Abstract
Chronic kidney disease (CKD), a damaged condition of the kidneys, is a global public health problem that can be caused by diabetes, hypertension, and other disorders. Recently, the MANBA gene was identified in CKD by integrating CKD-related variants and kidney expression quantitative trait loci (eQTL) data. This study evaluated the effects of MANBA gene variants on CKD and kidney function-related traits using a Korean cohort. We also analyzed the association of MANBA gene variants with kidney-related traits such as the estimated glomerular filtration rate (eGFR), and blood urea nitrogen (BUN), creatinine, and uric acid levels using linear regression analysis. As a result, 14 single nucleotide polymorphisms (SNPs) were replicated in CKD (p < 0.05), consistent with previous studies. Among them, rs4496586, which was the most significant for CKD and kidney function-related traits, was associated with a decreased CKD risk in participants with the homozygous minor allele (CC), increased eGFR, and decreased creatinine and uric acid concentrations. Furthermore, the association analysis between the rs4496586 genotype and MANBA gene expression in human tubules and glomeruli showed high MANBA gene expression in the minor allele carriers. In conclusion, this study demonstrated that MANBA gene variants were associated with CKD and kidney function-related traits in a Korean cohort.
1. Introduction
Chronic kidney disease (CKD) is an important health problem worldwide and increases mortality and morbidity by raising the risk of several diseases [1,2]. According to the Global Burden of Disease (GBD) study, the global prevalence of CKD in 2017 was estimated at 9.7%, with approximately 697.5 million people affected [3]. The prevalence of CKD in Korea in 2017 was 3% according to the chronic disease health statistics of the Korea Disease Control and Prevention Agency (KDCA) (https://health.cdc.go.kr/ (accessed on 13 April 2021)). In addition, the GBD study reported that fatality from CKD in 2017 was 1.2 million, the 12th primary cause of death worldwide. The common causes of impaired kidney function are diabetes, hypertension, and glomerulonephritis, which can be evaluated by the estimated glomerular filtration rate (eGFR) [4]. Previous studies have estimated the heritability of CKD to be 20–80% by measuring the contribution of genetic effects in a population of patients with CKD [5,6,7]. Therefore, since CKD is also affected by genetic factors, it is important to identify the risk factors associated with CKD development and genes in individuals with CKD.
A meta-analysis of genome-wide association studies (GWAS) on CKD and kidney function-related traits was performed in European, Asian, and African populations [8,9,10]. Previous studies have determined non-coding genetic variants related to CKD through GWAS but the elucidation of the underlying genes and mechanisms was limited. Therefore, a recent study performed expression quantitative trait loci (eQTL) analysis of renal glomeruli and tubular tissue and found 27 candidate genes that could be potential causes of kidney disease development [11]. Among them, MANBA, PGAP3, and CASP9 were confirmed to have functional roles in kidney disease development in previous animal studies [12,13,14]. Another study reported that variants of the MANBA gene identified using eQTL analysis and CKD-related variants identified using GWAS analysis showed statistically significant co-localization [12]. In addition, Gu et al. demonstrated the in vivo mechanism by which the MANBA gene affects kidney disease [15].
The β-mannosidase (MANBA) gene, encoding lysosome β-mannosidase, is located on human chromosome 4q24 [16,17]. Although many genetic variants associated with CKD have been identified, correlation analysis focusing on MANBA gene variants that directly affect kidney diseases are rare. Therefore, in this study, the association analysis of MANBA gene variants with CKD and kidney function-related traits was performed in the Korean Genome and Epidemiology Study (KoGES) cohort. We found 20 single nucleotide polymorphisms (SNPs) that showed a statistically significant association with CKD and kidney function-related traits among 229 SNPs of the MANBA gene. In addition, rs4496586, which had the highest significance for CKD, was associated with MANBA gene expression in renal tubules and glomeruli. These results replicate previous studies that functionally demonstrated the association between kidney disease and the MANBA gene.
2. Materials and Methods
2.1. Participants
The epidemiological data used in this study were obtained from the Health Examinee (HEXA) cohort of the Korean Genome and Epidemiology Study (KoGES). From 2004 to 2013, a total of 173,208 participants aged over 40 years were recruited in the HEXA cohort. Among them, genotype data on 58,700 participants were available. A more detailed description of the HEXA cohort has been described previously [18]. The Institutional Review Board (IRB) of the Korea Disease Control and Prevention Agency (KDCA, KBN-2021-003 (26 January 2021)) and Soonchunhyang University (202012-BR-086-01 (15 December 2020)) approved the study protocol. Written informed consent was obtained from all subjects. All methods were performed in accordance with the relevant guidelines and regulations.
2.2. Basic Characteristics
The parameters measured in this study included physical measurements such as height, and weight, and biochemical measurements such as uric acid, blood urea nitrogen (BUN), and serum creatinine levels. The characteristics of the participants in this study are shown in Table 1. Body mass index (BMI) was calculated by dividing body weight (kg) by height squared (m2). Blood samples were collected after an 8-h fast and all plasma samples were measured biochemically. Serum creatinine concentrations were measured by the Jaffe method using an automatic analyzer (Hitachi, Tokyo, Japan).

Table 1.
Characteristics of participants in the Korean population.
2.3. Definition of Chronic Kidney Disease
For the case-control analysis of CKD, control groups (30,813 participants) and cases (1130 participants) were classified according to the recommendations of the Kidney Disease Improving Global Outcome (KDIGO) guidelines. CKD was defined as an eGFR of <60 mL/min/1.72 m2 and a history of renal disease. Non-CKD was defined as an eGFR of ≥90 mL/min/1.72 m2. Known as the best estimate of kidney function, the eGFR was calculated through the creatinine-based modification of diet in renal disease (MDRD) equation. Kidney function was additionally assessed by BUN, uric acid, and serum creatinine levels.
2.4. Genotyping
The genotype data were provided by the Center for Genome Science, Korea National Institute of Health. A total of 58,700 genomic DNA samples isolated from peripheral blood were genotyped using the Affymetrix Axiom® Array (Affymetrix, Santa Clara, CA, USA). The genotype data were confirmed using the Korean-Chip (K-CHIP, Seoul, Korea) acquired by the K-CHIP consortium. Detailed information on Korean chips has been described previously [19]. Samples with less than 96%–99% genotyping accuracy, excessive heterozygosity, or sex inconsistency were excluded. Markers with missing genotype rates of <95%, a minor allele frequency of <1%, and Hardy-Weinberg equilibrium p-values of <1 × 10−6 were also excluded to control the quality of the genotyping results. After quality control, imputation analysis was performed using the 1000 genome phase 3 dataset (reference panel) in IMPUTE v2 software. A total of 8,056,211 SNPs were included in the study. This study selected SNPs significantly related to kidney disease in the MANBA gene. The location of the SNPs was identified using National Center for Biotechnology Information (NCBI) Human Genome Build 37 (hg19).
2.5. Statistical Analysis
Statistical analyses were conducted with PLINK version 1.90 beta (https://www.cog-genomics.org/plink2 (accessed on 13 April 2021)) [20] and PASW Statistics version 18.0 (SPSS Inc. Chicago, IL, USA). A total of 58,700 participants were classified as CKD cases and controls according to KDIGO guidelines, and logistic regression analysis was performed to calculate the odds ratios (ORs) and 95% confidence intervals (95% CIs). Linear regression analysis was used to analyze the association between BUN, uric acid, and creatinine levels, and eGFR, which are traits related to kidney function, and MANBA genes. Logistic and linear regression analyses were performed based on the additive genetic model after age and gender adjustments. The significance threshold (p < 6.76 × 10−4) was adjusted through Bonferroni correction. Regional plots were created using the LocusZoom program (http://locuszoom.org/ (accessed on 13 April 2021)). A conditional analysis was performed to identify secondary association signals. The publicly available Human Kidney eQTL database (http://susztaklab.com/eqtl (accessed on 13 April 2021)) was used to determine whether the variants significantly associated with CKD affected the expression level of the MANBA gene.
3. Results
3.1. Participant Characteristics
The clinical characteristics of the 58,700 participants (20,293 men, and 38,407 women) in this study are shown in Table 1. To analyze the association between CKD and variants in the MANBA gene, CKD was divided into cases and controls using the eGFR. BUN, uric acid, and creatinine levels, which are related to kidney function, were increased in the case group compared to the control group. The comparison between the CKD cases and controls using the Student’s t-test showed that all characteristics except BMI were significantly different.
3.2. Association Analysis of the MANBA Gene Variants with CKD and Kidney Function-Related Traits
The present study investigated the association of 229 SNPs in the MANBA gene with CKD and kidney function-related traits such as eGFR, BUN, creatinine, and uric acid levels. Before analyzing the association, we selected SNPs that tagged other SNPs with r2 < 0.8 to show the independently related SNPs of the MANBA gene. As a result, 74 of 229 SNPs were identified, and finally, 20 SNPs achieved significance of p < 6.76 × 10−4 (Bonferroni threshold: p = 0.05/74) for association with CKD or kidney function-related traits in the HEXA cohort (Table 2). Most of the 20 SNPs are significantly associated with kidney function-related traits such as eGFR, creatinine and uric acid levels. However, there was no association between SNPs in the MANBA gene and BUN levels. The odds ratio of CKD was 0.87 (95% CI 0.80–0.95, p = 2.61 × 10−3) for the rs4496586 which showed the highest association with CKD. The linear regression analysis showed a significant association of rs4496586with eGFR (β = 0.565, p = 1.43 × 10−9), creatinine (β = −0.004, p = 1.04 × 10−8), and uric acid levels (β = −0.019, p = 1.83 × 10−3). These results were consistent with the reduced risk of CKD in patients carrying the minor C allele of rs4496586.

Table 2.
Results of association analysis of the SNPs in the MANBA gene with chronic kidney disease and kidney function-related traits (r2 < 0.8).
Additionally, the Human Kidney eQTL database was used to analyze the association between the rs4496586 genotype, which had a high significance for CKD, and the MANBA gene expression in the renal tubules and glomeruli. The expression level of the MANBA gene was significantly increased in patients with the minor C allele of rs4496586 in the renal tubules and glomeruli (Figure 1).
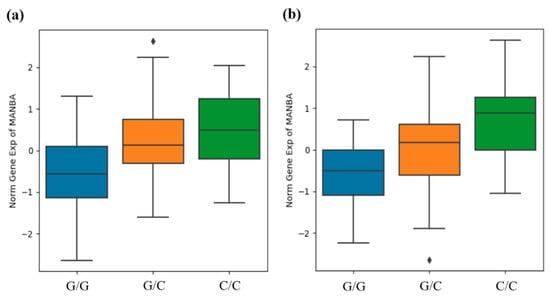
Figure 1.
Rs4496586 genotype and MANBA gene expression association in human kidney tubules (a) and glomeruli (b). The data are from the Human Kidney eQTL database (http://susztaklab.com/eqtl (accessed on 13 April 2021)). MANBA gene expression for the rs4496586 genotype in tubules (β = 0.636, p = 1.44 × 10−6) and glomeruli (β = 0.609, p = 2.49 × 10−6) was confirmed and statistically significant. p-value was calculated by linear regression. Center lines show medians, box limits indicate 25th and 75th percentiles, and whiskers extend to the 5th and 95th percentiles, outliers are represented by diamonds.
The SNPs of the MANBA gene, which were significant in the association analysis for CKD and kidney function-related traits, were shown using LocusZoom (http://csg.sph.umich.edu/locuszoom/ (accessed on 13 April 2021)), confirming the regional plots (Figure 2, Supplementary Figure S1). As a result, this study detected two independent signals through the regional plots. These SNPs of signal 1 (rs4496586) and signal 2 (rs223489) showed significantly high levels of CKD (p = 2.61 × 10−3 and p = 7.08 × 10−3), eGFR (p = 1.43 × 10−9 and p = 9.42 × 10−10), creatinine (p = 1.04 × 10−8 and p = 6.72 × 10−10), and uric acid (p = 1.83 × 10−3 and p = 8.61 × 10−3). To clearly identify secondary signals, this study performed conditional analyses, including the most significant SNPs of CKD, eGFR, creatinine, and uric acid in a stepwise manner. eGFR and creatinine found additional independent signals at the MANBA locus after conditioning for the most significant variants. In contrast, the conditional analyses for CKD and uric acid detected no conditionally independent signals (Supplementary Table S1).
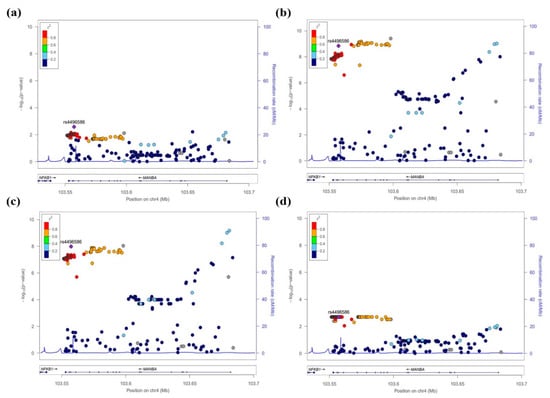
Figure 2.
Regional plots of the association results of MANBA SNPs with CKD (a) and kidney function-related traits such as eGFR (b), creatinine (c), and uric acid levels (d) in the HEXA cohort. The statistical significance (−log10 p-value) of the analyzed SNPs is plotted. The colors indicate the linkage disequilibrium (r2) between rs4496586 and the remaining SNPs. The genetic recombination rates are shown on the right y-axis. The plots were generated by the LocusZoom program (http://csg.sph.umich.edu/locuszoom (accessed on 13 April 2021)).
4. Discussion
CKD is a complex disease caused by impaired kidney function [21]. In addition, CKD is associated with serious complications such as cardiovascular disease, hyperlipidemia, hypertension, and metabolic bone disease [22,23,24]. The prevalence of CKD reported in Korea is lower than that confirmed in other countries [3,25]. However, the early diagnosis and treatment of CKD are important in preventing various complications [26]. This study performed an association analysis of variants in the MANBA gene with CKD and kidney function-related traits in a Korean cohort. The study found that 11 SNPs were not only statistically significant in CKD but were also significantly associated with kidney function-related traits such as creatinine and uric acid levels (Table 2). A previous study reported that the minor A allele of rs228611 was significantly associated with the eGFR (β = −0.0056, p = 3.58 × 10−12) [27]. Similar to previous results, this study showed that rs228611 with a minor G allele was associated with increases in the eGFR (β = 0.483, p = 2.51 × 10−7) (Supplementary Table S2). Another study identified rs223489 as a candidate eGFR variant (Effect allele = A, β = −0.0027, p = 2.6 × 10−17) by analyzing the human ortholog in the GWAS summary statistics (eGFR) for genes associated with abnormal kidney morphology in mice [8]. Therefore, it was suggested that rs223489, which was not previously identified as important, significantly affected the eGFR. Consistent with the previous study, our results also replicated the statistically significant association of rs223489 with the eGFR (Effect allele = G, β = 0.599, p = 2.6 × 10−10) in Koreans. In addition, rs223489 was detected as an important signal in CKD, eGFR, creatinine, and uric acid through the regional plots (Supplementary Figure S1).
Previous studies have reported a significant association between the eGFR and genetic variants in chromosome 4 through GWAS [9,28,29]. In addition, NFKB1 was suggested as a target gene for kidney function [27]. Unlike previous studies, Ko et al. identified candidate genes for CKD through an integrative analysis that combined CKD-associated variants and kidney eQTL results [12]. The expression of NFKB1 gene did not show any significance in the integrative analysis, and the MANBA gene, which showed statistically significant co-localization between CKD-associated variants and kidney eQTL results, was proposed as a potential target gene for the GWAS variant related to kidney function. Furthermore, the study revealed that kidney function was impaired when MANBA gene expression was suppressed in zebrafish. Thus, increasing the expression of the MANBA gene might be a potential new means to treat kidney dysfunction.
Qiu et al. suggested that many diseases are cell-type-specific, rather than organ-specific [11]. Thus, they performed eQTL analysis of the renal tubules and glomeruli and identified genes that may cause kidney disease, including the MANBA gene. In the present study rs4496586 had the highest significance in the association analysis between CKD and variants in the MANBA gene. Therefore, this study analyzed the association between the rs4496586 genotype and MANBA gene expression in the renal tubules and glomeruli. Our results showed that the expression level of the MANBA gene was significantly higher in samples with a minor allele of rs4496586 (Figure 1). In brief, rs4496586 increased the expression of the MANBA gene and decreased the risk of CKD in patients possessing a minor allele (C). These results are consistent with those of a previous study demonstrating the association between the expression of the MANBA gene and renal function in vivo [12].
Interestingly, a recent study conducted mechanistic experiments for the rs6847587 variant significantly associated with MANBA gene expression in renal tubules using mice and cells [15]. The results showed that a reduction in MANBA gene expression affected not only the structure and function of lysosomes, but also the endocytosis and autophagy pathways in vivo, leading to tubular damage, inflammation activation, and fibrosis. However, the study group mentioned the limitation of not using CKD-related variants. Our results from analyzing the association between CKD and MANBA gene variants showed that rs6847587 significantly reduced the risk of CKD in participants with the minor allele. The presence of the rs6847587 variant with the minor allele was associated with increased MANBA gene expression. Thus, these results are consistent with a previous study demonstrating the association of the MANBA gene in the development of kidney disease.
In summary, this study focused on the MANBA gene and confirmed the association of genetic variants with CKD and kidney function-related traits such as eGFR, BUN, creatinine, and uric acid levels based on KoGES. The MANBA gene variants showed significant associations with CKD, consistent with a recent study demonstrating that MANBA gene variants were related to kidney function through an integrative analysis of eQTL and CKD-related GWAS results. Moreover, this study confirmed MANBA gene expression according to genotypes through eQTL analysis and demonstrated that the MANBA gene variants affecting renal tubules and glomeruli were significantly related to CKD. However, in vivo studies are needed to determine the direct effects of MANBA gene variants, which are highly correlated with CKD, on kidney function. Future studies also need to evaluate the association between CKD and MANBA gene variants through population-specific analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jcm10112255/s1, Table S1: Conditional analyses of multiple SNPs at the MANBA locus. Table S2: Results of an association analysis of the SNPs in the MANBA gene with chronic kidney disease and kidney function-related traits. Figure S1: Regional plots of the association results of MANBA SNPs with CKD (a) and kidney function-related traits such as eGFR (b), creatinine (c), and uric acid levels (d) in the HEXA cohort. The statistical significance (−log10 p-value) of the analyzed SNPs is plotted. The colors indicate the linkage disequilibrium (r2) between rs223489 and the remaining SNPs. The genetic recombination rates are shown on the right y-axis. The plots were generated by the LocusZoom program (http://csg.sph.umich.edu/locuszoom/ (accessed on 13 April 2021)).
Author Contributions
Conceptualization, H.-R.K. and H.-S.J.; methodology, H.-S.J.; software, H.-R.K.; validation, H.-R.K., H.-S.J. and Y.-B.E.; investigation, H.-R.K.; resources, Y.-B.E.; data curation, H.-R.K. and H.-S.J.; writing—original draft preparation, H.-R.K. and H.-S.J.; writing— review and editing, H.-S.J. and Y.-B.E.; supervision, H.-S.J. and Y.-B.E.; project administration, Y.-B.E.; funding acquisition, Y.-B.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Soonchunhyang University research fund and a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [NRF-2020R1F1A1071977].
Institutional Review Board Statement
This study was approved by the Institutional Review Board of the Korea Disease Control and Prevention Agency (KDCA, KBN-2021-003 (26 January 2021)) and Soonchunhyang University (202012-BR-086-01 (15 December 2020)).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethnical concerns.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Levey, A.S.; Atkins, R.; Coresh, J.; Cohen, E.P.; Collins, A.J.; Eckardt, K.U.; Nahas, M.E.; Jaber, B.L.; Jadoul, M.; Levin, A.; et al. Chronic kidney disease as a global public health problem: Approaches and initiatives—A position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007, 72, 247–259. [Google Scholar] [CrossRef]
- Tonelli, M.; Wiebe, N.; Culleton, B.; House, A.; Rabbat, C.; Fok, M.; McAlister, F.; Garg, A.X. Chronic kidney disease and mortality risk: A systematic review. J. Am. Soc. Nephrol. 2006, 17, 2034–2047. [Google Scholar] [CrossRef]
- Carney, E.F. The impact of chronic kidney disease on global health. Nat. Rev. Nephrol. 2020, 16, 251. [Google Scholar] [CrossRef]
- Parmar, M.S. Chronic renal disease. BMJ 2002, 325, 85–90. [Google Scholar] [CrossRef]
- MacCluer, J.W.; Scavini, M.; Shah, V.O.; Cole, S.A.; Laston, S.L.; Voruganti, V.S.; Paine, S.S.; Eaton, A.J.; Comuzzie, A.G.; Tentori, F.; et al. Heritability of measures of kidney disease among zuni Indians: The Zuni Kidney Project. Am. J. Kidney Dis. 2010, 56, 289–302. [Google Scholar] [CrossRef]
- Langefeld, C.D.; Beck, S.R.; Bowden, D.W.; Rich, S.S.; Wagenknecht, L.E.; Freedman, B.I. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am. J. Kidney Dis. 2004, 43, 796–800. [Google Scholar] [CrossRef]
- Smyth, L.J.; Duffy, S.; Maxwell, A.P.; McKnight, A.J. Genetic and epigenetic factors influencing chronic kidney disease. Am. J. Physiol.-Ren. 2014, 307, F757–F776. [Google Scholar] [CrossRef]
- Wuttke, M.; Li, Y.; Li, M.; Sieber, K.B.; Feitosa, M.F.; Gorski, M.; Tin, A.; Wang, L.; Chu, A.Y.; Hoppmann, A.; et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2019, 51, 957–972. [Google Scholar] [CrossRef]
- Okada, Y.; Sim, X.; Go, M.J.; Wu, J.Y.; Gu, D.; Takeuchi, F.; Takahashi, A.; Maeda, S.; Tsunoda, T.; Chen, P.; et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat. Genet. 2012, 44, 904–909. [Google Scholar] [CrossRef]
- Morris, A.P.; Le, T.H.; Wu, H.; Akbarov, A.; van der Most, P.J.; Hemani, G.; Smith, G.D.; Mahajan, A.; Gaulton, K.J.; Nadkarni, G.N.; et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat. Commun. 2019, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Huang, S.; Park, J.; Park, Y.; Ko, Y.A.; Seasock, M.J.; Bryer, J.S.; Xu, X.X.; Song, W.C.; Palmer, M.; et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 2018, 24, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.A.; Yi, H.G.; Qiu, C.X.; Huang, S.Z.; Park, J.; Ledo, N.; Kottgen, A.; Li, H.Z.; Rader, D.J.; Pack, M.A.; et al. Genetic-Variation-Driven Gene-Expression Changes Highlight Genes with Important Functions for Kidney Disease. Am. J. Hum. Genet. 2017, 100, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.F.; Murakami, Y.; Pagnamenta, A.T.; Daumer-Haas, C.; Fischer, B.; Hecht, J.; Keays, D.A.; Knight, S.J.; Kolsch, U.; Kruger, U.; et al. Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 2014, 94, 278–287. [Google Scholar] [CrossRef]
- Araki, T.; Hayashi, M.; Nakanishi, K.; Morishima, N.; Saruta, T. Caspase-9 takes part in programmed cell death in developing mouse kidney. Nephron Exp. Nephrol. 2003, 93, e117–e124. [Google Scholar] [CrossRef]
- Gu, X.; Yang, H.; Sheng, X.; Ko, Y.A.; Qiu, C.; Park, J.; Huang, S.; Kember, R.; Judy, R.L.; Park, J.; et al. Kidney disease genetic risk variants alter lysosomal beta-mannosidase (MANBA) expression and disease severity. Sci. Transl. Med. 2021, 13, eaaz1458. [Google Scholar] [CrossRef]
- Bedilu, R.; Nummy, K.A.; Cooper, A.; Wevers, R.; Smeitink, J.; Kleijer, W.J.; Friderici, K.H. Variable clinical presentation of lysosomal beta-mannosidosis in patients with null mutations. Mol. Genet. Metab. 2002, 77, 282–290. [Google Scholar] [CrossRef]
- Sabourdy, F.; Labauge, P.; Stensland, H.M.; Nieto, M.; Garces, V.L.; Renard, D.; Castelnovo, G.; de Champfleur, N.; Levade, T. A MANBA mutation resulting in residual beta-mannosidase activity associated with severe leukoencephalopathy: A possible pseudodeficiency variant. BMC Med. Genet. 2009, 10, 84. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G.; KoGES Group. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, 1350. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Kim, Y.J.; Han, S.; Hwang, M.Y.; Shin, D.M.; Park, M.Y.; Lu, Y.; Yoon, K.; Jang, H.M.; Kim, Y.K.; et al. The Korea Biobank Array: Design and identification of coding variants associated with blood biochemical traits. Sci. Rep. 2019, 9, 1382. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Romagnani, P.; Remuzzi, G.; Glassock, R.; Levin, A.; Jager, K.J.; Tonelli, M.; Massy, Z.; Wanner, C.; Anders, H.J. Chronic kidney disease. Nat. Rev. Dis. Primers 2017, 3, 17088. [Google Scholar] [CrossRef]
- Thomas, R.; Kanso, A.; Sedor, J.R. Chronic kidney disease and its complications. Prim. Care 2008, 35, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Go, A.S.; Chertow, G.M.; Fan, D.; McCulloch, C.E.; Hsu, C.Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med. 2004, 351, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- National Kidney, F. K/DOQI clinical practice guidelines for bone metabolism and disease in chronic kidney disease. Am. J. Kidney Dis. 2003, 42, S1–S201. [Google Scholar] [CrossRef]
- Bikbov, B.; Purcell, C.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.K.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef]
- Fraser, S.D.; Blakeman, T. Chronic kidney disease: Identification and management in primary care. Pragmat. Obs. Res. 2016, 7, 21–32. [Google Scholar] [CrossRef]
- Pattaro, C.; Teumer, A.; Gorski, M.; Chu, A.Y.; Li, M.; Mijatovic, V.; Garnaas, M.; Tin, A.; Sorice, R.; Li, Y.; et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016, 7, 10023. [Google Scholar] [CrossRef]
- Pattaro, C.; Kottgen, A.; Teumer, A.; Garnaas, M.; Boger, C.A.; Fuchsberger, C.; Olden, M.; Chen, M.H.; Tin, A.; Taliun, D.; et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012, 8, e1002584. [Google Scholar] [CrossRef]
- Kottgen, A.; Glazer, N.L.; Dehghan, A.; Hwang, S.J.; Katz, R.; Li, M.; Yang, Q.; Gudnason, V.; Launer, L.J.; Harris, T.B.; et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 2009, 41, 712–717. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).