The Gut Microbiome, Seleno-Compounds, and Acute Myocardial Infarction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient and Public Involvement Statement
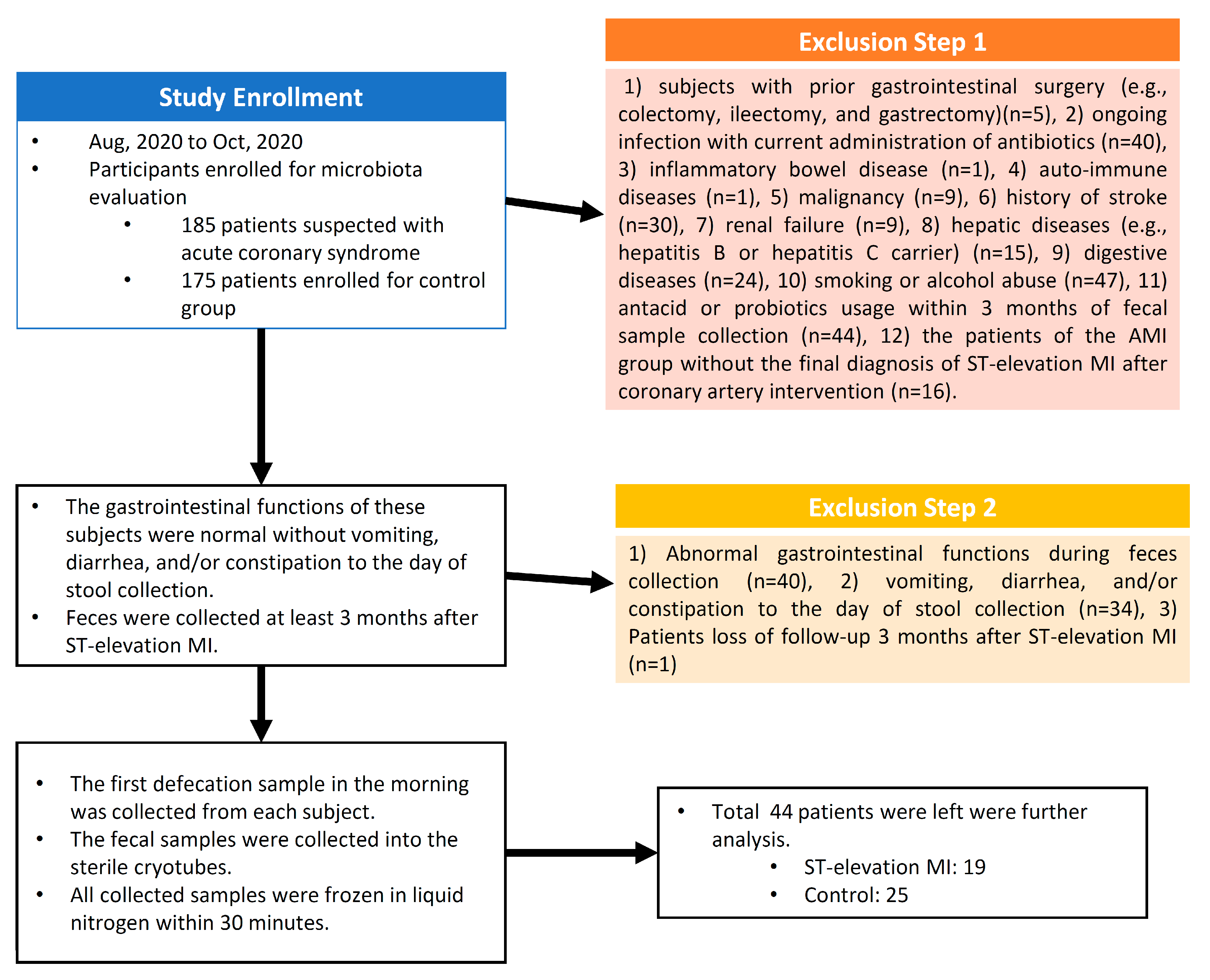
2.2. Study Setting and Participants
2.3. Fecal Collection and Processing
2.4. DNA Extraction
2.5. 16S rRNA Gene Sequencing
2.6. Comparison of the Gut Microbiome Composition
2.7. Statistical Analyses
3. Results
3.1. Basic Demographics
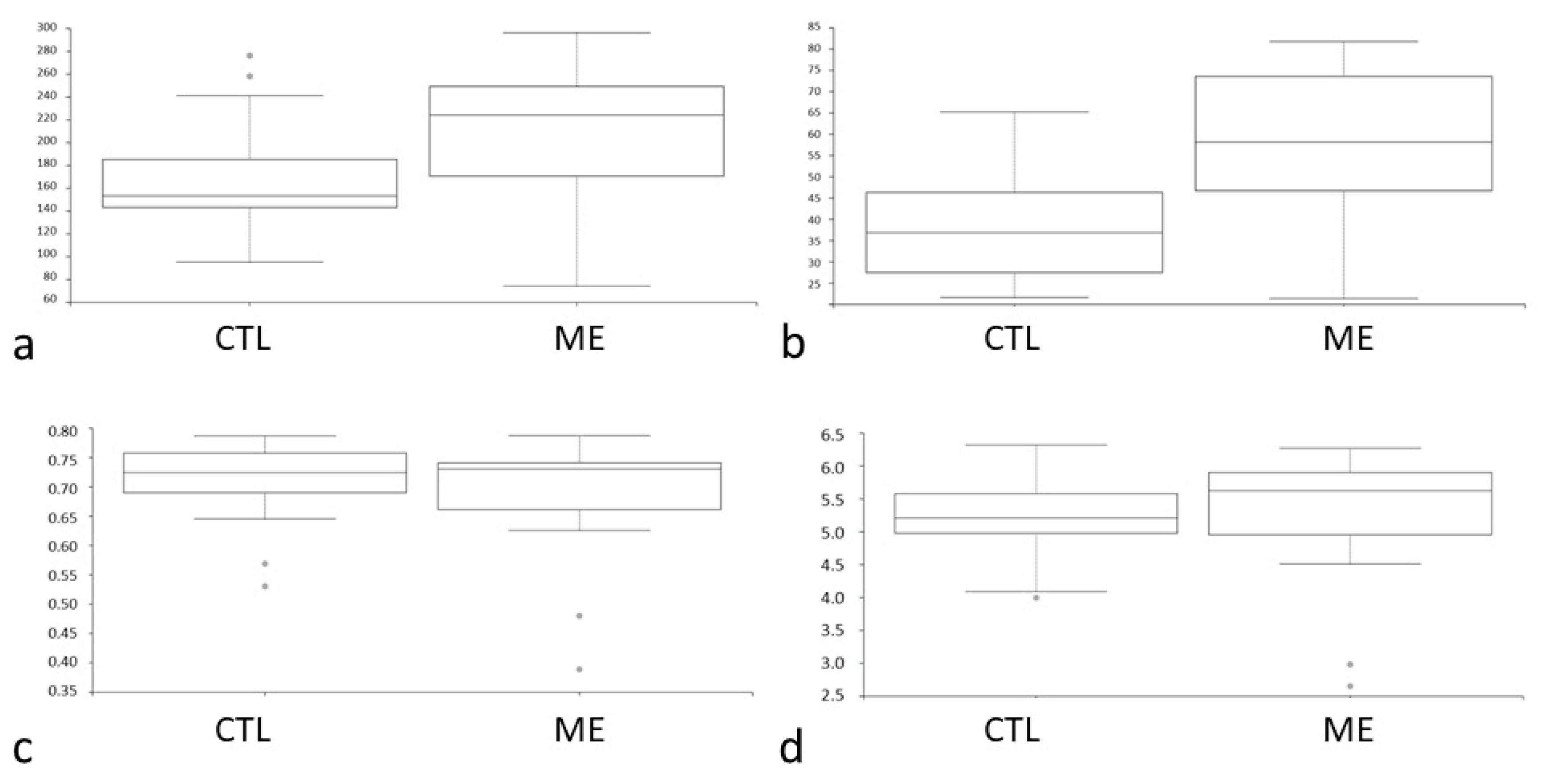
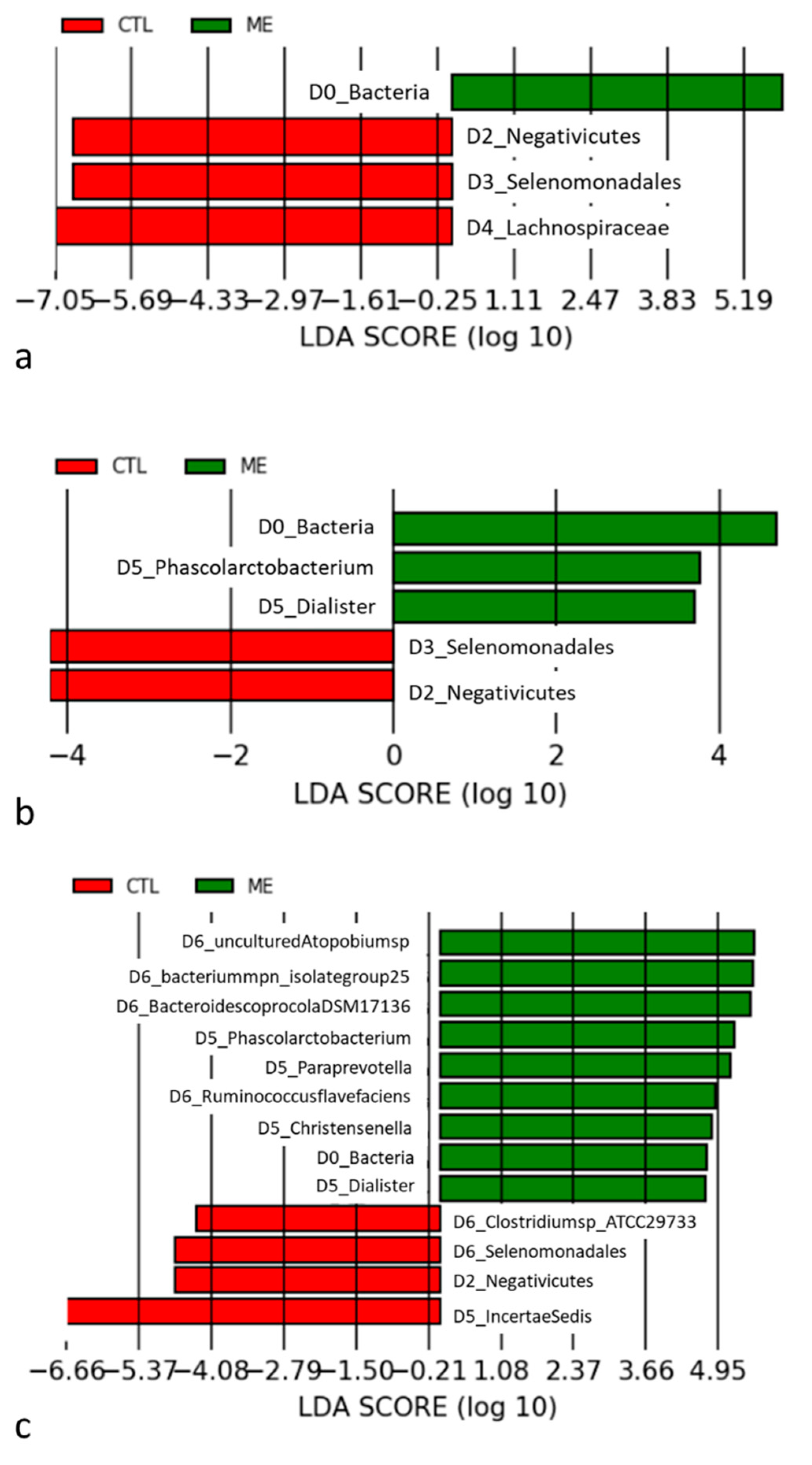
3.2. Analysis of Diversity between Groups
4. Discussion
4.1. Gut Microbiome Is Influenced in AMI Patients
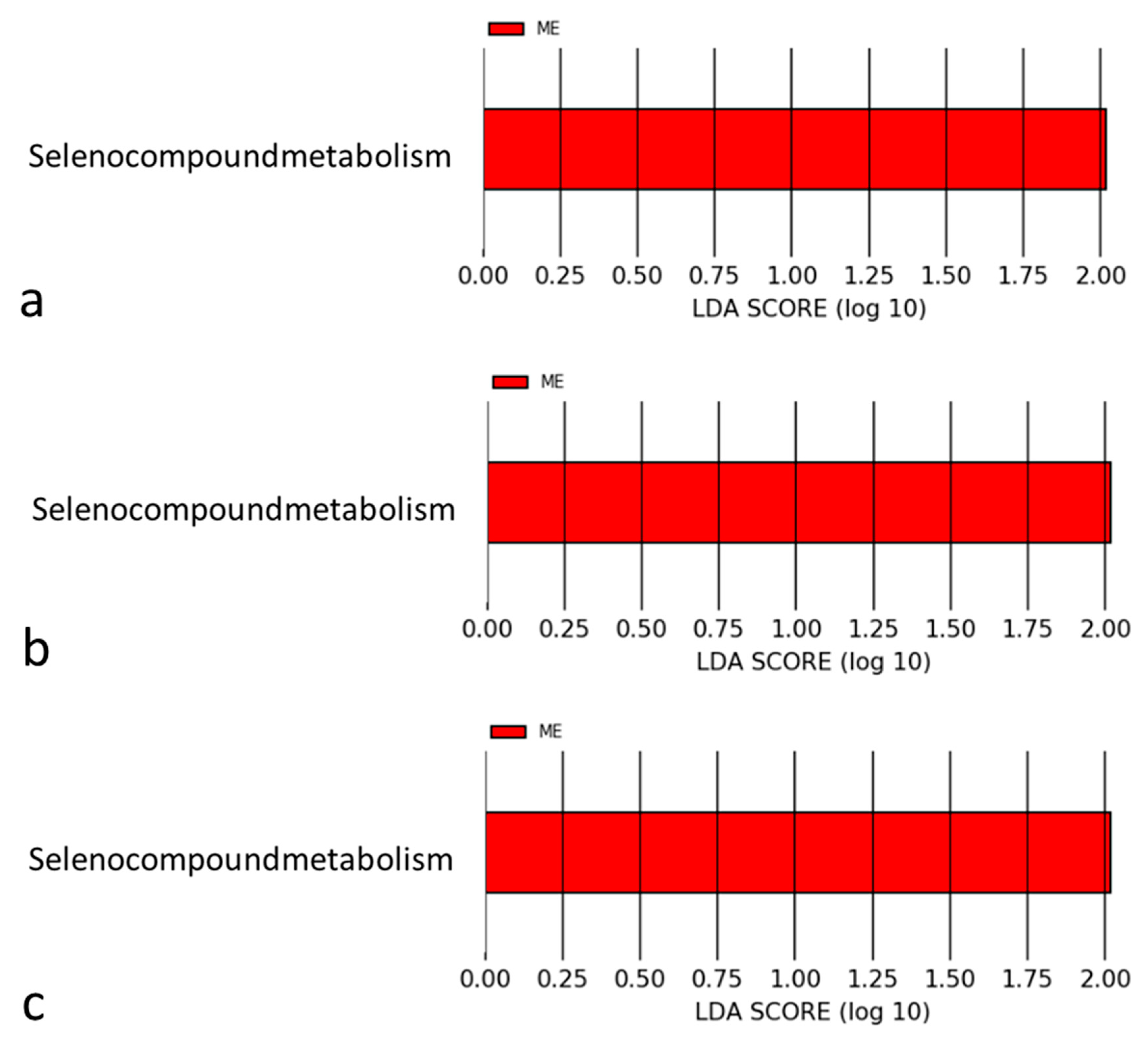
4.2. Mechanisms Associated with Selenomonadales and AMI
4.3. Selenium in the Pathophysiology of Cardiovascular Diseases
4.4. Mechanisms of Selenium for Preventing Coronary Atherosclerosis
4.5. Association between Selenium and AMI
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Akbar, H.; Foth, C.; Kahloon, R.A.; Mountfort, S. Acute Myocardial Infarction ST Elevation (STEMI). In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
- Lam, V.; Su, J.; Koprowski, S.; Hsu, A.; Tweddell, J.S.; Rafiee, P.; Gross, G.J.; Salzman, N.H.; Baker, J.E. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. 2012, 26, 1727–1735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, Y.; Tian, Y.; Huang, C.; Li, D.; Zhong, Q.; Ma, X. Interaction between Microbes and Host Intestinal Health: Modulation by Dietary Nutrients and Gut-Brain-Endocrine-Immune Axis. Curr. Protein Pept. Sci. 2015, 16, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Amar, J.; Lelouvier, B.; Servant, F.; Lluch, J.; Burcelin, R.; Bongard, V.; Elbaz, M. Blood Microbiota Modification After Myocardial Infarction Depends Upon Low-Density Lipoprotein Cholesterol Levels. J. Am. Heart Assoc. 2019, 8, e011797. [Google Scholar] [CrossRef] [PubMed]
- Kiouptsi, K.; Finger, S.; Garlapati, V.S.; Knorr, M.; Brandt, M.; Walter, U.; Wenzel, P.; Reinhardt, C. Hypoxia evokes increased PDI and PDIA6 expression in the infarcted myocardium of ex-germ-free and conventionally raised mice. Biol. Open 2019, 8, bio038851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisarenko, O.I.; Khlopkov, V.N.; Ruuge, E.K. A 1H NMR study of succinate synthesis from exogenous precursors in oxygen-deprived rat heart mitochondria. Biochem. Int. 1986, 12, 145–153. [Google Scholar]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [Green Version]
- Li, X.S.; Obeid, S.; Klingenberg, R.; Gencer, B.; Mach, F.; Räber, L.; Windecker, S.; Rodondi, N.; Nanchen, D.; Muller, O.; et al. Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: A prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur. Heart J. 2017, 38, 814–824. [Google Scholar] [CrossRef]
- Gao, J.; Yan, K.T.; Wang, J.X.; Dou, J.; Wang, J.; Ren, M.; Ma, J.; Zhang, X.; Liu, Y. Gut microbial taxa as potential predictive biomarkers for acute coronary syndrome and post-STEMI cardiovascular events. Sci. Rep. 2020, 10, 2639. [Google Scholar] [CrossRef]
- Lam, V.; Su, J.; Hsu, A.; Gross, G.J.; Salzman, N.H.; Baker, J.E. Intestinal Microbial Metabolites Are Linked to Severity of Myocardial Infarction in Rats. PLoS ONE 2016, 11, e0160840. [Google Scholar] [CrossRef]
- Lawton, J.S.; Tamis-Holland, J.E.; Bangalore, S.; Bates, E.R.; Beckie, T.M.; Bischoff, J.M.; Bittl, J.A.; Cohen, M.G.; DiMaio, J.M.; Don, C.W.; et al. 2021 ACC/AHA/SCAI Guideline for Coronary Artery Revascularization: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2022, 79, 197–215. [Google Scholar] [CrossRef]
- Benakis, C.; Brea, D.; Caballero, S.; Faraco, G.; Moore, J.; Murphy, M.; Sita, G.; Racchumi, G.; Ling, L.; Pamer, E.G.; et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nat. Med. 2016, 22, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Ascher, S.; Wilms, E.; Pontarollo, G.; Formes, H.; Bayer, F.; Müller, M.; Malinarich, F.; Grill, A.; Bosmann, M.; Saffarzadeh, M.; et al. Gut Microbiota Restricts NETosis in Acute Mesenteric Ischemia-Reperfusion Injury. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2279–2292. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Gong, Z.; Sun, G.; Xu, J.; Qi, C.; Sun, W.; Jiang, H.; Cao, P.; Ju, H. Dysbiosis of Gut Microbiota in Patients with Acute Myocardial Infarction. Front. Microbiol. 2021, 12, 680101. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.W.; Bäckhed, F.; Landmesser, U.; Hazen, S.L. Intestinal Microbiota in Cardiovascular Health and Disease: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 2089–2105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, X.; Liu, P.; Zhao, J.; Sun, J.; Guan, W.; Johnston, L.J.; Levesque, C.L.; Fan, P.; He, T.; et al. Dietary Clostridium butyricum Induces a Phased Shift in Fecal Microbiota Structure and Increases the Acetic Acid-Producing Bacteria in a Weaned Piglet Model. J. Agric. Food Chem. 2018, 66, 5157–5166. [Google Scholar] [CrossRef]
- Schwarz, K.; Foltz, C.M. Selenium as an integral part of factor 3 against dietary necrotic liver degeneration. J. Am. Chem. Soc. 1957, 79, 3292–3293, reprinted in Nutrition 1999, 15, 254–256. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Flores-Mateo, G.; Navas-Acien, A.; Pastor-Barriuso, R.; Guallar, E. Selenium and coronary heart disease: A meta-analysis. Am. J. Clin. Nutr. 2006, 84, 762–773. [Google Scholar] [CrossRef] [Green Version]
- Wójcicki, J.; Rózewicka, L.; Barcew-Wiszniewska, B.; Samochowiec, L.; Juźwiak, S.; Kadłubowska, D.; Tustanowski, S.; Juzyszyn, Z. Effect of selenium and vitamin E on the development of experimental atherosclerosis in rabbits. Atherosclerosis 1991, 87, 9–16. [Google Scholar] [CrossRef]
- Mehta, U.; Kang, B.P.; Kukreja, R.S.; Bansal, M.P. Ultrastructural examination of rabbit aortic wall following high-fat diet feeding and selenium supplementation: A transmission electron microscopy study. J. Appl. Toxicol. 2002, 22, 405–413. [Google Scholar] [CrossRef]
- Burk, R.F.; Hill, K.E. Selenoprotein P: An extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu. Rev. Nutr. 2005, 25, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohé, R.; Banning, A.; Schnurr, K. Selenium-dependent enzymes in endothelial cell function. Antioxid. Redox Signal. 2003, 5, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Bonomini, F.; Tengattini, S.; Fabiano, A.; Bianchi, R.; Rezzani, R. Atherosclerosis and oxidative stress. Histol. Histopathol. 2008, 23, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Stocker, R.; Keaney, J.F., Jr. Role of oxidative modifications in atherosclerosis. Physiol. Rev. 2004, 84, 1381–1478. [Google Scholar] [CrossRef]
- Forman, H.J.; Torres, M. Reactive oxygen species and cell signaling: Respiratory burst in macrophage signaling. Am. J. Respir. Crit. Care Med. 2002, 166, S4–S8. [Google Scholar] [CrossRef]
- Miller, S.; Walker, S.W.; Arthur, J.R.; Nicol, F.; Pickard, K.; Lewin, M.H.; Howie, A.F.; Beckett, G.J. Selenite protects human endothelial cells from oxidative damage and induces thioredoxin reductase. Clin. Sci. 2001, 100, 543–550. [Google Scholar] [CrossRef]
- Tang, R.; Liu, H.; Wang, T.; Huang, K. Mechanisms of selenium inhibition of cell apoptosis induced by oxysterols in rat vascular smooth muscle cells. Arch. Biochem. Biophys. 2005, 441, 16–24. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Bilgic, E.; Alili, L.; Sies, H.; Brenneisen, P. Selenoprotein P protects endothelial cells from oxidative damage by stimulation of glutathione peroxidase expression and activity. Free Radic. Res. 2006, 40, 936–943. [Google Scholar] [CrossRef]
- Forceville, X.; Aouizerate, P.; Guizard, M. Septic shock and selenium administration. Therapie 2001, 56, 653–661. [Google Scholar]
- Zhang, F.; Yu, W.; Hargrove, J.L.; Greenspan, P.; Dean, R.G.; Taylor, E.W.; Hartle, D.K. Inhibition of TNF-alpha induced ICAM-1, VCAM-1 and E-selectin expression by selenium. Atherosclerosis 2002, 161, 381–386. [Google Scholar] [CrossRef]
- Ahrens, I.; Ellwanger, C.; Smith, B.K.; Bassler, N.; Chen, Y.C.; Neudorfer, I.; Ludwig, A.; Bode, C.; Peter, K. Selenium supplementation induces metalloproteinase-dependent L-selectin shedding from monocytes. J. Leukoc. Biol. 2008, 83, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.T.; Zhou, L.N.; Huang, C.J.; Hua, X.; Jian, R.; Su, B.H.; Fang, F. Selenium inhibits high glucose- and high insulin-induced adhesion molecule expression in vascular endothelial cells. Arch. Med. Res. 2008, 39, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Rose, A.H.; Hoffmann, P.R. The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2012, 16, 705–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speckmann, B.; Steinbrenner, H. Selenium and selenoproteins in inflammatory bowel diseases and experimental colitis. Inflamm. Bowel Dis. 2014, 20, 1110–1119. [Google Scholar] [CrossRef]
- Bizerea, T.O.; Dezsi, S.G.; Marginean, O.; Stroescu, R.; Rogobete, A.; Bizerea-Spiridon, O.; Ilie, C. The Link Between Selenium, Oxidative Stress and Pregnancy Induced Hypertensive Disorders. Clin. Lab. 2018, 64, 1593–1610. [Google Scholar] [CrossRef]
- Büttner, P.; Obradovic, D.; Wunderlich, S.; Feistritzer, H.J.; Holzwirth, E.; Lauten, P.; Fuernau, G.; de Waha-Thiele, S.; Desch, S.; Thiele, H. Selenoprotein P in Myocardial Infarction With Cardiogenic Shock. Shock 2020, 53, 58–62. [Google Scholar] [CrossRef]
- Bor, M.V.; Cevìk, C.; Uslu, I.; Güneral, F.; Düzgün, E. Selenium levels and glutathione peroxidase activities in patients with acute myocardial infarction. Acta Cardiol. 1999, 54, 271–276. [Google Scholar]
- Navarro-Alarcón, M.; López-Garcia de la Serrana, H.; Pérez-Valero, V.; López-Martínez, C. Serum and urine selenium concentrations in patients with cardiovascular diseases and relationship to other nutritional indexes. Ann. Nutr. Metab. 1999, 43, 30–36. [Google Scholar] [CrossRef]
| AMI (n = 19) | Control (n = 25) | p-Value | |
|---|---|---|---|
| Mean ± SD | Mean ± SD | 95% CI | |
| Age (years) | 65.15 ± 11.53 | 73.20 ± 9.06 | <0.001 |
| DM | 4 (21%) | 3 (12%) | 0.43 |
| HTN | 14 (74%) | 15 (60%) | 0.43 |
| Hyperlipidemia | 5 (26%) | 6 (24%) | 0.86 |
| Aspirin | 19 (100%) | 3 (12%) | <0.001 |
| Plavix | 19 (100%) | 0 (0%) | <0.001 |
| ACEI/ARB | 17 (89%) | 14 (56%) | 0.01 |
| BB | 16 (84%) | 1 (4%) | <0.001 |
| BH (cm) | 166.08 ± 6.87 | 158.92 ± 6.62 | <0.001 |
| BW (kg) | 72.83 ± 13.04 | 61.08 ± 11.94 | <0.001 |
| SBP (mmHg) | 137.53 ± 4.45 | 135.76 ± 3.62 | 0.35 |
| DBP (mmHg) | 85.79 ± 3.16 | 83.52 ± 2.86 | 0.83 |
| WBC (103/μL) | 7.62 ± 3.28 | 6.52 ± 1.81 | 0.14 |
| Hb (g/dL) | 13.75 ± 2.57 | 13.00 ± 2.11 | 0.23 |
| GLU AC (mg/dL) | 114.62 ± 35.73 | 112.23 ± 36.97 | 0.81 |
| HbA1c (%) | 6.47 ± 1.34 | 6.23 ± 0.91 | 0.44 |
| GOT (U/L) | 36.46 ± 29.90 | 23.07 ± 9.13 | 0.04 |
| GPT (U/L) | 26.59 ± 16.43 | 17.61 ± 10.96 | 0.02 |
| Bil-T (mg/dL) | 0.92 ± 0.46 | 0.71 ± 0.38 | 0.22 |
| Bil-D (mg/dL) | 0.26 ± 0.26 | 0.15 ± 0.07 | 0.37 |
| ALP (U/L) | 115.55 ± 152.46 | 50.33 ± 12.90 | 0.49 |
| rGT (U/L) | 75.82 ± 84.90 | 17.67 ± 8.50 | 0.27 |
| BUN (mg/dL) | 21.54 ± 12.99 | 17.96 ± 5.78 | 0.29 |
| Cr (mg/dL) | 1.39 ± 2.08 | 0.96 ± 0.27 | 0.32 |
| eGFR (mL/min/1.73 m2) | 82.79 ± 47.78 | 79.32 ± 21.79 | 0.74 |
| UA (mg/dL) | 6.29 ± 1.60 | 5.70 ± 1.42 | 0.17 |
| Na (mmol/L) | 138.23 ± 5.80 | 138.31 ± 3.79 | 0.96 |
| K (mmol/L) | 4.20 ± 0.44 | 4.10 ± 0.58 | 0.46 |
| Ca (mmol/L) | 2.29 ± 0.11 | 2.29 ± 0.17 | 0.97 |
| Mg (mmol/L) | 0.80 ± 0.12 | 0.90 ± 0.05 | 0.12 |
| CHO (mg/dL) | 151.27 ± 39.44 | 166.79 ± 44.16 | 0.16 |
| TG (mg/dL) | 126.16 ± 55.33 | 149.96 ± 111.52 | 0.27 |
| LDL (mg/dL) | 93..06 ± 39.12 | 94.00 ± 29.19 | 0.93 |
| HDL (mg/dL) | 46.68 ± 12.56 | 46.70 ± 12.60 | 0.99 |
| CK (U/L) | 895.04 ± 1862.19 | 105.38 ± 20.33 | 0.04 |
| CK-MB (U/L) | 57.60 ± 118.37 | 2.69 ± 0.91 | 0.53 |
| Tn-I (ng/mL) | 16.73 ± 21.52 | 0.01 ± 0.00 | 0.52 |
| CRP (mg/dL) | 3.51 ± 4.59 | 0.06 ± 0.04 | 0.03 |
| NT-pro BNP (pg/mL) | 1994.68 ± 4037.07 | 1816.60 ± 3014.64 | 0.94 |
| LDH (U/L) | 221.50 ± 137.27 | 150.67 ± 52.77 | 0.41 |
| Lactic acid (mmol/L) | 1.56 ± 1.00 | 0.81 ± 0.16 | 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, F.-C.; Tsai, C.-F.; Huang, P.-S.; Shih, C.-Y.; Tsai, M.-H.; Hwang, J.-J.; Wang, Y.-C.; Chuang, E.Y.; Tsai, C.-T.; Chang, S.-N. The Gut Microbiome, Seleno-Compounds, and Acute Myocardial Infarction. J. Clin. Med. 2022, 11, 1462. https://doi.org/10.3390/jcm11051462
Chiu F-C, Tsai C-F, Huang P-S, Shih C-Y, Tsai M-H, Hwang J-J, Wang Y-C, Chuang EY, Tsai C-T, Chang S-N. The Gut Microbiome, Seleno-Compounds, and Acute Myocardial Infarction. Journal of Clinical Medicine. 2022; 11(5):1462. https://doi.org/10.3390/jcm11051462
Chicago/Turabian StyleChiu, Fu-Chun, Chin-Feng Tsai, Pang-Shuo Huang, Ching-Yu Shih, Mong-Hsun Tsai, Juey-Jen Hwang, Yi-Chih Wang, Eric Y. Chuang, Chia-Ti Tsai, and Sheng-Nan Chang. 2022. "The Gut Microbiome, Seleno-Compounds, and Acute Myocardial Infarction" Journal of Clinical Medicine 11, no. 5: 1462. https://doi.org/10.3390/jcm11051462
APA StyleChiu, F.-C., Tsai, C.-F., Huang, P.-S., Shih, C.-Y., Tsai, M.-H., Hwang, J.-J., Wang, Y.-C., Chuang, E. Y., Tsai, C.-T., & Chang, S.-N. (2022). The Gut Microbiome, Seleno-Compounds, and Acute Myocardial Infarction. Journal of Clinical Medicine, 11(5), 1462. https://doi.org/10.3390/jcm11051462







