Precision Assessment of Facial Asymmetry Using 3D Imaging and Artificial Intelligence
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample
2.2. Three-Dimensional Facial Surface Imaging
2.3. Measurements
2.3.1. Manual Analysis
Head Orientation
Manual Landmarks Identification
2.3.2. Artificial Intelligence-Based Analysis
Model and Datasets
Head Orientation
Landmark Identification
2.4. Evaluation of Facial Asymmetry
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
- The automated method proves notably more efficient than the manual technique for evaluating facial asymmetry using 3D facial images.
- The artificial intelligence-based software exhibits comparable reliability to the manual approach when calculating the asymmetry index based on 3D landmark coordinates.
- The disagreement observed between the automated and manual methods in a couple of the facial landmarks can be addressed through further improvement of the automated software. This may entail additional training of the software, considering the dynamic nature of soft tissues, and integrating updated 3D definitions of facial landmarks into the dataset.
- This automated technique is valuable for orthodontic practitioners and researchers, fostering progress toward an evidence-based practice with enhanced efficiency.
- Additionally, this method’s versatility suggests its potential extension for evaluating other facial features beyond asymmetry assessment.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ICC | Intraclass correlation |
| 2D | Two-dimensional |
| PA | Posteroanterior |
| CT | Computed tomography |
| CBCT | Cone beam computed tomography |
| MVLM | Deep multi-view learning model |
| VAM | VECTRA® 3D Analysis Module |
| MM | Millimeters |
| CLAIM | Checklist for Artificial Intelligence in Medical Imaging |
| BU-3DFE | Binghamton University 3D Facial Expression dataset |
| UPM-3DFE | Universiti Putra Malaysia Facial Expression Recognition Database |
| RGB | Red, Green, Blue color |
| CI | Confidence intervals |
References
- Pirttiniemi, P.M. Associations of mandibular and facial asymmetries—A review. Am. J. Orthod. Dentofac. Orthop. 1994, 106, 191–200. [Google Scholar] [CrossRef]
- Thiesen, G.; Gribel, B.F.; Freitas, M.P.M. Facial asymmetry: A current review. Dent. Press J. Orthod. 2015, 20, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Singh, H.; Mishra, S.; Sharma, P.; Kapoor, P.; Chandra, L. Facial asymmetry revisited: Part I-diagnosis and treatment planning. J. Oral Biol. Craniofacial Res. 2018, 8, 7–14. [Google Scholar] [CrossRef]
- Bishara, S.E.; Burkey, P.S.; Kharouf, J.G. Dental and facial asymmetries: A review. Angle Orthod. 1994, 64, 89–98. [Google Scholar] [PubMed]
- Nicot, R.; Hottenstein, M.; Raoul, G.; Ferri, J.; Horton, M.; Tobias, J.W.; Barton, E.; Gelé, P.; Sciote, J.J. Nodal pathway genes are downregulated in facial asymmetry. J. Craniofacial Surg. 2014, 25, e548. [Google Scholar] [CrossRef] [PubMed]
- Berssenbrügge, P.; Berlin, N.F.; Kebeck, G.; Runte, C.; Jung, S.; Kleinheinz, J.; Dirksen, D. 2D and 3D analysis methods of facial asymmetry in comparison. J. Cranio-Maxillofac. Surg. 2014, 42, e327–e334. [Google Scholar] [CrossRef]
- Agrawal, M.; Agrawal, J.A.; Nanjannawar, L.; Fulari, S.; Kagi, V. Dentofacial asymmetries: Challenging diagnosis and treatment planning. J. Int. Oral Health JIOH 2015, 7, 128. [Google Scholar]
- Katsumata, A.; Fujishita, M.; Maeda, M.; Ariji, Y.; Ariji, E.; Langlais, R.P. 3D-CT evaluation of facial asymmetry. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2005, 99, 212–220. [Google Scholar] [CrossRef]
- de Moraes, M.E.L.; Hollender, L.G.; Chen, C.S.; Moraes, L.C.; Balducci, I. Evaluating craniofacial asymmetry with digital cephalometric images and cone-beam computed tomography. Am. J. Orthod. Dentofac. Orthop. 2011, 139, e523–e531. [Google Scholar] [CrossRef]
- Akhil, G.; Kumar, K.P.S.; Raja, S.; Janardhanan, K. Three-dimensional assessment of facial asymmetry: A systematic review. J. Pharm. Bioallied Sci. 2015, 7 (Suppl. S2), S433. [Google Scholar] [CrossRef]
- Plooij, J.; Swennen, G.; Rangel, F.; Maal, T.; Schutyser, F.; Bronkhorst, E.; Kuijpers–Jagtman, A.M.; Bergé, S. Evaluation of reproducibility and reliability of 3D soft tissue analysis using 3D stereophotogrammetry. Int. J. Oral Maxillofac. Surg. 2009, 38, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Toma, A.M.; Zhurov, A.; Playle, R.; Ong, E.; Richmond, S. Reproducibility of facial soft tissue landmarks on 3D laser-scanned facial images. Orthod. Craniofacial Res. 2009, 12, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.O.; Morrison, C.S.; Linden, O.; Phillips, B.; Chang, J.; Byrne, M.E.; Sullivan, S.R.; Forrest, C.R. Quantitative facial asymmetry: Using three-dimensional photogrammetry to measure baseline facial surface symmetry. J. Craniofacial Surg. 2014, 25, 124–128. [Google Scholar] [CrossRef]
- Choi, J.W.; Park, H.; Kwon, S.-M.; Lee, J.Y. Surgery-first orthognathic approach for the correction of facial asymmetry. J. Cranio-Maxillofac. Surg. 2021, 49, 435–442. [Google Scholar] [CrossRef]
- Monill-González, A.; Rovira-Calatayud, L.; d’Oliveira, N.G.; Ustrell-Torrent, J.M. Artificial intelligence in orthodontics: Where are we now? A scoping review. Orthod. Craniofacial Res. 2021, 24, 6–15. [Google Scholar] [CrossRef]
- Mohammad-Rahimi, H.; Nadimi, M.; Rohban, M.H.; Shamsoddin, E.; Lee, V.Y.; Motamedian, S.R. Machine learning and orthodontics, current trends and the future opportunities: A scoping review. Am. J. Orthod. Dentofac. Orthop. 2021, 160, 170–192.e4. [Google Scholar] [CrossRef]
- Retrouvey, J.-M.; Conley, R.S. Decoding Deep Learning applications for diagnosis and treatment planning. Dent. Press J. Orthod. 2023, 27, e22spe5. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kong, D.; Tang, T.; Su, D.; Yang, P.; Wang, H.; Zhao, Z.; Liu, Y. Orthodontic treatment planning based on artificial neural networks. Sci Rep. 2019, 9, 2037. [Google Scholar] [CrossRef]
- Park, Y.-S.; Lek, S. Artificial neural networks: Multilayer perceptron for ecological modeling. In Developments in Environmental Modelling; Elsevier: Amsterdam, The Netherlands, 2016; Volume 28, pp. 123–140. [Google Scholar]
- Bichu, Y.M.; Hansa, I.; Bichu, A.Y.; Premjani, P.; Flores-Mir, C.; Vaid, N.R. Applications of artificial intelligence and machine learning in orthodontics: A scoping review. Prog. Orthod. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Dongare, A.; Kharde, R.; Kachare, A.D. Introduction to artificial neural network. Int. J. Eng. Innov. Technol. (IJEIT) 2012, 2, 189–194. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A comprehensive survey on transfer learning. Proc. IEEE 2020, 109, 43–76. [Google Scholar] [CrossRef]
- Hwang, H.-W.; Park, J.-H.; Moon, J.-H.; Yu, Y.; Kim, H.; Her, S.-B.; Srinivasan, G.; Aljanabi, M.N.A.; Donatelli, R.E.; Lee, S.-J. Automated identification of cephalometric landmarks: Part 2-Might it be better than human? Angle Orthod. 2020, 90, 69–76. [Google Scholar] [CrossRef]
- Song, Y.; Qiao, X.; Iwamoto, Y.; Chen, Y.-w. Automatic cephalometric landmark detection on X-ray images using a deep-learning method. Appl. Sci. 2020, 10, 2547. [Google Scholar] [CrossRef]
- Atici, S.F.; Ansari, R.; Allareddy, V.; Suhaym, O.; Cetin, A.E.; Elnagar, M.H. Fully automated determination of the cervical vertebrae maturation stages using deep learning with directional filters. PLoS ONE 2022, 17, e0269198. [Google Scholar] [CrossRef]
- Radwan, M.T.; Sin, Ç.; Akkaya, N.; Vahdettin, L. Artificial intelligence-based algorithm for cervical vertebrae maturation stage assessment. Orthod. Craniofacial Res. 2023, 26, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Misra, D.; Rodriguez, L.; Gill, M.; Liberton, D.K.; Almpani, K.; Lee, J.S.; Antani, S. Malocclusion classification on 3D cone-beam CT craniofacial images using multi-channel deep learning models. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1294–1298. [Google Scholar]
- Sin, Ç.; Akkaya, N.; Aksoy, S.; Orhan, K.; Öz, U. A deep learning algorithm proposal to automatic pharyngeal airway detection and segmentation on CBCT images. Orthod. Craniofacial Res. 2021, 24, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Lee, Y.-S.; Mo, S.-P.; Lim, K.; Jung, S.-K.; Kim, T.-W. Application of deep learning artificial intelligence technique to the classification of clinical orthodontic photos. BMC Oral Health 2022, 22, 454. [Google Scholar] [CrossRef]
- Rousseau, M.; Retrouvey, J.-M. Machine learning in orthodontics: Automated facial analysis of vertical dimension for increased precision and efficiency. Am. J. Orthod. Dentofac. Orthop. 2022, 161, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Rabiu, H.; Saripan, M.I.; Mashohor, S.; Marhaban, M.H. 3D facial expression recognition using maximum relevance minimum redundancy geometrical features. EURASIP J. Adv. Signal Process. 2012, 2012, 1–8. [Google Scholar] [CrossRef]
- Paulsen, R.R.; Juhl, K.A.; Haspang, T.M.; Hansen, T.; Ganz, M.; Einarsson, G. Multi-view consensus CNN for 3D facial landmark placement. In Proceedings of the Asian Conference on Computer Vision, Perth, Australia, 2–6 December 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 706–719. [Google Scholar]
- Farkas, L.G.; Cheung, G. Facial asymmetry in healthy North American Caucasians: An anthropometrical study. Angle Orthod. 1981, 51, 70–77. [Google Scholar]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G* Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- De Stefani, A.; Barone, M.; Hatami Alamdari, S.; Barjami, A.; Baciliero, U.; Apolloni, F.; Gracco, A.; Bruno, G. Validation of Vectra 3D imaging systems: A review. Int. J. Environ. Res. Public Health 2022, 19, 8820. [Google Scholar] [CrossRef]
- Hansson, S.; Östlund, E.; Bazargani, F. The Vectra M3 3-dimensional digital stereophotogrammetry system: A reliable technique for detecting chin asymmetry. Imaging Sci. Dent. 2022, 52, 43. [Google Scholar] [CrossRef]
- Huang, C.; Liu, X.; Chen, Y. Facial asymmetry index in normal young adults. Orthod. Craniofacial Res. 2013, 16, 97–104. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E., Jr. Checklist for artificial intelligence in medical imaging (CLAIM): A guide for authors and reviewers. Radiol. Soc. North Am. 2020, 2, e200029. [Google Scholar] [CrossRef]
- Collins, G.S.; Moons, K.G.; Dhiman, P.; Riley, R.D.; Beam, A.L.; Van Calster, B.; Ghassemi, M.; Liu, X.; Reitsma, J.B.; Van Smeden, M. TRIPOD+ AI statement: Updated guidance for reporting clinical prediction models that use regression or machine learning methods. bmj 2024, 385. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wei, X.; Sun, Y.; Wang, J.; Rosato, M.J. A 3D facial expression database for facial behavior research. In Proceedings of the 7th International Conference on Automatic Face and Gesture Recognition (FGR06), Southampton, UK, 10–12 April 2006; pp. 211–216. [Google Scholar]
- Habibu, R.; Syamsiah, M.; Hamiruce, M.M.; Iqbal, S.M. UPM-3D facial expression recognition Database (UPM-3DFE). In Proceedings of the PRICAI 2012: Trends in Artificial Intelligence: 12th Pacific Rim International Conference on Artificial Intelligence, Kuching, Malaysia, 3–7 September 2012; Proceedings 12. Springer: Berlin/Heidelberg, Germany, 2012; pp. 470–479. [Google Scholar]
- Walter, S.; Eliasziw, M.; Donner, A. Sample size and optimal designs for reliability studies. Stat. Med. 1998, 17, 101–110. [Google Scholar] [CrossRef]
- Han, X. On statistical measures for data quality evaluation. J. Geogr. Inf. Syst. 2020, 12, 178–187. [Google Scholar] [CrossRef]
- Menyhart, O.; Weltz, B.; Győrffy, B. MultipleTesting. com: A tool for life science researchers for multiple hypothesis testing correction. PLoS ONE 2021, 16, e0245824. [Google Scholar] [CrossRef]
- Ferring, V.; Pancherz, H. Divine proportions in the growing face. Am. J. Orthod. Dentofac. Orthop. 2008, 134, 472–479. [Google Scholar] [CrossRef]
- Ras, F.; Habets, L.L.; van Ginkel, F.C.; Prahl-Andersen, B. Method for quantifying facial asymmetry in three dimensions using stereophotogrammetry. Angle Orthod. 1995, 65, 233–239. [Google Scholar]
- Damstra, J.; Fourie, Z.; De Wit, M.; Ren, Y. A three-dimensional comparison of a morphometric and conventional cephalometric midsagittal planes for craniofacial asymmetry. Clin. Oral Investig. 2012, 16, 285–294. [Google Scholar] [CrossRef]
- de Menezes, M.; Rosati, R.; Ferrario, V.F.; Sforza, C. Accuracy and reproducibility of a 3-dimensional stereophotogrammetric imaging system. J. Oral Maxillofac. Surg. 2010, 68, 2129–2135. [Google Scholar] [CrossRef]
- Oyedotun, O.K.; Demisse, G.; El Rahman Shabayek, A.; Aouada, D.; Ottersten, B. Facial expression recognition via joint deep learning of rgb-depth map latent representations. In Proceedings of the IEEE International Conference on Computer Vision Workshops, Venice, Italy, 22–29 October 2017; pp. 3161–3168. [Google Scholar]
- Bronstein, M.M.; Bruna, J.; LeCun, Y.; Szlam, A.; Vandergheynst, P. Geometric deep learning: Going beyond euclidean data. IEEE Signal Process. Mag. 2017, 34, 18–42. [Google Scholar] [CrossRef]
- Fagertun, J.; Harder, S.; Rosengren, A.; Moeller, C.; Werge, T.; Paulsen, R.R.; Hansen, T.F. 3D facial landmarks: Inter-operator variability of manual annotation. BMC Med. Imaging 2014, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Gwilliam, J.R.; Cunningham, S.J.; Hutton, T. Reproducibility of soft tissue landmarks on three-dimensional facial scans. Eur. J. Orthod. 2006, 28, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Katina, S.; McNeil, K.; Ayoub, A.; Guilfoyle, B.; Khambay, B.; Siebert, P.; Sukno, F.; Rojas, M.; Vittert, L.; Waddington, J. The definitions of three-dimensional landmarks on the human face: An interdisciplinary view. J. Anat. 2016, 228, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Au, J.; Mei, L.; Bennani, F.; Kang, A.; Farella, M. Three-dimensional analysis of lip changes in response to simulated maxillary incisor advancement. Angle Orthod. 2020, 90, 118–124. [Google Scholar] [CrossRef]
- Paek, S.J.; Yoo, J.Y.; Lee, J.W.; Park, W.-J.; Chee, Y.D.; Choi, M.G.; Choi, E.J.; Kwon, K.-H. Changes of lip morphology following mandibular setback surgery using 3D cone-beam computed tomography images. Maxillofac. Plast. Reconstr. Surg. 2016, 38, 38. [Google Scholar] [CrossRef]
- Perrotti, G.; Goker, F.; Rossi, O.; Nowakowska, J.; Russillo, A.; Beltramini, G.; Tartaglia, G.; Testori, T.; Del Fabbro, M.; Mortellaro, C. 3D Computed Tomography vs. 2D radiography: Comparison of 3D direct anthropometry with 2D norm calculations and analysis of differences in soft tissue measurements. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 46–60. [Google Scholar]
- Alqattan, M.; Djordjevic, J.; Zhurov, A.I.; Richmond, S. Comparison between landmark and surface-based three-dimensional analyses of facial asymmetry in adults. Eur. J. Orthod. 2015, 37, 1–12. [Google Scholar] [CrossRef] [PubMed]


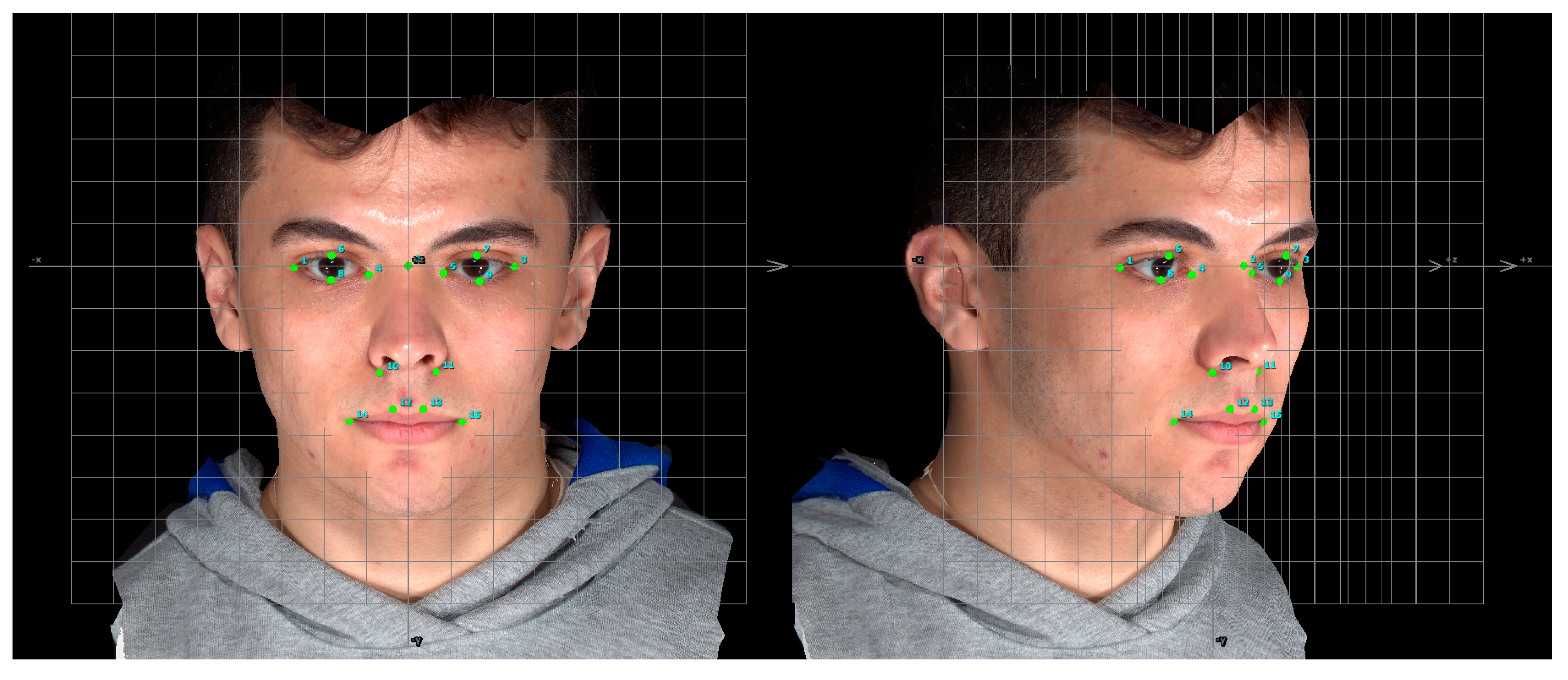
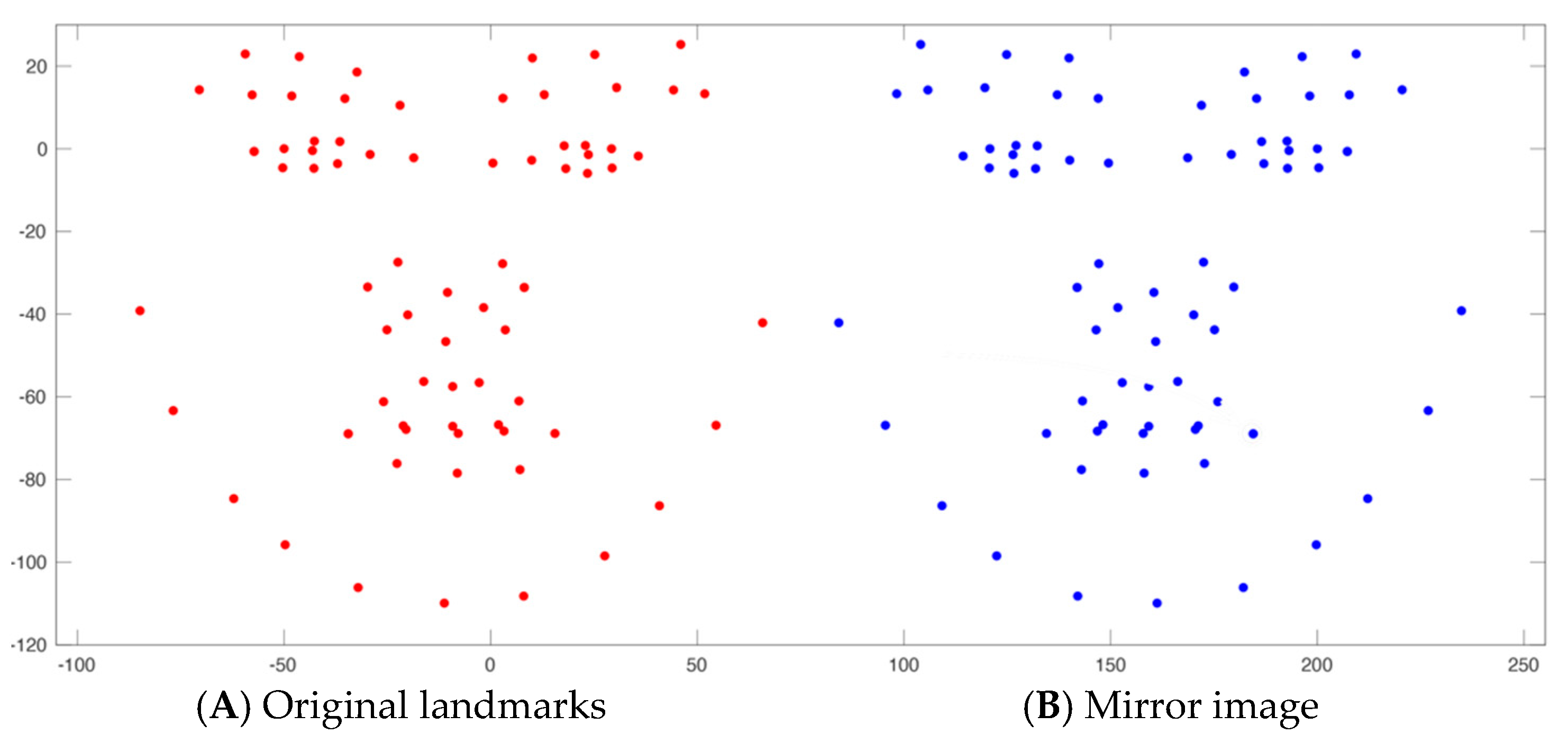
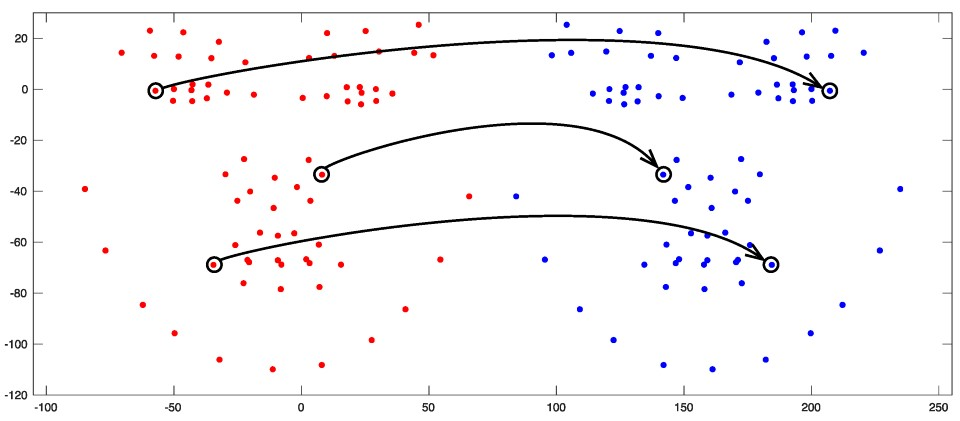
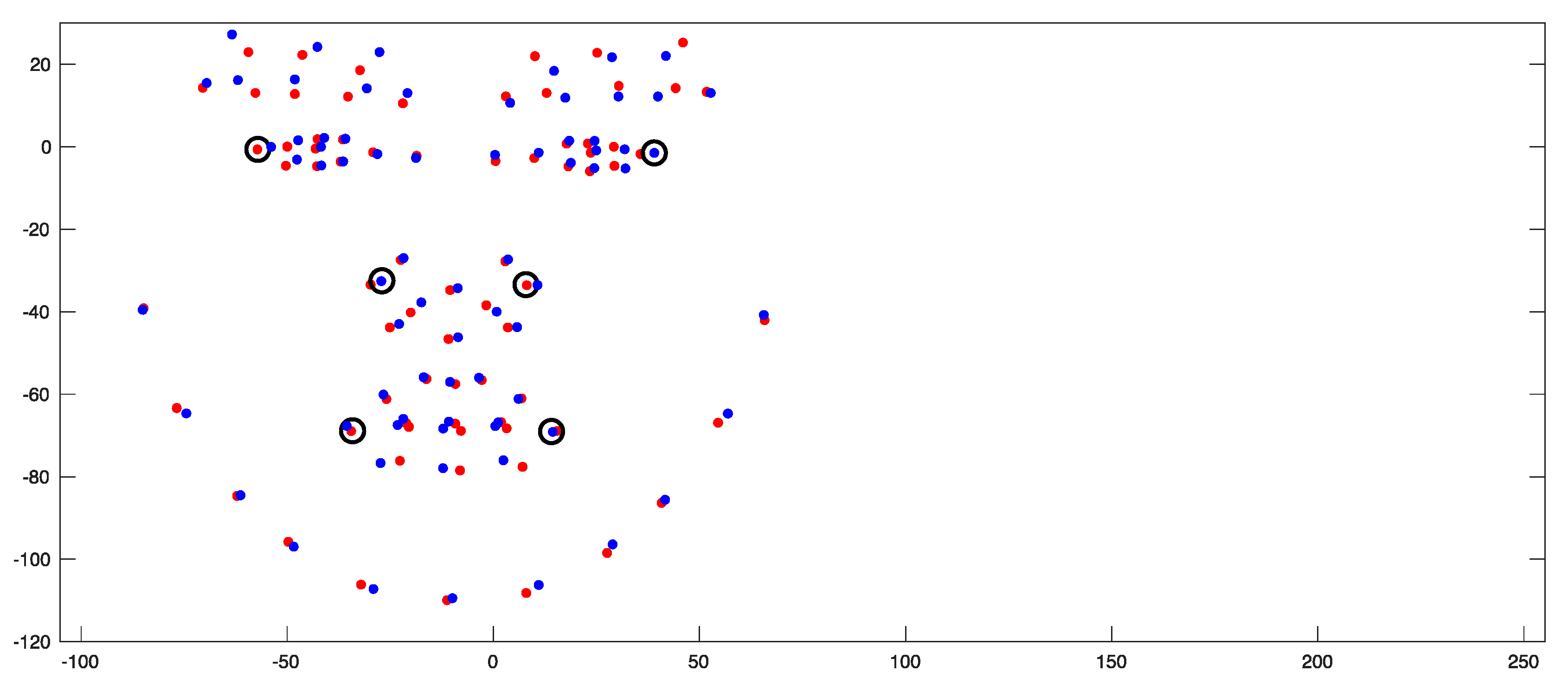
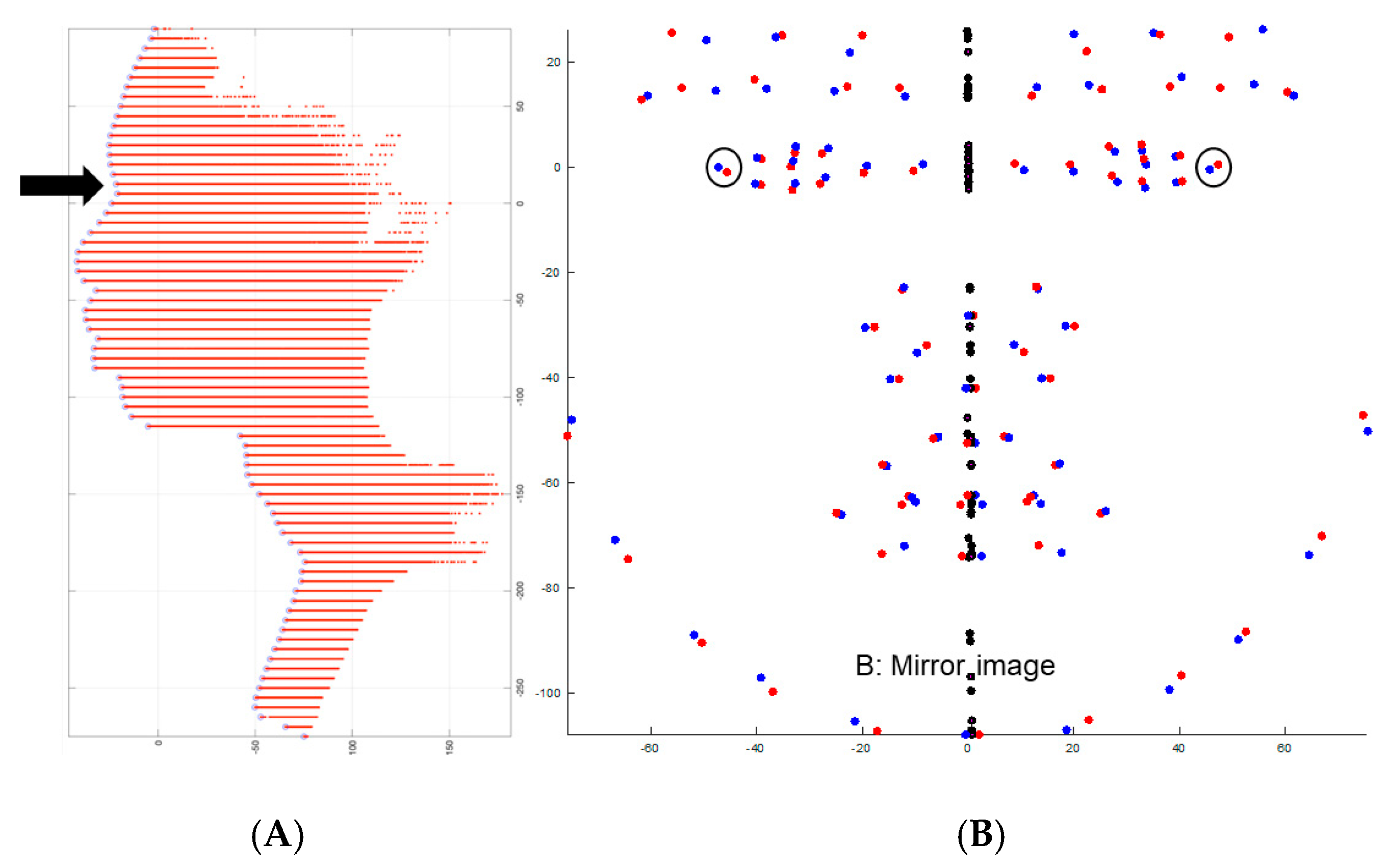
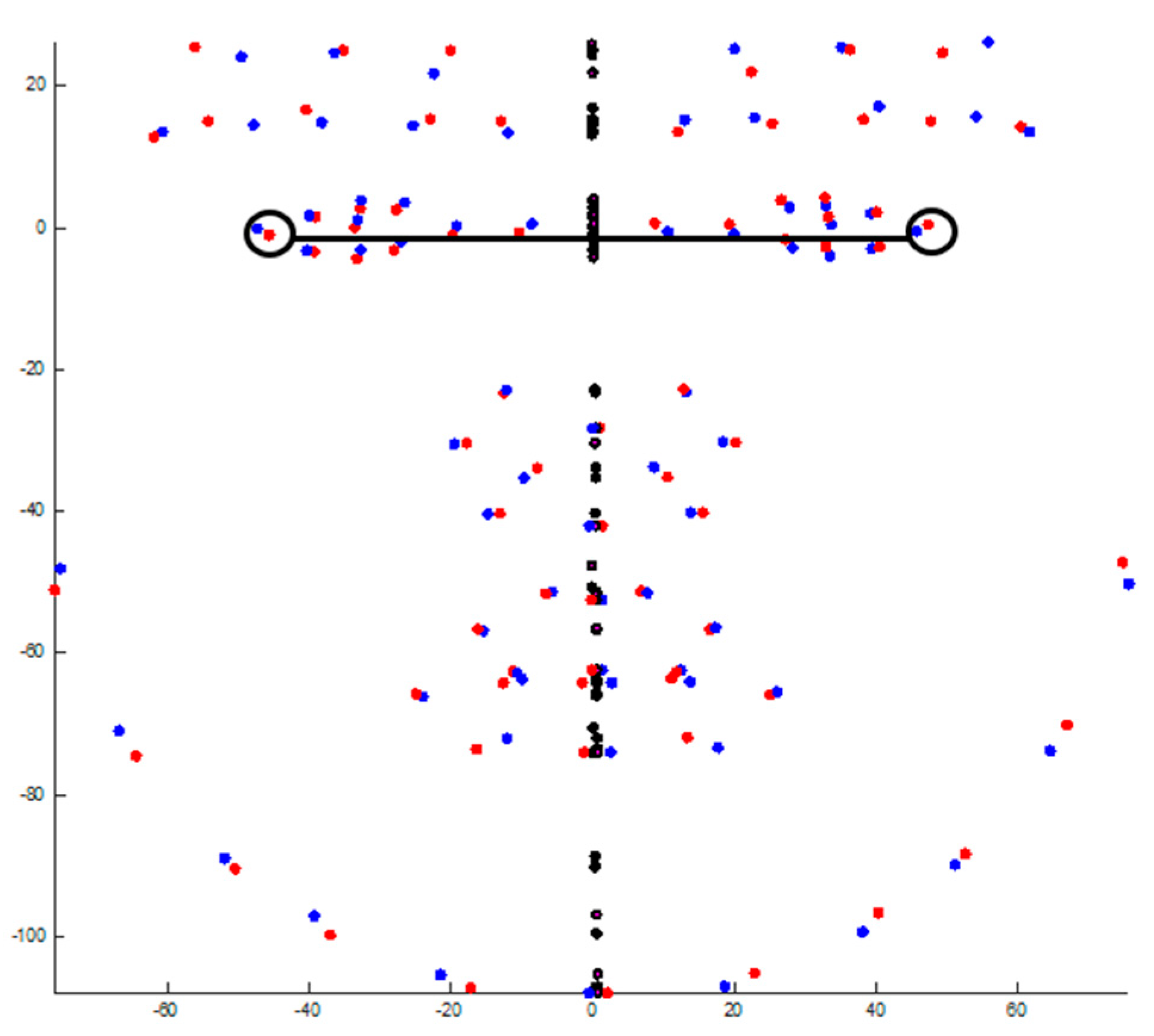
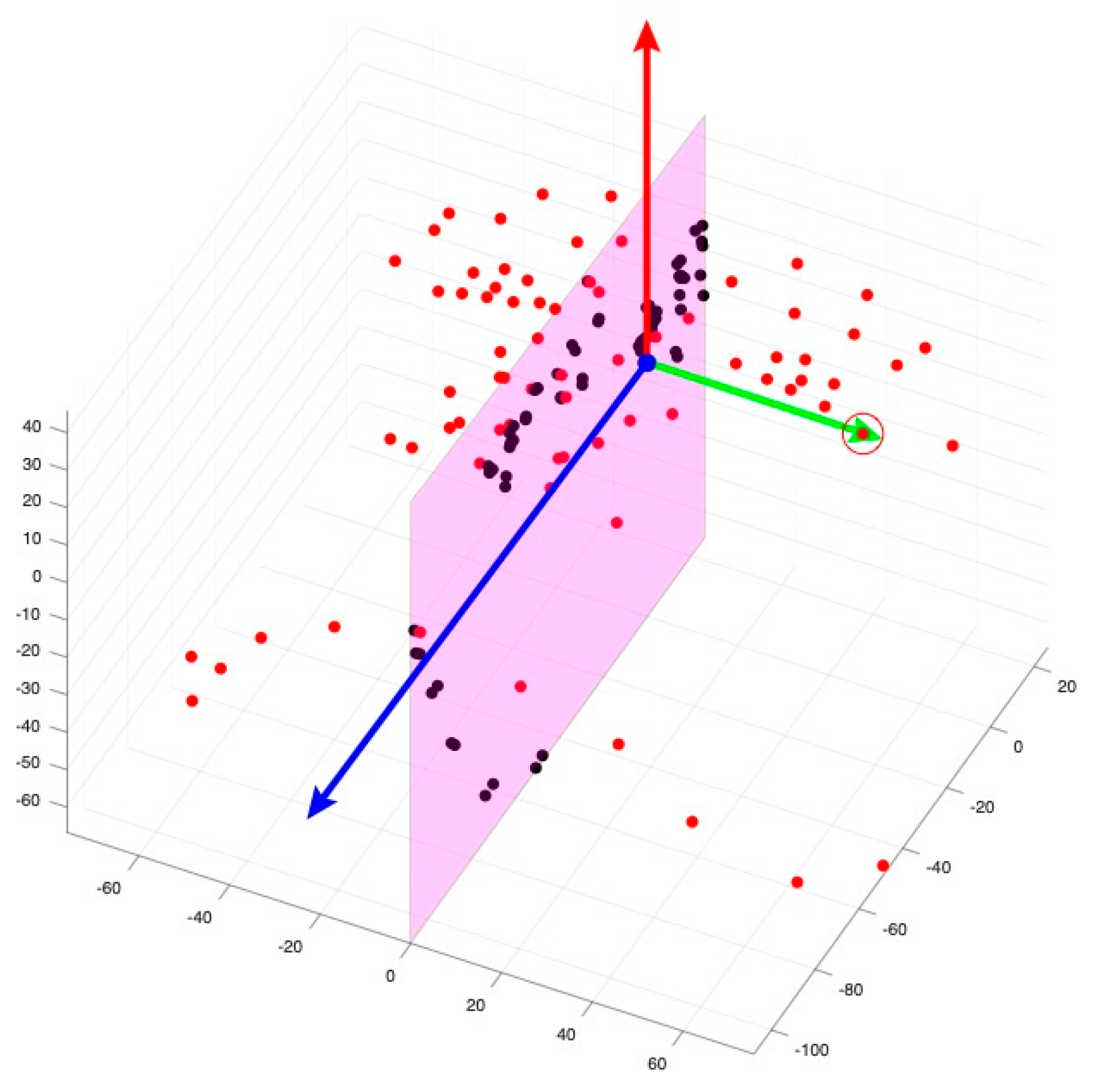
| Landmark * | Definition |
|---|---|
| Palpebrale superius 6,7 | Superior mid-portion of the free margin of upper eyelids |
| Palpebrale inferius 8,9 | Inferior mid-portion of the free margin of lower eyelids |
| Exocanthion 1,3 | The soft tissue point located at the outer commissure of each eye fissure |
| Endocanthion 4,5 | The soft tissue point located at the inner commissure of each eye fissure |
| Alare 10,11 | The most lateral point on each alar contour |
| Crista philtra 12,13 | The point at each crossing of the vermilion line and the elevated margin of the philtrum |
| Cheilion 14,15 | The point located at each labial commissure |
| Intrarater ICC (Main Observer) | 95% CI | Interrater ICC | 95% CI | |
|---|---|---|---|---|
| Palpebrale superius | 0.991 | 0.976–0.996 | 0.756 | 0.651–0.830 |
| Palpebrale inferius | 0.62 | 0.410–0.850 | 0.851 | 0.786–0.896 |
| Exocanthion | 0.607 | 0.008–0.845 | 0.719 | 0.598–0.804 |
| Endocanthion | 0.716 | 0.283–0.888 | 0.944 | 0.920–0.961 |
| Alare | 0.778 | 0.440–0.912 | 0.657 | 0.509–0.760 |
| Crista philtra | 0.624 | 0.051–0.851 | 0.855 | 0.793–0.899 |
| Cheilion | 0.963 | 0.906–0.985 | 0.959 | 0.942–0.972 |
| Manual Method | Automated Deep MVLM Method | Wilcoxon Signed-Rank Test p-Values | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Landmarks | Median | 25th Percentile (Q1) | 75th Percentile (Q3) | Mean | SD | Median | 25th Percentile (Q1) | 75th Percentile (Q3) | Mean | SD | Median Paired Difference | Hodges–Lehmann 95% CIs | p-Value | p-Value Adjusted * and Benjamini–Hochberg FDR Significance |
| Palpebrale superius | 2.65 | 1.75 | 3.79 | 2.85 | 1.48 | 2.51 | 1.64 | 3.48 | 3.24 | 5.96 | 0.11 | −3.81–3.87 | 0.4618 | 0.461 NS |
| Palpebrale inferius | 2.46 | 1.64 | 3.58 | 2.68 | 1.27 | 2.13 | 1.51 | 3.40 | 2.48 | 1.43 | 0.31 | −4.13–3.35 | 0.0565 | 0.132 NS |
| Exocanthion | 2.69 | 1.69 | 4.05 | 3.03 | 1.77 | 2.67 | 1.85 | 4.24 | 3.20 | 1.92 | −0.14 | −4.65–4.64 | 0.4064 | 0.462 NS |
| Endocanthion | 1.89 | 1.46 | 2.62 | 2.14 | 1.06 | 1.7 | 1.18 | 2.47 | 2.25 | 3.74 | 0.16 | −2.75–2.78 | 0.1223 | 0.214 NS |
| Alare | 2.05 | 1.31 | 2.74 | 2.15 | 1.11 | 1.54 | 1.09 | 2.09 | 1.70 | 0.96 | 0.39 | −1.64–3.21 | 0.0008 | 0.0056 SIG |
| Crista philtra | 1.42 | 0.87 | 2.54 | 1.91 | 1.56 | 1.33 | 0.85 | 2.04 | 1.62 | 1.26 | 0.18 | −3.37–4.38 | 0.2004 | 0.281 NS |
| Cheilion | 2.77 | 2.00 | 3.80 | 3.15 | 1.72 | 2.30 | 1.57 | 3.31 | 2.56 | 1.38 | 0.54 | −3.55–5.15 | 0.0023 | 0.0081 SIG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adel, M.; Hunt, K.J.; Lau, D.; Hartsfield, J.K.; Reyes-Centeno, H.; Beeman, C.S.; Elshebiny, T.; Sharab, L. Precision Assessment of Facial Asymmetry Using 3D Imaging and Artificial Intelligence. J. Clin. Med. 2025, 14, 7172. https://doi.org/10.3390/jcm14207172
Adel M, Hunt KJ, Lau D, Hartsfield JK, Reyes-Centeno H, Beeman CS, Elshebiny T, Sharab L. Precision Assessment of Facial Asymmetry Using 3D Imaging and Artificial Intelligence. Journal of Clinical Medicine. 2025; 14(20):7172. https://doi.org/10.3390/jcm14207172
Chicago/Turabian StyleAdel, Mohamed, Katie Jo Hunt, Daniel Lau, James K. Hartsfield, Hugo Reyes-Centeno, Cynthia S. Beeman, Tarek Elshebiny, and Lina Sharab. 2025. "Precision Assessment of Facial Asymmetry Using 3D Imaging and Artificial Intelligence" Journal of Clinical Medicine 14, no. 20: 7172. https://doi.org/10.3390/jcm14207172
APA StyleAdel, M., Hunt, K. J., Lau, D., Hartsfield, J. K., Reyes-Centeno, H., Beeman, C. S., Elshebiny, T., & Sharab, L. (2025). Precision Assessment of Facial Asymmetry Using 3D Imaging and Artificial Intelligence. Journal of Clinical Medicine, 14(20), 7172. https://doi.org/10.3390/jcm14207172







