4) construct a standard curve and determine amplification efficiency and detection limits; and (5) assess the applicability of the assay to mixed algal samples, simulating conditions typical of natural plankton communities. By filling the current methodological gap, this study provides the first molecular diagnostic tool for H. bohaiensis and lays a foundation for future ecological investigations, bloom monitoring, and potential early warning strategies.
2. Materials and Methods
2.1. Algal Cultures and Density Measurements
The dinoflagellate H. bohaiensis was obtained from the Guanghe Crab Company, Panjin, China. The strains were isolated from aquaculture ponds in Liaodong Bay and purified to sterile conditions using the capillary pipette method. In addition to H. bohaiensis, the following algal species were cultured for real-time PCR assay validation: Chlorella pyrenoidosa (green algae, marine strain), coexisting and isolated with H. bohaiensis in the same aquaculture ponds, and common HAB species in China’s coastal waters, including Phaeocystis globosa, Skeletonema costatum, and Alexandrium tamarense. Axenic cultures of P. globosa were obtained from the Guangxi Key Laboratory of Marine Environmental Change and Disaster Research, while axenic cultures of S. costatum and A. tamarense were sourced from the Marine Pollution Ecological Chemistry Laboratory of Ocean University of China. These strains are well-established reference species that are commonly used in laboratory studies.
Algal cultures were incubated in f/2 medium [21], prepared in nature seawater collected from the coastal area of Liaodong Bay with a salinity of 27~30 (filtered through 0.45 µm cellulose acetate Millipore membrane filters). All media and flasks were autoclaved at 121 °C for 20 min before assembly and operation. Cultures were maintained at 20 ± 1 °C with a 12 h/12 h light/dark cycle, and a 72 ± 5 μmol m−2 s−1 photon flux density was provided by a cool white fluorescent light (GXZ illumination incubator, Ningbo, China, Southeast Instrument Co., Ltd.). All cultures were shaken manually every day at set times to prevent an oxygen deficit and algal cells from adhering. To ensure axenic conditions, cultures were periodically checked microscopically for bacterial contamination and subcultured every 2–3 weeks by transferring exponentially growing cells into fresh medium.
Cell densities were measured under a microscope (N-300M, Ningbo, China) using a hemocytometer (MARIENFELD, Lauda-Königshofen, Germany). 1.5 mL of the algal solution was shaken well and taken into a 1.5 mL centrifuge tube, two drops of Lugol’s reagent were added to fix it, and the sample was mixed well on a homogenizer. Each sample was counted six times to derive an average value. Prior to DNA extraction, H. bohaiensis cells in the exponential growth phase were collected by centrifugation at 4000× g for 10 min and washed twice with sterile seawater to remove residual medium. Aliquots containing approximately 104–105 cells mL−1 were used for subsequent molecular analyses.
2.2. Primer Target Identification and Design for H. bohaiensis
A reference rDNA fragment from H. circularisquama (strain aihc559-kt; ITS1–5.8S rRNA–ITS2 with partial 28S; 1556 bp; NCBI) was aligned to the H. bohaiensis Trinity-assembled transcriptome (provided in the Supplementary Information S1) to delineate conserved segments and species-divergent regions. Sequence alignment and variant calling were implemented in Python (version 3.11; custom scripts; Smith–Waterman local alignment and sliding-window scans, and python codes were provided in https://doi.org/10.5281/zenodo.17783931). Variable positions within the ITS region were then exploited to design H. bohaiensis-specific primers using Primer-BLAST (BLAST: Basic Local Alignment Search Tool). Design criteria were (i) primer length of 15–30 nt with an expected amplicon size of 100–300 bp to balance specificity and extension efficiency; (ii) GC content of 40–60% with forward/reverse primers matched for similar melting temperatures (Tm 55–65 °C, ideally ~60 °C, ΔTm ≤ 4 °C); (iii) avoidance of adenine at the extreme 3′ end to reduce extension from 3′-terminal mismatches; and (iv) minimization of self- and cross-complementarity (no ≥4 contiguous complementary bases) to prevent secondary structures and primer–dimer formation that could reduce PCR efficiency and quantitative accuracy.
The selected primers were synthesized by Shanghai, China Bioengineering Co., Ltd. (Shanghai, China) and supplied as lyophilized powders requiring reconstitution prior to use. Because the dried oligonucleotides tend to adhere to the inner wall of the tube as a thin film, brief centrifugation before opening was performed to prevent material loss. After centrifugation, the tubes were carefully opened, and each primer was dissolved in the manufacturer-recommended volume of double-distilled water as indicated on the product label. The reconstituted stock concentration was 100 μM, which was subsequently diluted to a 10 μM working solution for use in real-time quantitative PCR assays. Experiments were performed with a sufficiently large number of replicates to ensure reliability (Figure 1).
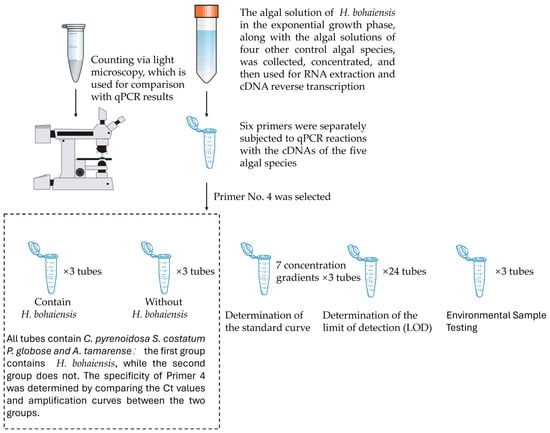
Figure 1.
Schematic workflow of the experiment. Some graphical elements were adapted from SciDraw (https://scidraw.io, accessed on 1 November 2025).
2.3. RNA Extraction and cDNA Synthesis
Centrifuge tubes and pipette tips used in RNA experiments were soaked in 0.1% diethylpyrocarbonate (DEPC) solution for 24 h, autoclaved at 120 °C for 30 min to remove residual DEPC, and dried in an oven. Algal samples were loaded into centrifuge tubes and centrifuged at 12,000 rpm for 10 min. The supernatant was discarded, and the algal pellet was transferred to a 5 mL tube and frozen for later use. RNA was extracted from 100 to 200 mg of algal paste using a Plant RNA Extraction Kit (Yeasen Biotechnology, Shanghai, China). Total RNA was used without rRNA depletion, and the extracted RNA was quantified for concentration and assessed for quality using NanoDrop 2000 (Thermo Fisher Scientific, Shanghai, China). Absorbances at A260 and A280 were measured; if the A260/A280 ratio ranged between 1.8 and 2.1, the RNA was stored in a −20 °C refrigerator for subsequent use. Otherwise, further purification or re-extraction was required. The extracted RNA was reverse-transcribed using a cDNA Synthesis Kit (BBI Life Sciences, Shanghai, China).
2.4. qPCR Assay Development
Real-time quantitative PCR was performed using SYBR Green fluorescence, with three replicates for each primer pair. A ThermoFisher 7500 real-time PCR instrument was used for reactions. The reaction mixture contained 10 μL of 2× Q5 SYBR qPCR Master Mix, 0.5 μL of each forward and reverse primer (10 μmol/L), 1.5–2 μL of cDNA template, and double-distilled water to a final volume of 20 μL. The thermal cycling conditions were as follows: initial denaturation at 95 °C for 3 min; 41 cycles of denaturation at 95 °C for 10 s, and annealing at 60 °C for 30 s.
2.5. Primers’ Specificity Validation
The specificity of Primer No. 4 (designed in Section 2.2) was evaluated by qPCR using cDNA from H. bohaiensis, C. pyrenoidosa, P. globosa, S. costatum, and A. tamarense. Two validation strategies were employed: (i) individual reactions using Primer No. 4 with each algal cDNA separately, and (ii) a mixed reaction using Primer No. 4 with pooled cDNA from all five species. All assays were performed in triplicate to ensure reproducibility.
The mixed algal community used for specificity testing was composed of H. bohaiensis and four other algal species that are commonly found in coastal environments. These species were chosen to represent a range of common HAB organisms and co-occurring algae in China’s coastal waters. The community included: H. bohaiensis (the target species); C. pyrenoidosa (green algae, marine strain), which coexists with H. bohaiensis in aquaculture ponds; P. globosa, a common HAB species in coastal waters; S. costatum, a common diatom found in marine environments; and A. tamarense, a well-known harmful dinoflagellate species. The mixed algal community was constructed by combining H. bohaiensis, C. pyrenoidosa, S. costatum, P. globosa, and A. tamarense at a defined volume ratio of 1:3:3:3:3, creating a controlled laboratory matrix for specificity testing. This mixture was used to evaluate the potential for cross-reactivity and performance of the H. bohaiensis specific qPCR assay under conditions resembling natural, mixed algal populations.
2.6. Standard Curve Preparation
A 1 L culture of H. bohaiensis was centrifuged, and total RNA was extracted and reverse-transcribed into cDNA. The resulting cDNA was serially diluted in 10-fold gradients (1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 10, and 1 cell equivalents) and subjected to qPCR using Primer No. 4, with three replicates for each dilution. Following amplification, a standard curve was generated by linear regression analysis, plotting the logarithm (base 10) of the microscopically determined cell number on the x-axis against the corresponding mean Ct value on the y-axis. An amplification efficiency of 90–110% and a correlation coefficient (R2) greater than 0.98 were considered indicative of a reliable quantitative assay.
2.7. Determination of Limit of Detection and Method Reliability
The concentration of H. bohaiensis cDNA was equivalent to an algal density of 5 cells, and qPCR was performed using primer No. 4 with 24 parallel samples. The limit of detection (LOD) was calculated as:
where SD is the standard deviation of the Ct values measured from the 5-cell equivalents samples.
To assess the reliability of the qPCR quantification method, H. bohaiensis cultures grown in the laboratory were diluted to obtain a 1 L suspension. A 1.5 mL aliquot was counted microscopically, yielding a cell density of 9 × 104 cells/mL. Total RNA was extracted from the remaining culture, and its concentration and purity were determined using a NanoDrop spectrophotometer. The RNA was then reverse-transcribed into cDNA using a digital thermostatic water bath. The resulting cDNA served as the template for qPCR using Primer No. 4, with three technical replicates performed for each reaction.
Using the Ct value from the qPCR assay, the predetermined standard curve was applied to quantify the population of H. bohaiensis algae in a laboratory-cultured sample. Error analysis was performed by comparing these results with the cell counts obtained via microscope. The method was considered reliable as long as the error remained below 20% of the allowable margin for error in the National Standards of the People’s Republic of China [22].
2.8. Spiked Environmental Sample Testing
Detect the algal density of H. bohaiensis cultured in the laboratory using an optical microscope. Based on the obtained algal density, add 0.1 mL of the original algal solution to 2 L of seawater collected from the Bohai Sea to prepare a spiked environmental sample. Centrifuge and concentrate the sample to obtain a small amount of algal sludge, which is used for qPCR detection. Finally, compare the values obtained from qPCR with those from the optical microscope detection.
3. Results
3.1. Primer Specificity Validation for H. bohaiensis
The primer sequences used in this study are listed in Table 1. Primers 1–3 are dinoflagellate (including Heterocapsa triquetra) sequences reported in previous studies [23,24], and were used to amplify relatively long fragments of the ITS–LSU rDNA region for sequencing and phylogenetic analyses. These primers are universal rDNA primers and were not designed to target specific ITS subregions. In contrast, primer pairs 4–6 were specifically designed for quantitative PCR detection of H. bohaiensis according to the methods in Section 2.2. These primers amplify short fragments (134–163 bp) located within the ITS1 and ITS2 regions, which exhibit high interspecific variability among Heterocapsa species and therefore provide high specificity for species-level detection.

Table 1.
Primer sets used for gene amplification and species-specific detection of H. bohaiensis.
Amplification was considered successful when the Ct value was below 35 and the melting curve exhibited a single sharp peak at a consistent Tm. Reactions with Ct > 40 or multiple melting peaks were regarded as negative or non-specific.
The specificity of the qPCR assay developed was evaluated using cDNA from both target and non-target algal species. In reactions with H. bohaiensis cDNA, primers No. 1, 4, 5, and 6 showed potential specificity based on the average Ct values.No.2 did not exhibit specific amplification for H. bohaiensis, and Primer 3 failed to achieve amplification for all five algal species (Table 2 and Figure 2). Melting curves further revealed that primers No. 1 and 4 produced single, well-overlapped peaks without any secondary signals, with average melting temperatures (Tm) of 89.69 °C and 88.84 °C, respectively, confirming specific amplification of H. bohaiensis cDNA. In contrast, primer No. 5 generated multiple melting peaks, and primer No. 6 displayed double peaks, indicating non-specific amplification. Therefore, primers No. 5 and No. 6 were excluded from subsequent analyses.

Table 2.
Ct results of qPCR for H. bohaiensis, C. pyrenoidosa, S. costatum, P. globose and A. tamarense. The numbers and colors in the table correspond to the primer numbers and their respective amplification curves, respectively.
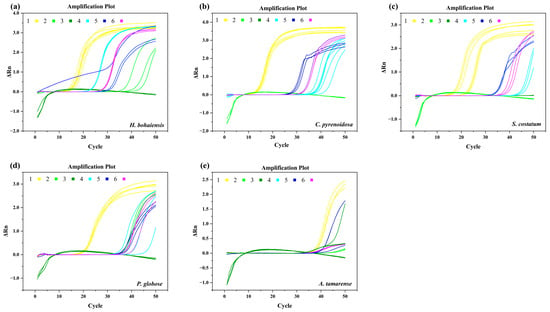
Figure 2.
Amplification curves of cDNA samples of (a) H. bohaiensis (b) C. pyrenoidosa (c) S. costatum (d) P. globose (e) A. tamarense (6 kinds of primers).
In reactions with non-target algal cDNA templates, several primers showed weak or non-specific amplification. For C. pyrenoidosa (Table 2), primers No. 1 and No. 6 produced amplification signals with single, well-defined melting peaks (Tm = 89.50 °C and 89.40 °C, respectively), indicating specific amplification of Chlorella cDNA. Primer No. 5 also yielded a single melting peak (Tm = 92.49 °C), but the higher Tm value suggested non-specific amplification.
For S. costatum, P. globose, and A. tamarense, qPCR reactions were performed with all six primer pairs. No consistent amplification or specific melting peaks corresponding to H. bohaiensis were observed. Only primer No. 1 produced a stable single peak (Tm = 88.62 °C) in S. costatum, confirming non-specific amplification of its cDNA. Primer No. 6 occasionally showed weak or multiple melting peaks in A. tamarense, further indicating non-specific binding.
Overall, these results demonstrate that among all tested primers, only primers No. 4 showed strong and specific amplification exclusively for H. bohaiensis, whereas other primers exhibited varying degrees of non-specific amplification with non-target algal templates.
3.2. Primer Performance in Detecting H. bohaiensis in Mixed Algal Samples
In mixed algal samples containing H. bohaiensis, qPCR using Primer No. 4 produced clear amplification signals (Figure 3a), with a mean Ct value of 25.07 across eight replicates (Table 3). The melting curves showed high consistency among replicates, with Tm values ranging from 88.20 °C to 88.71 °C (mean = 88.52 °C; ΔTm = 0.52 °C). The uniform melting profiles and narrow Tm range indicate stable and specific amplification of H. bohaiensis targets within the mixed algal matrix, confirming that Primer No. 4 can accurately detect the target species even in the presence of other algal DNAs.
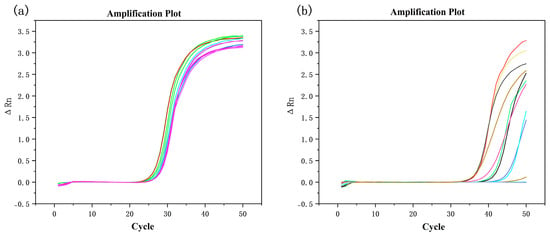
Figure 3.
Amplification curves of mixed algal cDNA (a) containing H. bohaiensis (b) without H. bohaiensis using Primer 4.

Table 3.
Values of qPCR for mixed algal cDNA using Primer 4 (n = 8 replicates).
In contrast, when qPCR was performed on mixed algal samples lacking H. bohaiensis (Figure 3b), no amplification signal was detected within 40 cycles using Primer No. 4. The amplification plots showed no exponential phase, and the melting curve displayed only background noise without a defined peak. These findings confirm that Primer No. 4 does not cross-react with non-target algal DNA in complex mixtures, further supporting its high specificity and reliability for the detection of H. bohaiensis in environmental or multi-species samples.
3.3. Standard Curve and Quantification
The SYBR Green real-time qPCR was performed on H. bohaiensis cDNA across seven tenfold serial dilutions using Primer No. 4, with three replicates per dilution. The melting curves (Figure 4) showed single, well-overlapped peaks, confirming specific amplification of H. bohaiensis cDNA.
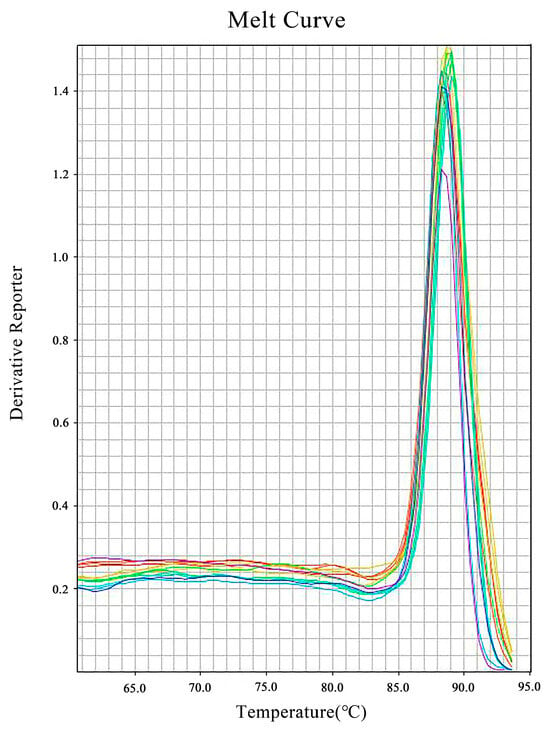
Figure 4.
Solubilization curves of 7 dilution gradients of cDNA samples (No.4 primer).
A standard curve was generated by plotting the logarithm (base 10) of algal cell number against the corresponding mean Ct values (Table 4 and Figure 5). The resulting regression equation was y = −3.2623x + 44.674 with an R2 value of 0.9868, corresponding to an amplification efficiency of 102.5%. These results demonstrate that the qPCR assay exhibits excellent linearity and amplification performance, providing a reliable basis for quantitative detection of H. bohaiensis.

Table 4.
Standard curve PCR test results.
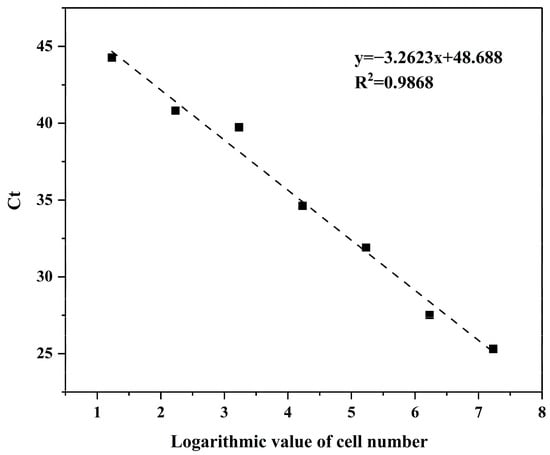
Figure 5.
H. bohaiensis qPCR counting standard curve (p < 0.05).
3.4. Limits of Detection and Method Reliability
To determine the LOD, qPCR reactions were performed using cDNA corresponding to an algal density of 5 cells, with 24 technical replicates. The Ct values ranged from 35.69 to 45.17, yielding a mean Ct of 40.61 and a standard deviation (SD) of 2.53. Using the slope of −3.2623 from the standard curve, the calculated LOD was approximately 2.3 cells, indicating that the developed qPCR assay can reliably detect as few as 2–3 H. bohaiensis cells in a reaction. To further validate this result, the assay was tested using cDNA equivalent to 2.7 cells, which produced clear, reproducible amplification curves consistent with the standard-curve predictions (Figure 6). The relatively low standard deviation among replicates further supports the assay’s high reproducibility and sensitivity.
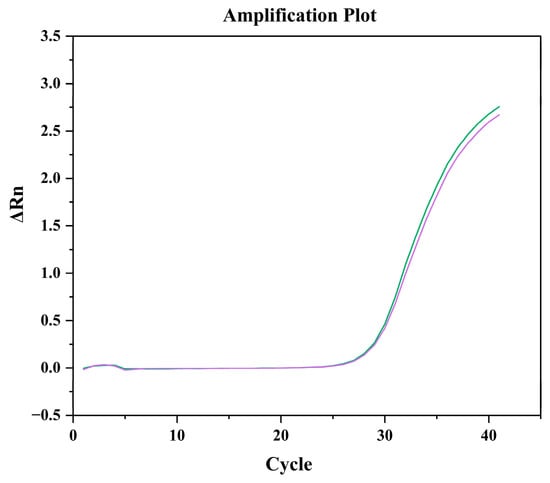
Figure 6.
Amplification curves of cDNA samples of H. bohaiensis (2.7 cells/L).
The qPCR reactions produced consistent amplification signals with Ct values of 28.48, 28.87, and 27.92 and Tm of 88.18, 88.34, and 88.50 °C (mean Tm = 88.34 °C), confirming specific amplification of H. bohaiensis. By substituting the Ct values into the standard curve equation, the calculated algal densities were 9.23 × 104, 7.01 × 104, and 1.37 × 105 cells/mL, with a mean value of 9.97 × 104 cells/mL. Compared with the microscopic count of 9 × 104 cells/mL, the deviation was 10.79%, which is well within the ±20% allowable error specified for plankton surveys (National Standards of the People’s Republic of China [22]). These results demonstrate that the developed qPCR assay provides accurate and reliable quantification of H. bohaiensis in culture samples.
3.5. Verification of Method Reliability Using Actual Seawater Samples
The density of the original algal solution determined by the microscopic counting method was 1.30 × 106 cells/mL. After dilution with natural seawater, the target algal density was adjusted to 6.5 × 104 cells/L. This algal density is similar to that in natural water bodies [25]. The large-volume natural samples were concentrated, and then RNA was extracted for qPCR detection. The qPCR reactions produced consistent amplification signals with Ct values of 42.78 and 42.37 and Tm of 92.70 and 89.96 (Figure 7). By substituting the Ct values into the standard curve equation, the calculated algal densities were 6.46 × 104 cells/L and 8.71 × 104 cells/L, with a mean value of 7.59 × 104 cells/L. Compared with the result of microscopic count, the deviation was 16.77%. It is demonstrated that this method can be applied to the detection of samples in natural seawater.
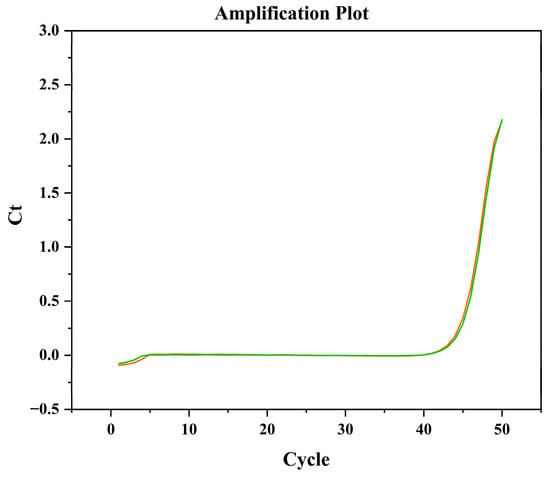
Figure 7.
Amplification curves of cDNA samples of H. bohaiensis in natural seawater.
4. Discussion
4.1. Comparison to Existing Studies
In this experiment, specific primers for H. bohaiensis (F: 5′-CCATCGAACCAGAACTCCGT-3′; R: 5′-AGTGTAGTGCACCGCATGTC-3′) were designed, and a SYBR Green-based real-time quantitative PCR assay was developed for the rapid detection and quantification of H. bohaiensis. Based on the literature reviewed to date, this work provides the first report of specific primer development for H. bohaiensis.
The results of this study demonstrate that the qPCR assay developed for H. bohaiensis represents a substantial improvement over traditional microscopy-based identification methods. Conventional morphological identification, though foundational for phytoplankton taxonomy, remains limited by its low sensitivity, time-consuming nature, and reliance on expert judgment [5,26]. In contrast, the qPCR assay provides rapid, specific, and quantitative detection that can be completed within several hours from RNA extraction to final quantification [27,28]. Compared with microscopic enumeration, which typically requires more than a day for accurate counting, the developed assay enables reliable quantification with high precision. The strong linear correlation between Ct values and algal cell numbers (R2 = 0.9868) and the amplification efficiency exceeding 100% indicate that the method performs comparably to or better than previously developed qPCR assays for other harmful algal species such as Karenia mikimotoi and Prorocentrum minimum [18,29].
In addition to the microscopy method, we also compared the qPCR assay with other alternative detection approaches, including loop-mediated isothermal amplification (LAMP), DNA metabarcoding, and direct toxin analysis (Table 5). Comparison of available detection methods indicates that qPCR provides a suitable balance of sensitivity, specificity, and quantitative performance for targeted species monitoring. Other approaches are valuable for toxin confirmation or community-level assessments but are less optimal for routine quantification of a predefined species. Accordingly, qPCR is well suited for the detection of H. bohaiensis in the Bohai Sea.

Table 5.
Comparison of Four Methods: qPCR, LAMP, DNA Metabarcoding, and Direct Toxin Analysis (LC-MS/MS).
The qPCR assay achieved a calculated limit of detection (LOD) of approximately 2–3 cell equivalents per reaction, consistent with or surpassing the sensitivity reported in earlier molecular detection studies [36,37]. This value was derived from replicate (n = 24) reactions using 5-cell equivalents and verified through additional tests at 2.7-cell concentrations, which produced consistent amplification curves and stable melting peaks. When compared with previously reported assays for other harmful dinoflagellates, the sensitivity achieved in this study is comparable to or slightly better than most existing methods. For example, Galluzzi L et al. [5] reported a detection limit of about 10 cells per reaction for Alexandrium minutum using a SYBR Green-based qPCR assay, while Yuan J et al. [26] achieved an LOD of 3–10 cells per reaction for Prorocentrum donghaiense employing a TaqMan probe system. The LOD obtained for H. bohaiensis (~2–3 cells per reaction) thus falls well within this high-sensitivity range, underscoring the reliability and efficiency of the developed method.
In addition to its analytical performance, the developed qPCR assay exhibits excellent specificity and applicability to complex algal community samples. The combination of primer specificity, high amplification efficiency (102.5%), and stable melting temperature profiles (~88.5 °C) ensures high quantitative accuracy while minimizing the risk of false positives caused by co-occurring phytoplankton. Consistent with earlier findings that qPCR can reliably quantify target taxa within mixed phytoplankton assemblages [38], our results confirmed that H. bohaiensis can be specifically and accurately detected even in the presence of other algal species. This capability is particularly valuable for field monitoring, where mixed blooms and low target abundance frequently complicate microscopic identification. Moreover, the high sensitivity of this method provides a distinct advantage for early warning and environmental surveillance, allowing the detection of H. bohaiensis at the initial stages of bloom development when cell densities are still low.
Overall, the developed qPCR assay offers a rapid, sensitive, and highly specific molecular approach for the detection and quantification of H. bohaiensis. By significantly improving efficiency and reducing subjectivity compared with microscopy, this method provides a valuable tool for routine monitoring of H. bohaiensis harmful blooms in coastal ecosystems.
4.2. Advantages and Limitations of This Study
The qPCR assay developed here shows high sensitivity (LOD ~2–3 cells), strong specificity, and reliable quantification, and its results were consistent with microscopic counts. Despite these strengths, the study has certain limitations. Since H. bohaiensis has not been reported in field surveys in recent years, the reliability of the detection method can only be verified by simulating actual environmental samples through spiking experiments. A potential limitation of the present assay is the intragenomic variability of ITS regions, which has been reported in many eukaryotic microorganisms, including dinoflagellates [39]. Sequence heterogeneity among ITS copies within a single genome may affect primer binding efficiency and introduce bias in qPCR quantification. Therefore, ITS-based qPCR results should be interpreted with caution, particularly when absolute quantification is required [40]. Furthermore, this detection method can only quantitatively detect H. bohaiensis at the genetic level, but cannot distinguish the species of H. bohaiensis or determine whether it is a toxic strain; in subsequent work, methods such LC-MS can be used for further toxin detection experiments.
4.3. Applications in HABs Monitoring
Given its capacity to form harmful blooms and disrupt aquaculture systems [11], H. bohaiensis represents an emerging concern for phytoplankton monitoring programs in the Bohai Sea and other Pacific coastal regions. Despite its ecological and economic relevance, specific and sensitive detection tools for H. bohaiensis have been lacking. The present study demonstrates that real-time quantitative PCR can provide a rapid, sensitive, and species-specific approach for both qualitative and quantitative detection of H. bohaiensis. The identification of a highly specific primer pair offers a practical molecular tool for early warning, routine monitoring, and future ecological studies of this species. As H. bohaiensis continues to expand its geographic presence, the qPCR assay developed here will support more accurate surveillance of emerging HAB events and contribute to improved management of coastal and aquaculture ecosystems. Collectively, these findings highlight the value of the developed qPCR assay as a practical and reliable tool for advancing early detection, ecological assessment, and management of H. bohaiensis HABs events.
The qPCR assay, showing high sensitivity and strong specificity, makes it suitable for rapid field deployment and for integration into existing coastal monitoring programs as a species-specific diagnostic tool [41]. Because the assay is compatible with conventional qPCR platforms, it can be readily incorporated into multi-tiered surveillance frameworks that combine microscopy and molecular diagnostics [17]. In addition, the validated primer set provides a promising basis for eDNA-based applications, where early detection of rare or cryptic taxa is critical [42]. Adapting this assay to high-throughput eDNA workflows or digital PCR would further enhance its utility for tracking bloom initiation and assessing spatiotemporal dynamics. More broadly, the methodological approach used here, linking targeted sequence analysis with rigorous specificity testing, offers a transferable framework for developing molecular assays for other emerging HAB species.
5. Conclusions
In this study, a SYBR Green qPCR assay was developed and laboratory-validated for the rapid and sensitive detection of H. bohaiensis. A specific primer pair was designed and validated, demonstrating excellent amplification efficiency, a low detection limit of approximately 2–3 cells per reaction, and no cross-reactivity with other commonly occurring algal species. The assay produced reliable quantitative results that were closely agreed with microscopic counts, confirming its robustness and analytical accuracy under controlled laboratory conditions. A spiked experiment using cultured cells in natural seawater demonstrated that the qPCR method can reliably detect low-density algae in environmental matrices, with results comparable to those obtained by microscopic counting (deviation 16.77%). This study establishes a reliable laboratory framework for the molecular detection and quantification of H. bohaiensis. This assay holds promise for future environmental monitoring applications, pending further validation with field-collected samples. Continued application and refinement of this assay will support more timely and informed management of coastal and aquaculture environments threatened by H. bohaiensis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jmse14010098/s1, S1: H. bohaiensis Trinity-assembled transcriptome.
Author Contributions
Conceptualization, Y.Z.; Data curation, M.C.; Formal analysis, R.J. and M.C.; Funding acquisition, Y.Z. and J.Z.; Investigation, R.J. and M.C.; Methodology, R.J.; Supervision, J.Z.; Validation, Y.Z.; Writing—review and editing, M.C. and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. U24A20582).
Data Availability Statement
The Python code used for primer design in this study is openly available in Zenodo at: https://doi.org/10.5281/zenodo.17783931.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| HABs | Harmful algal blooms |
| DEPC | Diethylpyrocarbonate |
| LAMP | Loop-mediated isothermal amplification |
References
- Anderson, D.M.; Wells, M.L.; Trainer, V.L.; Suddleson, M.; Claridge, K.; Coyne, K.J.; Dortch, Q.; Gobler, C.J.; Heil, C.A.; Inaba, N. Controlling harmful algal blooms (HABs) in marine waters: Review of current status and future prospects. Harmful Algae 2025, 150, 102989. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Chen, J.; Anderson, D.M. Modified local sands for the mitigation of harmful algal blooms. Harmful Algae 2011, 10, 381–387. [Google Scholar] [CrossRef]
- Fernandes-Salvador, J.A.; Davidson, K.; Sourisseau, M.; Revilla, M.; Schmidt, W.; Clarke, D.; Miller, P.I.; Arce, P.; Fernández, R.; Maman, L. Current status of forecasting toxic harmful algae for the north-east Atlantic shellfish aquaculture industry. Front. Mar. Sci. 2021, 8, 666583. [Google Scholar] [CrossRef]
- Oh, J.-W.; Pushparaj, S.S.C.; Muthu, M.; Gopal, J. Review of harmful algal blooms (HABs) causing marine fish kills: Toxicity and mitigation. Plants 2023, 12, 3936. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Penna, A.; Bertozzini, E.; Vila, M.; Garcés, E.; Magnani, M. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 2004, 70, 1199–1206. [Google Scholar] [CrossRef]
- Wang, Z.; Peng, L.; Xie, C.; Wang, W.; Zhang, Y.; Xiao, L.; Tang, Y.; Yang, Y. Metabarcoding of harmful algal bloom species in sediments from four coastal areas of the southeast China. Front. Microbiol. 2022, 13, 999886. [Google Scholar] [CrossRef]
- Xiao, J.; Sun, N.; Zhang, Y.; Sun, P.; Li, Y.; Pang, M.; Li, R. Heterocapsa bohaiensis sp. nov. (Peridiniales: Dinophyceae): A novel marine dinoflagellate from the Liaodong Bay of Bohai Sea, China. Acta Oceanol. Sin. 2018, 37, 18–27. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, G.; Yang, Y.; Lu, D.; Liu, L.; Wei, Y.; Sun, N.; Su, Y. Interspecific competition between the bloom-causing dinoflagellates Hetrocapsa bohaiensis and the local species Chlorella pyrenoidosa. Mar. Environ. Res. 2023, 184, 105855. [Google Scholar] [CrossRef]
- Hanifah, A.H.; Teng, S.T.; Law, K.; Abdullah, N.; Chiba, S.U.A.; Lum, W.M.; Tillmann, U.; Lim, P.T.; Leaw, C.P. Six marine thecate Heterocapsa (Dinophyceae) from Malaysia, including the description of three novel species and their cytotoxicity potential. Harmful Algae 2022, 120, 102338. [Google Scholar] [CrossRef]
- Arana-Garcia, J.; Hernández-Becerril, D.U.; Escarcega-Bata, A.; Ruíz-García, U.; Zamudio-Resendiz, M.E. Species of the thecate dinoflagellate genus Heterocapsa (Dinophyceae; Heterocapsaceae) from the tropical Mexican Pacific, with special reference to two new records in the area and the ultrastructure of Heterocapsa borneoensis. Bot. Sci. 2025, 103, 160–175. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, T.; Qu, J.; Sun, N.; Liu, L. Toxicity and haemolytic activity of a newly described dinoflagellate, Heterocapsa bohainensis to the rotifer Brachionus plicatilis. Harmful Algae 2019, 84, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guo, H. The Preliminary Study on the Growth and Toxicity of Heterocapsa sp. and the Effects of Different Cultural Density on Calanus sinicus Growth. Master’s Thesis, Dalian Ocean University, Dalian, China, 2016. [Google Scholar]
- Holland, M.M.; Artigas, L.F.; Atkinson, A.; Best, M.; Bresnan, E.; Devlin, M.; Eerkes-Medrano, D.; Johansen, M.; Johns, D.G.; Machairopoulou, M. Mind the gap—The need to integrate novel plankton methods alongside ongoing long-term monitoring. Ocean. Coast. Manag. 2025, 262, 107542. [Google Scholar] [CrossRef]
- Esenkulova, S.; Sutherland, B.J.; Tabata, A.; Haigh, N.; Pearce, C.M.; Miller, K.M. Comparing metabarcoding and morphological approaches to identify phytoplankton taxa associated with harmful algal blooms. Facets 2020, 5, 784–811. [Google Scholar] [CrossRef]
- Park, T.-G.; Park, Y.-T.; Lee, Y. Development of a SYTO9 based real-time PCR probe for detection and quantification of toxic dinoflagellate Karlodinium veneficum (Dinophyceae) in environmental samples. Phycologia 2009, 48, 32–43. [Google Scholar] [CrossRef]
- Bowers, H.A.; Tengs, T.; Glasgow, H.B., Jr.; Burkholder, J.M.; Rublee, P.A.; Oldach, D.W. Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related dinoflagellates. Appl. Environ. Microbiol. 2000, 66, 4641–4648. [Google Scholar] [CrossRef]
- Pearson, L.A.; D’Agostino, P.M.; Neilan, B.A. Recent developments in quantitative PCR for monitoring harmful marine microalgae. Harmful Algae 2021, 108, 102096. [Google Scholar] [CrossRef]
- Yuan, J.; Mi, T.; Zhen, Y.; Yu, Z. Development of a real-time PCR method (Taqman) for rapid identification and quantification of Prorocentrum donghaiense. J. Ocean. Univ. China 2012, 11, 366–374. [Google Scholar] [CrossRef]
- Kim, S.; Cho, M.; Yoo, J.; Park, B.S. Application of a quantitative PCR to investigate the distribution and dynamics of two morphologically similar species, Karenia mikimotoi and K. papilionacea (Dinophyceae) in Korean coastal waters. Toxins 2023, 15, 469. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hustedt, I.C.N.; Confervacea, D. Studies of Marine Planktonic Diatoms. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar]
- GB/T 12763.6-2007; Specifications for Oceanographic Survey. Part 6: Marine Biological Survey. Third Institute of Oceanography: Xiamen, China, 2007.
- Zhang, Z.; Green, B.R.; Cavalier-Smith, T. Phylogeny of ultra-rapidly evolving dinoflagellate chloroplast genes: A possible common origin for sporozoan and dinoflagellate plastids. J. Mol. Evol. 2000, 51, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Ahn, G.; Park, G.-Y.; Park, D.-Y.; Jeong, O.C.; Kim, Y.-H.; Ahn, J.-Y. Microalgae direct extract reagent for Heterocapsa triquetra. Toxicol. Environ. Health Sci. 2019, 11, 73–78. [Google Scholar] [CrossRef]
- Razali, R.; Mustapa, N.; Ku Yaacob, K.K.; Yusof, F.; Teng, S.T.; Hanafiah, A.; Hii, K.; Mohd Din, M.; Gu, H.; Leaw, C.P.; et al. Diversity of Heterocapsa (Dinophyceae) and the algal bloom event in the mariculture areas of Johor Strait, Malaysia. Plankton Benthos Res. 2022, 17, 290–300. [Google Scholar] [CrossRef]
- Yuan, J.; Mi, T.; Zhen, Y.; Yu, Z. Development of a rapid detection and quantification method of Karenia mikimotoi by real-time quantitative PCR. Harmful Algae 2012, 17, 83–91. [Google Scholar] [CrossRef]
- Grivalský, T.; Střížek, A.; Přibyl, P.; Lukavský, J.; Čegan, R.; Hobza, R.; Hrouzek, P. Comparison of various approaches to detect algal culture contamination: A case study of Chlorella sp. contamination in a Phaeodactylum tricornutum culture. Appl. Microbiol. Biotechnol. 2021, 105, 5189–5200. [Google Scholar] [CrossRef]
- Lv, Y.; Zhen, Y.; Cen, J.; Lu, S.; Li, M.; Liu, Y.; Chi, X.; Yuan, J.; Wang, J. Kareniaceae in focus: A molecular survey of harmful algal dinoflagellates in the South China Sea. Harmful Algae 2025, 145, 102863. [Google Scholar] [CrossRef]
- Zhang, F.; Shi, Y.; Jiang, K.; Song, W.; Ma, C.; Xu, Z.; Ma, L. Rapid detection and quantification of Prorocentrum minimum by loop-mediated isothermal amplification and real-time fluorescence quantitative PCR. J. Appl. Phycol. 2014, 26, 1379–1388. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Wang, L.; Li, L.; Alam, M.; Geng, Y.; Li, Z.; Yamasaki, S.; Shi, L. Loop-mediated isothermal amplification method for rapid detection of the toxic dinoflagellate Alexandrium, which causes algal blooms and poisoning of shellfish. FEMS Microbiol. Lett. 2008, 282, 15–21. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- He, L.; Yu, Z.; Xu, X.; Zhu, J.; Yuan, Y.; Cao, X.; Song, X. Metabarcoding analysis identifies high diversity of harmful algal bloom species in the coastal waters of the Beibu Gulf. Ecol. Evol. 2023, 13, e10127. [Google Scholar] [CrossRef]
- Hungerford, J.M.; Wekell, M.M. Analytical methods for marine toxins. In Handbook of Natural Toxins; Routledge: London, UK, 2019; pp. 415–473. [Google Scholar]
- Gerssen, A.; Pol-Hofstad, I.E.; Poelman, M.; Mulder, P.P.; Van den Top, H.J.; De Boer, J. Marine toxins: Chemistry, toxicity, occurrence and detection, with special reference to the Dutch situation. Toxins 2010, 2, 878–904. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.F.; De Salas, M.; Adamson, J.; Rhodes, L.L. Rapid and accurate identification by real-time PCR of biotoxin-producing dinoflagellates from the family Gymnodiniaceae. Mar. Drugs 2014, 12, 1361–1376. [Google Scholar] [CrossRef] [PubMed]
- Kon, N.F.; Lau, W.L.; Hii, K.S.; Law, I.K.; Teng, S.T.; Lim, H.C.; Takahashi, K.; Gu, H.; Lim, P.T.; Leaw, C.P. Quantitative real-time PCR detection of a harmful unarmoured dinoflagellate, Karlodinium australe (D inophyceae). Phycol. Res. 2017, 65, 291–298. [Google Scholar] [CrossRef]
- Park, T.-G.; Park, G.-H.; Park, Y.-T.; Kang, Y.-S.; Bae, H.-M.; Kim, C.-H.; Jeong, H.-J.; Lee, Y. Identification of the dinoflagellate community during Cochlodinium polykrikoides (Dinophyceae) blooms using amplified rDNA melting curve analysis and real-time PCR probes. Harmful Algae 2009, 8, 430–440. [Google Scholar] [CrossRef]
- Thornhill, D.J.; Lajeunesse, T.C.; Santos, S.R. Measuring rDNA diversity in eukaryotic microbial systems: How intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol. Ecol. 2007, 16, 5326–5340. [Google Scholar] [CrossRef]
- Lindner, D.L.; Banik, M.T. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia 2011, 103, 731–740. [Google Scholar] [CrossRef]
- Zhang, Q.C.; Chen, Z.F.; Zhao, J.Y.; Yu, R.C.; Qiu, L.M.; Kong, F.Z.; Wang, Y.F.; Yan, T.; Zhou, M.J. Development of a sensitive qPCR method for the detection of pelagophyte Aureococcus anophagefferens. Limnol. Oceanogr. Methods 2020, 18, 41–51. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.





