Abstract
Annual vaccination is considered as the main preventive strategy against seasonal influenza. Due to the highly variable nature of major viral antigens, such as hemagglutinin (HA) and neuraminidase (NA), influenza vaccine strains should be regularly updated to antigenically match the circulating viruses. The influenza virus nucleoprotein (NP) is much more conserved than HA and NA, and thus seems to be a promising target for the design of improved influenza vaccines with broad cross-reactivity against antigenically diverse influenza viruses. Traditional subunit or recombinant protein influenza vaccines do not contain the NP antigen, whereas live-attenuated influenza vaccines (LAIVs) express the viral NP within infected cells, thus inducing strong NP-specific antibodies and T-cell responses. Many strategies have been explored to design broadly protective NP-based vaccines, mostly targeted at the T-cell mode of immunity. Although the NP is highly conserved, it still undergoes slow evolutionary changes due to selective immune pressure, meaning that the particular NP antigen selected for vaccine design may have a significant impact on the overall immunogenicity and efficacy of the vaccine candidate. In this review, we summarize existing data on the conservation of the influenza A viral nucleoprotein and review the results of preclinical and clinical trials of NP-targeting influenza vaccine prototypes, focusing on the ability of NP-specific immune responses to protect against diverse influenza viruses.
1. Introduction
Respiratory viral infections are frequent human pathologic conditions caused predominantly by seasonal viruses [1]. Although the course of these infections is usually mild, there are dangerous complications associated with opportunistic diseases that can exacerbate the course of comorbid conditions, especially in the elderly, young children, and immunocompromised individuals [2]. One of the most dangerous respiratory viral infections is influenza, which poses a serious threat to the global economy and public health and requires the development of effective means of prevention and treatment, particularly taking into account the variability of pathogens [3].
The most effective measure of prevention against influenza is vaccination, which provides individual protection and serves as the basis for herd immunity [4]. At present, there exist several types of licensed influenza vaccines, including inactivated, live-attenuated, and recombinant protein vaccines. In addition, various alternative vaccine platforms, such as mRNA, DNA, viral vectors, and recombinant nanoparticles, have been investigated [5]. Viral surface proteins are most commonly used as targets for vaccine development as they mediate the interaction of the virus with the target cell receptors, the fusion of the viral and cell envelopes, and the entry of the pathogen into the host cell. Indeed, the abundance of these molecules on the virion surface, along with their usually high immunogenicity and active expression within infected cells, enables their effective presentation to the immune system, as manifested by pronounced humoral and T-cell responses during subsequent infection. However, this property of viral surface proteins also makes them a major target for viral evolution toward host defense evasion and increases their susceptibility to escape mutations [6]. This variability of surface antigens requires an almost annual update of influenza vaccine strain compositions and often leads to reduced vaccine effectiveness when there is an antigenic mismatch between circulating and vaccine strains [7,8]. Therefore, the development of influenza vaccines with broader and more durable protection against seasonal and potentially pandemic influenza strains is of high priority [9]. Various strategies have been explored to develop more broadly protective influenza vaccines, where highly conserved viral antigens may be used as a target for the induction of immune responses [10,11]. In particular, one of the most common conserved viral antigens used for universal influenza vaccine design is nucleoprotein (NP), a highly abundant internal protein involved in the transcription and replication of the viral genome, influencing the host specificity and virulence of viruses [12,13]. As NP sequences are conserved not only between different strains within the same subtype but also between different influenza A virus subtypes or between two influenza B virus lineages, their use as a target antigen has become one of the main strategies for the development of cross-protective influenza virus vaccines [14,15,16].
It has been generally recognized that the key mechanism of the immune system response to this antigen is the activation of the T-cell mode of immunity (CD8+ cytotoxic T-lymphocytes) through presentation by infected cells of NP-derived peptides in complex with major histocompatibility complex-I (MHC-I) molecules, followed by their killing [17]. In addition, an infected organism typically produces a significant amount of anti-NP antibodies with high affinity that do not exhibit neutralizing activity but are associated with a reduction in disease severity [18]. Although this antigen is not exposed on the surface of virions, it has been detected on the surface of virus-infected cells [19,20], making it a potential target for antibody-mediated immune responses, such as antibody-dependent cellular cytotoxicity (ADCC) or complement activation [21,22].
Although NP is considered to be a highly conserved protein of influenza viruses, there is a slow accumulation of mutations in this antigen as a result of genetic evolution and positive selection by host immunity [23]. In this review, we summarize current knowledge on the conservation of influenza A viral nucleoprotein and review the results of preclinical and clinical trials considering NP-targeted influenza vaccine prototypes, focusing on the ability of NP-specific immune responses to protect against diverse influenza viruses.
2. Nucleoprotein Structure and Functions
In general, the organization of the NP–genome complex and domain structure of the NP molecule are similar to those of the majority of ssRNA(-) respiratory viruses. The NP protomer includes two folded domains: an N-terminal domain (NTD) and a C-terminal core domain (CTD). It is widely accepted that the site of nucleic acid binding is usually located between the NTD and CTD lobes, forming a clamp for the viral antigenome [24] (Figure 1). The influenza virus NP molecule of influenza virus binds to 24 nucleotides, and this number is fixed [25,26]. As the NP protein tends to bind the host RNA in the absence of viral RNA, its protomers are chaperoned by phosphoprotein to maintain the monomeric state and prevent the formation of empty or defective virions [27].
The crystal structures of recombinant NP–RNA complexes have been described as regularized formations [28,29], resulting from a spontaneous assembly of multimeric forms by the interactions of the corresponding domains [14,30]. In the infected cell, NP occurs in a number of forms: its monomers are complexed with a phosphoprotein; NP oligomers are bound with RNA and organized into ribonucleocapsid, and multimeric NP constitutes the relaxed structure making the RNA accessible for polymerase [31].
The multiple functions of the NP protein in the viral life cycle include the following: (i) protection of viral RNA from degradation by cellular enzymes (by physical RNA separation and masking from immune recognition, (ii) fitting the helical structure of RNP, (iii) modulation of the transcription and replication of viral RNA templates in a histone-like manner, and (iv) induction of immunosuppression (inhibition of effector cytokine synthesis and FcR-medicated signaling) upon infection [32].
NP was initially not considered a priority target for influenza vaccine development, as this antigen is not exposed on the surface of the viral particle, and anti-NP antibodies do not possess neutralizing activity. However, further studies have indicated that a large number of virus-specific T cells are produced to NP epitopes and that anti-NP antibodies could mediate cellular protective reactions. Since the 1980s, this antigen has become an attractive target for the development of universal influenza vaccines, mainly due to the high conservation rate of the NP among antigenically distant viruses.
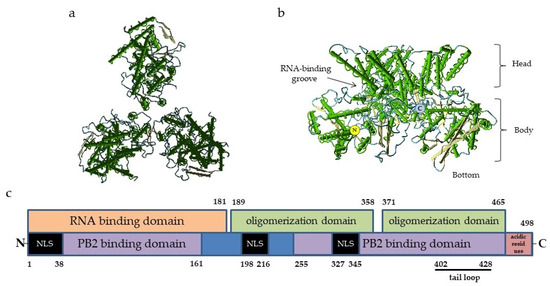
Figure 1.
Structure of influenza virus NP: (a)—Top view of nucleoprotein trimer along the nucleocapsid axis, PDB ID 2IQH. (b)—Side view of a nucleoprotein trimer; the structures of the molecule are indicated, PDB ID 2IQH; and (c)—the scheme of NP molecule domains involved in the binding of RNA and other proteins. The numbers refer to the positions of amino acid residues. The C-terminal acidic residues (indicated in red) serve as repressors of PB2 and NP binding. The black bar indicates the tail loop region responsible for the oligomerization. NLS, nuclear localization signal. The locations of the NP functional domains are given according to [28,31,33,34].
In general, the NP antigen is known to mainly elicit both cytotoxic CD4+ and CD8+ T-cell (CTL) responses, as NP-derived peptides may be synthesized by the infected cell or produced as a result of exogenous RNP phagocytosis, which then may be presented in complex with both MHC class I or II, respectively [35]. These two cell populations are able to distinguish virus-infected cells from intact cells and similarly perform specific cytolysis by granules containing perforin and granzymes [36], thus playing a pivotal role in pathogen elimination. Therefore, targeted vaccination inducing T-cell responses against conserved NP epitopes is considered to be a promising strategy for the establishment of cross-protective antiviral immunity.
Unlike another internal influenza antigen, PB1, which is equally targeted by CD4+ and CD8+ T cells [37], NP is believed to be a major target for CD8+ CTLs [17,38]. Nevertheless, CD4+ CTLs constitute a minor population of CD4+ T cells (mainly represented by T helpers) [39] and have been described as primarily targeting NP and M antigens upon infection with the influenza virus [40,41]. This fact underpins the feasibility of the development of new vaccination strategies for the elderly using vaccines based on CD4+ CTL NP epitopes, as it is well known that with age, the number of CD4+ CTLs increases, but the activity of these cells upon influenza vaccination remains unchanged [36].
As NP has been detected at the membranes of infected epitheliocytes [19], it may be targeted by the humoral immune mode. Unfortunately, little is known about the patterns of generation, mechanism of action, and protective potential of anti-NP antibodies, which are actively produced in response to infection or vaccination [21,42]. In theory, they may contribute to viral clearance, mediating innate killing reactions, such as ADCC or antibody-dependent cellular endocytosis [43]. This hypothesis is supported by data on their ability to activate natural killer (NK) cells ex vivo [21] and by the killing of influenza-infected cells in vitro induced by sera from individuals with intensive anti-NP humoral responses but low anti-HA and anti-NA titers [22]. The function of anti-NP antibodies is most likely mediated by FcεRI receptors [44].
Notably, although the major components of the immune system involved in the anti-NP response are known, additional studies are needed to clarify the details that should be taken into account when developing effective epitope-based universal influenza vaccines.
3. Development of NP-Based Broadly Protective Vaccine Prototypes
The nucleoprotein of the influenza virus has been considered an attractive target for the design of broadly protective influenza vaccines for almost four decades [45], due to the T-cell responses elicited with respect to this antigen after infection, as well as after immunization with whole-virion vaccines, being highly cross-reactive. Indeed, an early study found that cytotoxic T cells produced in mice primed with the influenza virus of a particular subtype could recognize transfectant mouse L cells expressing a NP of a different subtype, whereas no recognition was observed in heterologous HA-expressing L cells [46].
The first attempt to use nucleoprotein as a vaccine antigen was reported by Wraith et al. in 1985. In their study, a modified X31 (H3N2) influenza virus was treated with bromelain four times to release the majority of the surface glycoproteins [47]. The resulting core was further treated with ammonium deoxycholate and centrifuged at 10,000× g for 30 min. The supernatant was highly enriched with NP, although residual HA protein was also present. This NP/HA preparation was injected i.p. (50 µg) into BALB/c mice, and 3 weeks later, high levels of X31 virus-specific cytotoxic memory T cells were detected in mouse splenocytes, suggesting that NP was a major inducer of T-cell immunity [47]. To further test this hypothesis, the same group of investigators prepared more pure NP protein extract (containing less than 3% HA impurity) from the X31 virus and used it for immunogenicity and cross-protection studies in mice [48]. The main finding of this study was that 75% of the NP-primed mice survived a lethal challenge with the heterologous H1N1 virus, although the clinical manifestation of the disease was not significantly improved. These data suggest that the induced NP-specific cytotoxic T cells contributed to viral clearance but did not prevent the infection [48].
A further attempt to improve the NP-based vaccine was made by Tite et al., who expressed NP from the A/NT/60/68 (H3N2) influenza virus in Salmonella typhimurium and studied this protein as a vaccine antigen in mice. Similar to the studies with purified NPs, i.p. immunized mice were protected against lethality caused by a heterologous H1N1 virus, but still displayed considerable morbidity before recovery. Importantly, when these mice were boosted i.n. with the same antigen, improved protection was observed including accelerated clearance of the virus from the lungs. Furthermore, these i.n. boosted mice also rapidly cleared an antigenically unrelated influenza B virus [49].
A key mechanism of NP-mediated protection is thought to involve the triggering of T-cell immune responses. To make this possible, the antigen should be produced inside the target cell on the matrix of the exogenous mRNA/DNA delivered by the viral vector or lipid nanoparticles. mRNA platforms seem to be a promising tool for the development of NP-based vaccines due to their low risk of insertional mutagenesis, high potency, accelerated development cycles, and cost-effectiveness [50]. Some vaccine candidates comprising mRNA-expressing NP coated with a lipid layer have been proposed [18,51,52]; however, the best results regarding the stimulation of T-cell responses and protection against viral challenges were achieved when NP mRNA was used in combination with mRNAs encoding other influenza antigens.
Several attempts have been made to create vaccines based on NP-encoding DNA, which was intended to be administered in an adjuvant-free manner via the i.m. route. In their early study, Ulmer et al. demonstrated that an NP-based DNA vaccine evoked T-cell responses mediated by antigenic presentation through MHC-I and provided cross-protection against lethal influenza viruses in a murine challenge model [53]. Furthermore, the authors demonstrated the lymphoproliferative and Th1-type cytokine-secreting effects of the NP DNA injection. In particular, increases in the levels of IFNγ and IL-2 mediated by CD4+ T cells as a result of restimulation of splenocytes with the NP antigen in vitro were shown. T-cell depletion followed by a viral challenge revealed the effector action of CD4+ and CD8+ lymphocytes generated in response to immunization with NP DNA [54]. Epstein et al. found that mice primed with NP DNA and boosted with NP encoded by the adenoviral vector showed higher titers of specific serum IgG, as well as a significant increase in the number of effector T cells, and were better protected against the lethal challenge with various influenza variants—including the highly pathogenic H5N1 virus—than mice vaccinated with NP DNA alone [55]. Furthermore, Lo et al. used DNA and adenoviral vectors expressing NP, M2, or NP+M2 antigens to demonstrate that “DNA prime-adenoviral vector boost” vaccination induces systemic humoral and cellular responses, as well as protection against a homologous challenge in the same manner as immunization with cold-adapted viruses. However, only vaccination with DNA and the adenoviral vector encoding both NP and M2 antigens could confer protection against a challenge with the H5N1 virus [56]. Rao et al. tested the efficacy of a similar prime-boost vaccination regimen in mouse and ferret models. According to their results, HA-, NP-, and M2-bearing DNA- or adenovirus-based vaccines were able to elicit specific humoral responses, but only vaccination with HA-encoding or mixed DNAs and adenoviral vectors fully protected the animals from a lethal dose of a pathogenic H5N1 virus [57].
As a tool for delivering the NP gene into cells, several vector platforms have been proposed. In this line, a recombinant vector MVA-NP+M1 bearing the influenza antigens NP and M1 and propagated in chicken embryo fibroblasts or AGE1.CR.pIX cells was developed by Berthoud et al. [58]. To date, this vaccine has passed phases I and IIa of the clinical trials and shown an ability to stimulate a rapid T-cell response across different age groups and to boost pre-existing levels of specific T cells in aged individuals [59]. However, there were no statistically significant improvements in the outcomes of the disease caused by i.n. infection of vaccinated subjects [60]. Furthermore, to boost pre-existing cross-reactive T cells capable of protection against heterosubtypic influenza A viruses, MVA-NP+M1 was combined with a licensed inactivated influenza vaccine and tested in people over 65 years of age in phase IIb [61,62]. Despite an increase in the magnitude of the T-cell responses, the study could not demonstrate an increase in hemagglutination-inhibiting antibody titers or an association of disease outcomes with the number of IFNγ+ T cells. A pentavalent NP-encoding vaccine developed on the backbone of the Wyeth strain of vaccinia (a vector similar to MVA) presented immunogenicity and protective potential in an H5N1 challenge study [63].
Later, Dicks et al. constructed a novel replication-deficient safe vector for the expression of vaccine antigens based on the chimpanzee adenovirus ChAdOx1 [64]. The vaccine candidate ChAdOx1-NP+M1 turned out to be safe when administered to volunteers and was able to stimulate T-cell immune responses [65]. A similar vector was used by Arunkumar et al., who demonstrated the efficacy of priming with ChAdOx1-NP+M1 and boosting with MVA-NP+M1 for inducing the vigorous expansion of IFNγ+ CD4+ T cells. At the same time, the best protection against a challenge with H3N2, H7N9, and H10N8 viruses was achieved in the case of vaccination with the vectors simultaneously expressing chimeric HA, NP, and M1 [66]. Further study in a ferret model confirmed the efficacy of immunization with these ChAdOx1- and MVA-based triple-expressing vectors in a prime-boost regimen to abrogate replication of the H3N2 virus in the respiratory tract [67]. The effects of prime-boost immunization of pigs with ChAdOx2- and MVA-vectored vaccines expressing NP, M1, and NA antigens depended on the administration route: i.m. injection evoked systemic antibody responses, while aerosol immunization led to mucosal formation of specific T cells and IgGs. Notably, shedding of the H3N2 virus challenge and lung damage were reduced in both cases [68].
Adenovirus-associated virus (AAV)-vectored vaccines expressing H1, M1, and NP have been constructed by Sipo et al., which were shown to evoke potent specific T-cell and serum IgG formation in mice. However, only immunization with a trivalent vaccine improved the survival of mice challenged with the virus belonging to the same subtype [69]. In contrast, the results of Demminger et al. suggested that immunization of mice with AAV-based vaccines expressing NA, HA, or cHA alone allows not only for the induction of pronounced systemic and mucosal humoral responses, but also protects the animals against a homologous challenge [70]. The study of Lee et al. described the protective properties of the NPs of two influenza B lineages encoded in the replication-deficient Ad5 vector. The authors revealed the induction of cross-recognizing antibodies and CD8+ T-cell responses, as well as cross-protection against heterologous viruses in a mouse challenge model [16]. To develop a tool for the control of influenza in swine, the parapox orf virus was used as a vector for the expression of NP and/or HA of a swine influenza virus. The resultant vaccine candidates expressing HA and NP or HA alone were tested in pigs and were found to elicit cross-neutralizing antibodies accompanied by robust virus-specific T-cell responses. Interestingly, immunization with the vector containing both HA and NP genes led to the induction of a larger number of CD8+ T cells, the development of a Th2-biased immune response, and incremental protection against challenge infection [71].
In theory, the immunogenicity of recombinant NP may be improved with the addition of costimulatory adjuvants or by fusion with functional exogenic or oligomerization domains. Zheng et al. described the testing of a universal influenza vaccine based on the recombinant NP protein expressed in a bacterial system and administered to mice three times in combination with or without the C48/80 adjuvant. Although high serum IgG titers were detected in all of the NP-immunized mice, the challenge with different influenza strains at a lethal dose revealed the best protection in the case of i.n. administration of NP in complex with C48/80 [72]. Similar results have been obtained by Guo et al., who revealed the ability of an i.n. administered vaccine, based on the combination of recombinant NP of the A/PR/8/34 (H1N1) influenza virus and cholera toxin, to provide complete protection against the homologous virus challenge and partial protection against the heterologous H5N1 and H9N2 avian influenza viruses [73]. Another promising adjuvant for the recombinant NP protein, bis-(3′,5′)-cyclic dimeric adenosine monophosphate (c-di-AMP), has been proposed by Sanchez et al. The formulation was administered i.n. and was found to stimulate NP-specific serum and mucosal antibodies, as well as promoting robust Th1-type responses accompanied by increases in INFγ and IL-2 levels. The protective efficacy of the vaccine against weight loss and lung tissue damage was also described [74]. In a study by Cookenham et al., recombinant NP was administered to young and aged mice in complex with a novel squalene-based adjuvant SLA-SE. The study revealed a modest increase in the numbers of NP-specific CD4+ and CD8+ T cells in splenocytes and intensive generation of anti-NP IgG2c antibodies, as well as viral clearance intensification and increased survival rates; however, the authors found no significant changes in the number of memory CD8+ T cells in vaccinated mice [75]. Li et al. reported that the combination of NP and M1 recombinant influenza proteins provoked cytotoxic anti-NP (but not anti-M1) T-cell responses and conferred protection against a homologous virus challenge only when mice were immunized with proteins supplemented with a physical radiofrequency adjuvant (RFA). However, the RFA-adjuvanted antigens did not increase the levels of anti-NP and anti-M1 antibodies, compared to proteins mixed with the AddaVax adjuvant, and did not increase the serum level of the proinflammatory cytokine IL-6. Thus, RFA likely acts as a mild T-cell response stimulator [76]. Furthermore, the authors managed to enhance NP-induced CD8+ T-cell responses and improve the protection against lethal viral challenge infections by using the CpG 1018 adjuvant, instead of the AddaVax. This change in adjuvant also led to a switching of the type of immune response to a Th1-biased (major antiviral effector pathway mediated by IFNγ-secreting cells) instead of a Th1/Th2-balanced response (mediated by IL4-producing T lymphocytes) [77].
The induction of high levels of serum and mucosal anti-NP antibodies and a robust T-cell response have been observed after the i.n. immunization of mice with recombinant NP fused with a cell-penetrating trans-activator (TAT) protein from the human immunodeficiency virus-1 (HIV-1). This easy-to-deliver vaccine variant provided effective protection against the PR8, H9N2, and H3N2 viruses in a murine challenge study [78]. Using the same strategy, Tan et al. created a fusion protein consisting of recombinant NP as a displaying component and M2e antigen, both to study the changes in antigenicity of these proteins caused by point mutations and to propose a universal influenza vaccine candidate. While NP substitutions affected the binding of this antigen with the anti-NP antibody, mutated M2e continued to be recognized by anti-M2e IgG [79].
As another tool for more effective delivery and antigenic presentation of recombinant NP, the i.n. administration of the chitosan-based nanoparticles containing an inner NP(H9N2) core and outer HA1(H9N2) layer has been described. These stable nanobodies were generated using the ionic gelation method. Higher titers of anti-HA and anti-NP antibodies and an increased number of activated CD4+/CD8+ T cells were found in chickens immunized with HA1/NP nanoparticles compared with inactivated vaccine administration, which resulted in reduced viral shedding upon challenge [80]. Other novel double-layered nanoparticles, composed of HA- or M2-enriched shells and an NP-containing core that were able to induce neutralizing anti-HA and anti-M2 antibodies and ADCC activity, have been proposed by Ma et al. as a new candidate for influenza prophylaxis. In addition, immunization with these conglomerates stimulated robust NP-specific T-cell responses. As expected, the best protection of mice against virus challenges was achieved when using a trivalent vaccine combining HA, NA, and NP antigens, as evidenced by a histopathological analysis of the lungs and survival dynamics [81]. In another study, Wei et al. developed nanoparticles consisting of recombinant NP encapsulated by an apoferritin (AFt) nanocage conjugated with HA molecules exposed on the outer surface of the AFt. This vaccine prototype induced cross-reactive anti-HA and NP-specific antibodies in mice and provided complete protection against a homologous PR8 H1N1 and a heterologous A/FM/1/47 (H1N1) virus challenge, whereas the variant lacking NP conferred only 60% protection against the heterologous strain. Such a conjugation strategy may be potentially used not only for the creation of nonadjuvanted vaccines, but also as a tool for targeted drug delivery [82].
An attempt to improve the immunogenicity of recombinant NP from the A/WSN/1933 (H1N1) virus by antigenic multimerization has been made by Del Campo et al., who supplemented the NP sequence with the oligomerization domain OVX313 and developed the NP heptamer named OVX836, which consists of seven copies of the NP antigen [83]. This polymer appeared to be significantly more immunogenic—due to the more active uptake by dendritic cells, compared to wild-type NP—and stimulated robust NP-specific CD4+ and CD8+ T-cell responses, particularly lung-localized CD8+ tissue-resident memory (TRM) cells in mice [84]. In a phase 1 clinical trial, OVX836 administered intramuscularly was found to be safe and immunogenic at a dose of up to 180 µg [85]. In a recent phase IIa clinical trial, higher doses of OVX836 (300 or 480 μg) were compared to the 180 μg dose in terms of safety, immunogenicity, and protection against seasonal influenza-like illnesses in a particular influenza season. All vaccine doses demonstrated a good safety profile and the induction of pronounced cellular responses in a dose-dependent manner, which was evidenced by increases in the numbers of IFNγ-secreting cells in peripheral blood as early as a week after immunization [86]. Importantly, this study demonstrated an approximately 80% efficacy against seasonal H3N2 influenza viruses, indicating the ability of the induced T cells to protect vaccinated individuals from natural influenza infection. However, this study did not investigate the role of anti-NP antibodies, and this mode of immunity could also contribute to protection against infection.
Table 1 summarizes the most significant findings from the preclinical studies of NP-based universal influenza vaccine prototypes based on various vaccine platforms.

Table 1.
Examples of NP-based influenza vaccines.
NP-based vaccines are not commercially available, but many of the prototypes have been assessed in clinical trials (Table 2). In addition to the candidates described above, the results of clinical trials considering several other vaccine prototypes have been published. For example, the vaccine Multimeric-001, which is a recombinant protein comprising the conserved linear epitopes of HA, NP, and M1 antigens mixed with Montanide ISA-51 oil adjuvant, was shown to be safe and immunogenic in phases I and II of the clinical trials [97]. In addition, Multimeric-001 has been studied in the elderly as a priming tool prior to administration of the trivalent inactivated influenza vaccine; in this age group, such a vaccination regimen provided intensified antiviral antibody generation, compared to unprimed individuals [98,99]. Despite these encouraging results, this vaccine did not facilitate a reduction in influenza severity and thus failed to induce specified outcomes in a phase III clinical trial in older adults (NCT03450915). A possible cause of vaccine inefficiency could be weak epitope-specific T-cell formation, which was insufficient for protection against the disease.

Table 2.
Clinical trials of NP-based influenza vaccines.
The same oil-based compound, Montanide ISA-51, has been used as an adjuvant in a synthetic influenza vaccine (FLU-v) containing the peptides of the NP, M1, and M2 viral proteins. Being s.c. administered, this vaccine induced vigorous virus-specific CTL responses in healthy participants in a phase I clinical trial [107]. A positive correlation of this effect with a milder disease course and reduced viral loads was observed in a following human challenge with a wild-type H3N2 influenza virus [106]. The immunogenicity of FLU-v in healthy volunteers has been further revealed in a phase 2b clinical trial [111]. A H1N1 human challenge study [109], as well as a phase 3 trial study [110], confirmed its efficacy for influenza prevention.
Little is known about the clinical efficacy of the FP-01.1 vaccine, consisting of six fluorocarbon-modified 35-mer peptides comprising the conserved epitopes of NP, M1, PB1, and PB2, which are able to trigger CD4+ and CD8+ T cells. This candidate was proposed to prime and boost vaccination against different influenza A variants and has demonstrated safety and the ability to stimulate cross-reactive IFNγ-secreting CD4+ and CD8+ T cells at a dose of 150 μg/peptide in a phase I trial [113]. The results of further clinical trials on this vaccine have not yet been published (Table 2).
4. Variability of NP Sequences and Implications for the Performance of NP-Based Vaccines
Although it is widely accepted that the NP protein is highly conserved among different subtypes of the influenza A virus, this antigen undergoes slow evolutionary changes as a result of the virus escaping recognition by the CTLs [113]. Several previous studies have demonstrated that a number of critical amino acid substitutions occur in the NP protein and that these mutations are associated with escape from human CTLs [114,115,116,117]. It has been shown that the substitutions in epitopes including the 103–111, 251–259, 380–388, 383–391, and 418–426 amino acid residues—in particular, mutations at positions 103 (K203R), 259 (S259L), 384 (R384K/G), 421 (D421E), 423 (P423S), and 425 (I425M)—may abrogate the antigenic presentation mediated by MHC-I and, thus, allow the virus to avoid recognition by CD8+ CTLs [116,118]. Such changes may lead to a complete inability of effector T cells generated in response to infection with previous influenza variants to recognize the viruses belonging to recent strains [118].
A possible consequence of these mutations in T-cell epitopes is that CTLs targeted at nonmutated epitopes will inefficiently recognize cells infected with the mutated virus. To demonstrate the variability in the NP sequence, we aligned the amino acid sequences of NPs that are most often used as vaccine antigens, along with recent seasonal H1N1 and H3N2 influenza viruses, specifically, A/WSN/1933 (H1N1) and A/PR8/34 (H1N1) (Figure 2 and Figure S1). We also included NP sequences of master donor viruses of a licensed live-attenuated influenza vaccine (LAIV), as they are known to induce robust NP-specific T-cell responses [119,120] (Figure S2). As shown in Figure 2, a number of mutations had occurred in the NP sequences of more recent viruses, suggesting that T cells targeted to some NP epitopes of the developed NP-based vaccine candidates, as well as to that of the licensed LAIVs, may not recognize the corresponding proteins of currently circulating influenza A viruses. For example, an immunodominant NP418–426 epitope has up to four mutated residues in recent H3N2 viruses, compared to the original model viruses. Although CTLs specific to this particular epitope are characterized by high functional avidity [121], it is very unlikely that T cells produced in response to vaccination with the nonmutated NP will recognize NP418–426 epitopes with three or four mismatched residues.
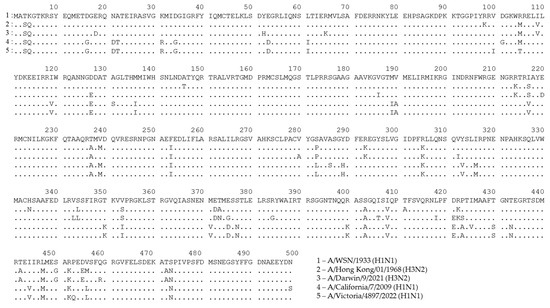
Figure 2.
Alignment of NP sequences for A/WSN/1933 (H1N1) influenza virus and recent H1N1 and H3N2 seasonal influenza strains. This figure was generated using GISAID data and the uGene v33.0 software [122].
Earlier, we performed a simple immune epitope conservancy analysis of the NPs of currently circulating H1N1 and H3N2 viruses, relative to the NPs originating from the master donor virus of a licensed LAIV [123]. The analysis revealed that 8 out of 18 (44.4%) A/Leningrad/134/17/57 (H2N2)-derived NP epitopes had been conserved among recent H1N1 and H3N2 viruses, suggesting that most of the predicted NP-specific immunodominant CTL epitopes of the vaccine prototypes developed from older influenza A viruses are no longer present in the vast majority of circulating influenza viruses. These findings may have at least two negative consequences: (i) the massive implementation of the NP-based vaccines designed on the backbone of obsolete influenza viruses will result in the stimulation of CTL clones that will fail to recognize cells infected with recent seasonal influenza viruses, thereby unnecessarily overloading the immune system of vaccinees; and (ii) many of the new NP epitopes that have emerged in recent influenza viruses will not be recognized by vaccine-induced T cells, as these epitopes were not yet present in the older vaccine viruses.
It is well known that, in addition to the T-cell response, the NP antigen can induce the production of NP-specific antibodies. A number of studies describing the protective properties not only of recombinant NP but also of passively transferred immune sera from NP-immunized animals have demonstrated the efficacy of such vaccination in a heterosubtypic influenza virus challenge. In a study by Carragher et al., immunization of mice with recombinant NP resulted in high titers of specific IgG and reduced replication of the virus challenge, but not in antibody-deficient animals. Similarly, passive immunization of naive mice devoid of B cells or intact mice with anti-NP serum significantly reduced virus titers in the lungs [124]. LaMere et al. have shown that anti-NP antibodies administered to mice with depleted B cells provided partial heterologous protection and a reduced viral load through mechanisms mediated by FcRs and CD8+ T cells. Notably, FcR-deprived mice immunized with anti-NP antibodies appeared to be less-protected, suggesting an FcR-mediated effect of anti-NP antibodies in vivo [44]. The high immunogenicity of the NP protein and the ability of anti-NP monoclonal antibodies of influenza patients to mediate Fc-associated protective responses have been demonstrated by Varderven et al. [125]. In contrast, passive immunization of mice with human anti-NP mAbs was insufficient to protect against the virus challenge. These studies suggest that effective antiviral protection in vivo may be provided not only by anti-NP antibodies, but also by other influenza-specific defense factors [125]. In the study of Fujimoto et al., transgenic mice expressing anti-NP-antibody genes from B cells of H5N1 convalescents were exposed to a lethal dose of this highly pathogenic avian virus. The resulting antibodies had no neutralizing activity, but provided resistance to the animals against infection by both H5N1 and mouse-adapted H1N1. These data suggest that anti-NP antibodies are involved in cross-protection against influenza, probably through indirect antiviral reactions, such as ADCC or complement cascade activation [126]. Overall, NP-specific antibodies may contribute significantly to cross-protection induced by NP-based vaccines, such that this mode of immunity should not be disregarded. Although NP-specific antibodies are considered highly cross-reactive, the use of anti-NP monoclonal antibodies revealed some variations in the antigenicity of this protein between evolutionarily diverse influenza viruses [127]. In addition, several mAbs have been described that can only bind to a specific subtype of influenza A virus, suggesting a critical influence of some NP residues on antibody binding [128]. In the study of Bodewes et al., human monoclonal anti-NP antibody bound diverse H1N1 and H3N2 viruses with different intensities, indicating variations in the affinity of the antibody for the respective NPs [19]. All of these data suggest that slow evolutionary changes in the NP protein may also affect its binding to anti-NP antibodies, which should also be considered during the development of broadly reactive NP-based influenza vaccines.
5. Approaches to Improve the Performance of Influenza Virus Vaccines
There are several ways to reduce NP epitope mismatches between the vaccine antigen and currently circulating seasonal influenza viruses. First, for vaccines based on recombinant proteins, viral vectors, and nucleic acid platforms, more recent NP antigens should be considered in the development of vaccine prototypes. There are no clear obstacles preventing the replacement of the NP antigen in the experimental vaccine candidates with that of a recent influenza A virus, other than conducting additional experiments to confirm its stability and safety. However, NPs of seasonal influenza viruses of H1N1 and H3N2 differ quite significantly; therefore, further studies may be needed to assess the rational design of a universal NP antigen that includes all of the most important immunodominant B- and T-cell epitopes of both virus subtypes. For example, consensus sequences of influenza virus hemagglutinin molecules have been successfully generated using a COBRA approach (computationally optimized broadly reactive antigens), and these consensus HA vaccines demonstrated broad protection against heterologous influenza viruses within a particular subtype [129,130,131]. As the NP protein is inherently much more conserved than the HA antigen, the use of COBRA algorithms is likely to generate some NP consensus sequences that would induce broadly protective antibody and cell-mediated immunity. Furthermore, the performance of NP-based vaccines can be significantly improved by using new potent adjuvants and delivery platforms (as reviewed in [132,133,134]).
For live-attenuated influenza vaccines, the situation is somewhat different. Currently licensed LAIVs are based on the cold-adapted backbone viruses A/Leningrad/134/17/57 (Len/17) and A/Ann Arbor/6/60 (AA), which were isolated in 1957 and 1960, respectively [135,136]. These viruses were serially passaged at low temperatures and acquired unique mutations in the internal protein genes, which made the viruses temperature-sensitive and cold-adapted in vitro and attenuated in vivo. These mutations have been clearly identified in both master donor viruses [137,138] and, although no common mutations were found in the AA and Len/17 viruses, all substitutions affected the viral RNA synthesis machinery [139]. Therefore, a reasonable idea arose to develop vaccine strains against various human and animal influenza A viruses by simply introducing these attenuating mutations into the genes of the virus polymerase complex [140,141,142]. A side-by-side comparison of the Len/17 and AA mutations introduced into the A/PR8/34 backbone virus demonstrated that the Len/17 mutations resulted in higher attenuation of the virus compared with the AA mutations, although the immunogenic and cross-protective properties were comparable [143]. However, for some wild-type viruses, introduction of the full set of ts signatures from the cold-adapted AA virus was not enough to render the virus sufficiently attenuated; this depended mainly on the source of the PB2 gene [144]. Importantly, a recent study has found that the addition of a mutation L319Q in the PB1 protein significantly improved the attenuation of the H1N1pdm-based LAIV candidate carrying AA-specific attenuating mutations [145].
It should be noted that the strategy of introducing attenuating mutations directly into the genome of wild-type viruses results in LAIVs with the most relevant repertoire of B- and T-cell epitopes across the viral proteome, and such vaccines should obviously provide the highest possible level of protection against circulating influenza viruses [146]. However, the procedure for preparing such vaccine candidates is laborious and time-consuming, as vaccine strains against both H1N1 and H3N2 seasonal influenza viruses must be prepared through side-directed mutagenesis. Therefore, it is unlikely that LAIVs against seasonal influenza viruses will be routinely prepared in this way. Moreover, there will always be the possibility of the emergence of wild-type virus variants whose pathogenicity cannot be compensated for by a standard set of attenuating mutations, as was the case with the 2009 H1N1 pandemic variant [140].
In the meantime, there is a simple and straightforward way to improve the immunogenicity and cross-protection of currently licensed LAIVs by simply replacing the NP gene of the cold-adapted master donor virus with that of a wild-type influenza virus, such as switching from a 6:2 genome composition to a 5:3 composition [147]. In this case, there is no need to design a “universal” NP protein that includes the most important immunodominant B- and T-cell epitopes of both virus subtypes, as traditional LAIVs contain both the H1N1 and H3N2 vaccine strains. Given that nucleoprotein is a major target antigen for influenza-specific cross-reactive cytotoxic T cells [17,148], updating epitopes even in this protein alone is expected to lead to more broadly protective influenza vaccine variants. Of note, the NP gene of the Len/17 strain does not contain attenuating mutations and can be replaced by the wild-type NP without safety concerns [137]. Indeed, a panel of H3N2 5:3 LAIV candidates developed on the Len/17 backbone were equally safe and immunogenic in a ferret model, when compared to classical H3N2 6:2 LAIVs, confirming the feasibility of switching to 5:3 LAIV genome compositions [149]. It should be noted that the differences in NP-specific T-cell responses between 5:3 and 6:2 LAIVs were not tested in ferrets; however, a series of experiments on C57BL/6J mice demonstrated that NP366 epitope-specific CTL responses differed significantly between the 6:2 and 5:3 LAIVs of the H7N9 subtype. In particular, the 6:2 LAIV induced a robust CTL response (both in splenocytes and in lungs) to the homologous Len/17 NP366 epitope, whereas very low reactivity was observed for the H7N9 NP366 epitope [123,150], suggesting that classical LAIV induces an excessive level of T-cell responses to outdated NP epitopes, and these T cells unnecessarily overwhelm the immune system as they no longer recognize evolutionarily diverse viruses. Overall, animal experiments have demonstrated that the Len/17-based 5:3 LAIVs of different subtypes afforded better protection against a heterologous viral challenge than standard 6:2 LAIVs, prompting their further evaluation in clinical trials [123,150,151,152]. Importantly, experiments on PBMCs isolated from HLA-typed blood donors have confirmed that 5:3 LAIVs could stimulate human influenza CD8+ T cells more relevant to current infections than classical 6:2 LAIVs [153].
For LAIVs based on the A/Ann Arbor/6/60 backbone, switching to the 5:3 genome composition may be complicated as one of the ts mutations was mapped to the NP protein (D34G) [138]. However, recent H1N1 viruses have naturally acquired this mutation (Figure 1); therefore, at least for this subtype, 5:3 LAIVs may be developed without safety concerns. However, further studies are needed to confirm this assumption.
6. Concluding Remarks
It is generally recognized that T cells have a major contribution to immunity against influenza and other respiratory pathogens by helping to eliminate pathogens from an infected individual [154,155]. This type of immunity is typically more broadly reactive and lasts longer than antibody responses to major viral antigenic determinants. Therefore, it is not surprising that the influenza virus nucleoprotein has attracted much attention as a target antigen for the development of broadly protective influenza vaccines, as a high proportion of T cells elicited by the infection recognize NP-specific epitopes. Clearly, NP is one of the most widely used antigens for designing universal influenza vaccines, along with the ectodomain of matrix 2 protein (M2e) [156].
To date, many experimental NP-based influenza vaccines aimed at inducing a cross-reactive T-cell responses, with the overall goal of protection against multiple influenza A subtypes, have been submitted to clinical or preclinical testing. These vaccines have been developed using different vaccine platforms, such as recombinant proteins fused to different adjuvants or oligomerizing domains, DNA/mRNA vaccines, or viral vector-based vaccines. Almost all published studies have demonstrated a broader spectrum of action of experimental vaccines; however, not all vaccines that have reached clinical trials have shown efficacy in humans. One possible reason for the insufficient degree of protection against current wild viruses is the mismatch between the epitope composition of the NP protein in the vaccine virus and the current circulating strain(s), as most vaccines are developed on the basis of well-characterized NP proteins from viruses isolated almost 90 years ago. During this period, despite a fairly high degree of NP conservation, a large number of mutations have arisen, some of which can be categorized as immunogenic T-cell epitopes. Furthermore, it is well known that even a single amino acid mismatch in a T-cell epitope can dramatically alter the immunogenicity of a peptide [157].
Although live-attenuated influenza vaccines stand apart from broad-spectrum NP-based vaccines, it is well known that they induce a potent T-cell response, and a major target for this response is also a nucleoprotein. Backbone viruses for LAIV were isolated more than 60 years ago, and there is also a serious mismatch between the NP epitopes of vaccine virus and that of wild-type strains. Preliminary findings in LAIVs expressing NPs of recent influenza viruses suggest that the source of the NP antigen significantly affects the magnitude of the T-cell response to the NP epitopes of current influenza viruses; therefore, the 6:2 LAIV genome composition should be switched to the 5:3 formula, with the NP derived from wild-type viruses, along with HA and NA.
In addition to cell-mediated immunity, the NP antigen generates high levels of anti-NP antibodies. This mode of immunity has been undeservedly ignored in preclinical studies of NP-based influenza vaccine prototypes, perhaps because anti-NP antibodies do not possess a neutralizing activity. However, these antibodies can bind with virus-infected cells and mediate protection through some Fc-dependent functions [22,124,158]. It is reasonable to suggest that slow evolutionary changes in the NP can also affect the antigenicity of the protein and lead to the reduced binding of anti-NP antibodies raised to immunization with the developed NP-based vaccines with the NP antigen on the cells infected with contemporary influenza viruses. Similar to influenza NP, the SARS-CoV-2 nucleocapsid (N) protein slowly evolves, and although the N-based serologic tests developed on the ancestral strain are still sensitive enough to detect antibody responses to evolved SARS-CoV-2 variants [159], these mutations may lead to substantial changes in the immunogenicity and antigenicity of the NP protein [160].
In conclusion, this review summarized recent advances in the design of broad-spectrum vaccines employing the nucleoprotein protein as the vaccine antigen. We showed that the particular antigen choice for vaccine design can have important implications for vaccine efficacy against current circulating influenza viruses and that even minor epitope mismatches between the vaccine and circulating viruses can significantly reduce vaccine performance. Finally, it is essential to conduct side-by-side analyses of some vaccine prototypes with different sources of NP proteins to determine more precisely whether it is really necessary to use NPs from more recent influenza viruses to design NP-based vaccines, or whether the cross-reactivity of vaccines based on older proteins yields a comparable performance to vaccine candidates with updated NPs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vaccines11121747/s1, Figure S1: Alignment of NP sequences of A/PR/8/34 (H1N1) influenza virus and recent H1N1 and H3N2 seasonal influenza strains; Figure S2: Alignment of NP sequences of A/Leningrad/134/17/57 (H2N2) influenza virus and recent H1N1 and H3N2 seasonal influenza strains.
Author Contributions
Conceptualization, A.R. and I.I.-S.; methodology, A.R.; investigation, A.R.; resources, A.R., L.R. and I.I.-S.; writing—original draft preparation, A.R. and I.I.-S.; writing—review and editing, L.R.; visualization, A.R.; supervision, L.R.; project administration, I.I.-S., L.R.; funding acquisition, I.I.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the RSCF grant 21-75-30003.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of Respiratory Viral Infections. Annu. Rev. Virol. 2020, 7, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Xu, D.; Zhang, Y.; Wang, T.; Zhang, L.; Gu, W.; Shen, M. Epidemiological characteristics of four common respiratory viral infections in children. Virol. J. 2021, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Rémy, V.; Largeron, N.; Quilici, S.; Carroll, S. The Economic Value of Vaccination: Why Prevention is Wealth. J. Mark. Access Health Policy 2015, 3, 29284. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Elbahesh, H.; Reperant, L.A.; Rimmelzwaan, G.F.; Osterhaus, A. Influenza Vaccines: Successes and Continuing Challenges. J. Infect. Dis. 2021, 224, S405–S419. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Holmes, E.C. The Ecology and Evolution of Influenza Viruses. Cold Spring Harb. Perspect. Med. 2020, 10, a038489. [Google Scholar] [CrossRef] [PubMed]
- Awadalla, M.E.; Alkadi, H.; Alarjani, M.; Al-Anazi, A.E.; Ibrahim, M.A.; Alohali, T.A.; Enani, M.; Alturaiki, W.; Alosaimi, B. Moderately Low Effectiveness of the Influenza Quadrivalent Vaccine: Potential Mismatch between Circulating Strains and Vaccine Strains. Vaccines 2023, 11, 1050. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Wan, X.F.; Ye, Z.; Plant, E.P.; Zhao, Y.; Xu, Y.; Li, X.; Finch, C.; Zhao, N.; Kawano, T.; et al. H3N2 Mismatch of 2014-15 Northern Hemisphere Influenza Vaccines and Head-to-head Comparison between Human and Ferret Antisera derived Antigenic Maps. Sci. Rep. 2015, 5, 15279. [Google Scholar] [CrossRef]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- Isakova-Sivak, I.; Stepanova, E.; Mezhenskaya, D.; Matyushenko, V.; Prokopenko, P.; Sychev, I.; Wong, P.F.; Rudenko, L. Influenza vaccine: Progress in a vaccine that elicits a broad immune response. Expert Rev. Vaccines 2021, 20, 1097–1112. [Google Scholar] [CrossRef]
- Wang, W.C.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Progress towards the Development of a Universal Influenza Vaccine. Viruses 2022, 14, 1684. [Google Scholar] [CrossRef]
- Scholtissek, C.; Burger, H.; Kistner, O.; Shortridge, K.F. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 1985, 147, 287–294. [Google Scholar] [CrossRef]
- Wei, Y.; Zeng, Y.; Zhang, X.; Xu, S.; Wang, Z.; Du, Y.; Zhang, B.; Lei, C.Q.; Zhu, Q. The Nucleoprotein of H7N9 Influenza Virus Positively Regulates TRAF3-Mediated Innate Signaling and Attenuates Viral Virulence in Mice. J. Virol. 2020, 94, 01640-20. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sneyd, H.; Dekant, R.; Wang, J. Influenza A Virus Nucleoprotein: A Highly Conserved Multi-Functional Viral Protein as a Hot Antiviral Drug Target. Curr. Top. Med. Chem. 2017, 17, 2271–2285. [Google Scholar] [CrossRef] [PubMed]
- Donchet, A.; Oliva, J.; Labaronne, A.; Tengo, L.; Miloudi, M.; Gerard, F.C.A.; Mas, C.; Schoehn, G.; Ruigrok, R.W.H.; Ducatez, M.; et al. The structure of the nucleoprotein of Influenza D shows that all Orthomyxoviridae nucleoproteins have a similar NP(CORE), with or without a NP(TAIL) for nuclear transport. Sci. Rep. 2019, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kang, J.O.; Chang, J. Nucleoprotein vaccine induces cross-protective cytotoxic T lymphocytes against both lineages of influenza B virus. Clin. Exp. Vaccine Res. 2019, 8, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Grant, E.; Wu, C.; Chan, K.F.; Eckle, S.; Bharadwaj, M.; Zou, Q.M.; Kedzierska, K.; Chen, W. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+ T-cell responses. Immunol. Cell Biol. 2013, 91, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.A.; Weber, T.; Cejas, P.J.; Cox, K.S.; Touch, S.; Austin, L.A.; Ou, Y.; Citron, M.P.; Luo, B.; Gindy, M.E.; et al. Characterization of humoral and cell-mediated immunity induced by mRNA vaccines expressing influenza hemagglutinin stem and nucleoprotein in mice and nonhuman primates. Vaccine 2022, 40, 4412–4423. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Geelhoed-Mieras, M.M.; Wrammert, J.; Ahmed, R.; Wilson, P.C.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. In vitro assessment of the immunological significance of a human monoclonal antibody directed to the influenza a virus nucleoprotein. Clin. Vaccine Immunol. CVI 2013, 20, 1333–1337. [Google Scholar] [CrossRef]
- Virelizier, J.L.; Allison, A.C.; Oxford, J.S.; Schild, G.C. Early presence of ribonucleoprotein antigen on surface of influenza virus-infected cells. Nature 1977, 266, 52–54. [Google Scholar] [CrossRef]
- Vanderven, H.A.; Ana-Sosa-Batiz, F.; Jegaskanda, S.; Rockman, S.; Laurie, K.; Barr, I.; Chen, W.; Wines, B.; Hogarth, P.M.; Lambe, T.; et al. What Lies Beneath: Antibody Dependent Natural Killer Cell Activation by Antibodies to Internal Influenza Virus Proteins. EBioMedicine 2016, 8, 277–290. [Google Scholar] [CrossRef]
- Jegaskanda, S.; Co, M.D.T.; Cruz, J.; Subbarao, K.; Ennis, F.A.; Terajima, M. Induction of H7N9-Cross-Reactive Antibody-Dependent Cellular Cytotoxicity Antibodies by Human Seasonal Influenza A Viruses that are Directed Toward the Nucleoprotein. J. Infect. Dis. 2017, 215, 818–823. [Google Scholar] [CrossRef]
- Machkovech, H.M.; Bedford, T.; Suchard, M.A.; Bloom, J.D. Positive Selection in CD8+ T-Cell Epitopes of Influenza Virus Nucleoprotein Revealed by a Comparative Analysis of Human and Swine Viral Lineages. J. Virol. 2015, 89, 11275–11283. [Google Scholar] [CrossRef]
- Reguera, J.; Cusack, S.; Kolakofsky, D. Segmented negative strand RNA virus nucleoprotein structure. Curr. Opin. Virol. 2014, 5, 7–15. [Google Scholar] [CrossRef]
- Tang, Y.S.; Xu, S.; Chen, Y.W.; Wang, J.H.; Shaw, P.C. Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction. Nucleic Acids Res. 2021, 49, 4144–4154. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.; Martin-Benito, J.; Zurcher, T.; Valpuesta, J.M.; Carrascosa, J.L.; Ortin, J. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 2000, 74, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Turrell, L.; Lyall, J.W.; Tiley, L.S.; Fodor, E.; Vreede, F.T. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat. Commun. 2013, 4, 1591. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Krug, R.M.; Tao, Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 2006, 444, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Diefenbacher, M.; Tan, T.J.C.; Bauer, D.L.V.; Stadtmueller, B.M.; Wu, N.C.; Brooke, C.B. Interactions between Influenza A Virus Nucleoprotein and Gene Segment Untranslated Regions Facilitate Selective Modulation of Viral Gene Expression. J. Virol. 2022, 96, e0020522. [Google Scholar] [CrossRef] [PubMed]
- Prokudina, E.N.; Semenova, N.; Chumakov, V.; Stitz, L. An antigenic epitope of influenza virus nucleoprotein (NP) associated with polymeric forms of NP. Virol. J. 2008, 5, 37. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Santak, M.; Matic, Z. The Role of Nucleoprotein in Immunity to Human Negative-Stranded RNA Viruses-Not Just Another Brick in the Viral Nucleocapsid. Viruses 2022, 14, 521. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Castrucci, M.R.; Kawaoka, Y. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 1997, 71, 9690–9700. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sankhala, R.S.; Florio, T.J.; Zhou, L.; Nguyen, N.L.T.; Lokareddy, R.K.; Cingolani, G.; Pante, N. Synergy of two low-affinity NLSs determines the high avidity of influenza A virus nucleoprotein NP for human importin alpha isoforms. Sci. Rep. 2017, 7, 11381. [Google Scholar] [CrossRef]
- Kobayashi, K.S.; van den Elsen, P.J. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 2012, 12, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Juno, J.A.; van Bockel, D.; Kent, S.J.; Kelleher, A.D.; Zaunders, J.J.; Munier, C.M. Cytotoxic CD4 T Cells-Friend or Foe during Viral Infection? Front. Immunol. 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Bui, H.H.; Sidney, J.; Zhang, Q.; Glenn, J.; Oseroff, C.; Mbawuike, I.N.; Alexander, J.; Newman, M.J.; Grey, H.; et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 2008, 82, 12241–12251. [Google Scholar] [CrossRef]
- Wu, C.; Zanker, D.; Valkenburg, S.; Tan, B.; Kedzierska, K.; Zou, Q.M.; Doherty, P.C.; Chen, W. Systematic identification of immunodominant CD8+ T-cell responses to influenza A virus in HLA-A2 individuals. Proc. Natl. Acad. Sci. USA 2011, 108, 9178–9183. [Google Scholar] [CrossRef]
- Soghoian, D.Z.; Streeck, H. Cytolytic CD4(+) T cells in viral immunity. Expert Rev. Vaccines 2010, 9, 1453–1463. [Google Scholar] [CrossRef]
- Wilkinson, T.M.; Li, C.K.; Chui, C.S.; Huang, A.K.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef]
- Chen, L.; Zanker, D.; Xiao, K.; Wu, C.; Zou, Q.; Chen, W. Immunodominant CD4+ T-cell responses to influenza A virus in healthy individuals focus on matrix 1 and nucleoprotein. J. Virol. 2014, 88, 11760–11773. [Google Scholar] [CrossRef]
- de Boer, G.F.; Back, W.; Osterhaus, A.D. An ELISA for detection of antibodies against influenza A nucleoprotein in humans and various animal species. Arch. Virol. 1990, 115, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Sedova, E.S.; Scherbinin, D.N.; Lysenko, A.A.; Alekseeva, S.V.; Artemova, E.A.; Shmarov, M.M. Non-neutralizing Antibodies Directed at Conservative Influenza Antigens. Acta Nat. 2019, 11, 22–32. [Google Scholar] [CrossRef] [PubMed]
- LaMere, M.W.; Lam, H.T.; Moquin, A.; Haynes, L.; Lund, F.E.; Randall, T.D.; Kaminski, D.A. Contributions of antinucleoprotein IgG to heterosubtypic immunity against influenza virus. J. Immunol. 2011, 186, 4331–4339. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Luo, J.; Chen, Z. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection 2014, 42, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Townsend, A.R.; McMichael, A.J.; Carter, N.P.; Huddleston, J.A.; Brownlee, G.G. Cytotoxic T cell recognition of the influenza nucleoprotein and hemagglutinin expressed in transfected mouse L cells. Cell 1984, 39, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Wraith, D.C.; Askonas, B.A. Induction of influenza A virus cross-reactive cytotoxic T cells by a nucleoprotein/haemagglutinin preparation. J. Gen. Virol. 1985, 66 Pt 6, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Wraith, D.C.; Vessey, A.E.; Askonas, B.A. Purified influenza virus nucleoprotein protects mice from lethal infection. J. Gen. Virol. 1987, 68 Pt 2, 433–440. [Google Scholar] [CrossRef]
- Tite, J.P.; Hughes-Jenkins, C.; O’Callaghan, D.; Dougan, G.; Russell, S.M.; Gao, X.M.; Liew, F.Y. Anti-viral immunity induced by recombinant nucleoprotein of influenza A virus. II. Protection from influenza infection and mechanism of protection. Immunology 1990, 71, 202–207. [Google Scholar]
- Le, T.; Sun, C.; Chang, J.; Zhang, G.; Yin, X. mRNA Vaccine Development for Emerging Animal and Zoonotic Diseases. Viruses 2022, 14, 401. [Google Scholar] [CrossRef]
- McMahon, M.; O’Dell, G.; Tan, J.; Sarkozy, A.; Vadovics, M.; Carreno, J.M.; Puente-Massaguer, E.; Muramatsu, H.; Bajusz, C.; Rijnink, W.; et al. Assessment of a quadrivalent nucleoside-modified mRNA vaccine that protects against group 2 influenza viruses. Proc. Natl. Acad. Sci. USA 2022, 119, e2206333119. [Google Scholar] [CrossRef] [PubMed]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, J.B.; Donnelly, J.J.; Parker, S.E.; Rhodes, G.H.; Felgner, P.L.; Dwarki, V.J.; Gromkowski, S.H.; Deck, R.R.; DeWitt, C.M.; Friedman, A.; et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 1993, 259, 1745–1749. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, J.B.; Fu, T.M.; Deck, R.R.; Friedman, A.; Guan, L.; DeWitt, C.; Liu, X.; Wang, S.; Liu, M.A.; Donnelly, J.J.; et al. Protective CD4+ and CD8+ T cells against influenza virus induced by vaccination with nucleoprotein DNA. J. Virol. 1998, 72, 5648–5653. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.L.; Kong, W.P.; Misplon, J.A.; Lo, C.Y.; Tumpey, T.M.; Xu, L.; Nabel, G.J. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 2005, 23, 5404–5410. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.Y.; Wu, Z.; Misplon, J.A.; Price, G.E.; Pappas, C.; Kong, W.P.; Tumpey, T.M.; Epstein, S.L. Comparison of vaccines for induction of heterosubtypic immunity to influenza A virus: Cold-adapted vaccine versus DNA prime-adenovirus boost strategies. Vaccine 2008, 26, 2062–2072. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Kong, W.P.; Wei, C.J.; Van Hoeven, N.; Gorres, J.P.; Nason, M.; Andersen, H.; Tumpey, T.M.; Nabel, G.J. Comparative efficacy of hemagglutinin, nucleoprotein, and matrix 2 protein gene-based vaccination against H5N1 influenza in mouse and ferret. PLoS ONE 2010, 5, e9812. [Google Scholar] [CrossRef]
- Berthoud, T.K.; Hamill, M.; Lillie, P.J.; Hwenda, L.; Collins, K.A.; Ewer, K.J.; Milicic, A.; Poyntz, H.C.; Lambe, T.; Fletcher, H.A.; et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 52, 1–7. [Google Scholar] [CrossRef]
- Antrobus, R.D.; Lillie, P.J.; Berthoud, T.K.; Spencer, A.J.; McLaren, J.E.; Ladell, K.; Lambe, T.; Milicic, A.; Price, D.A.; Hill, A.V.; et al. A T cell-inducing influenza vaccine for the elderly: Safety and immunogenicity of MVA-NP+M1 in adults aged over 50 years. PLoS ONE 2012, 7, e48322. [Google Scholar] [CrossRef]
- Lillie, P.J.; Berthoud, T.K.; Powell, T.J.; Lambe, T.; Mullarkey, C.; Spencer, A.J.; Hamill, M.; Peng, Y.; Blais, M.E.; Duncan, C.J.; et al. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2012, 55, 19–25. [Google Scholar] [CrossRef]
- Swayze, H.; Allen, J.; Folegatti, P.; Yu, L.M.; Gilbert, S.; Hill, A.; Ellis, C.; Butler, C.C. A phase IIb study to determine the safety and efficacy of candidate INfluenza Vaccine MVA-NP+M1 in combination with licensed Ina CTivated infl Uenza vaccine in adult S aged 65 years and above (INVICTUS): A study protocol. F1000Research 2019, 8, 719. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.; Ellis, C.; Folegatti, P.M.; Swayze, H.; Allen, J.; Bussey, L.; Bellamy, D.; Lawrie, A.; Eagling-Vose, E.; Yu, L.M.; et al. Efficacy and Safety of a Modified Vaccinia Ankara-NP+M1 Vaccine Combined with QIV in People Aged 65 and Older: A Randomised Controlled Clinical Trial (INVICTUS). Vaccines 2021, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.; Leung, Y.H.; Nicholls, J.M.; Perera, P.Y.; Lichy, J.H.; Yamamoto, M.; Waldmann, T.A.; Peiris, J.S.; Perera, L.P. Vaccinia virus-based multivalent H5N1 avian influenza vaccines adjuvanted with IL-15 confer sterile cross-clade protection in mice. J. Immunol. 2009, 182, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Dicks, M.D.; Spencer, A.J.; Edwards, N.J.; Wadell, G.; Bojang, K.; Gilbert, S.C.; Hill, A.V.; Cottingham, M.G. A novel chimpanzee adenovirus vector with low human seroprevalence: Improved systems for vector derivation and comparative immunogenicity. PLoS ONE 2012, 7, e40385. [Google Scholar] [CrossRef] [PubMed]
- Antrobus, R.D.; Coughlan, L.; Berthoud, T.K.; Dicks, M.D.; Hill, A.V.; Lambe, T.; Gilbert, S.C. Clinical assessment of a novel recombinant simian adenovirus ChAdOx1 as a vectored vaccine expressing conserved Influenza A antigens. Mol. Ther. 2014, 22, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Asthagiri Arunkumar, G.; McMahon, M.; Pavot, V.; Aramouni, M.; Ioannou, A.; Lambe, T.; Gilbert, S.; Krammer, F. Vaccination with viral vectors expressing NP, M1 and chimeric hemagglutinin induces broad protection against influenza virus challenge in mice. Vaccine 2019, 37, 5567–5577. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Asthagiri Arunkumar, G.; Liu, W.C.; Stadlbauer, D.; Albrecht, R.A.; Pavot, V.; Aramouni, M.; Lambe, T.; Gilbert, S.C.; Krammer, F. Vaccination with Viral Vectors Expressing Chimeric Hemagglutinin, NP and M1 Antigens Protects Ferrets Against Influenza Virus Challenge. Front. Immunol. 2019, 10, 2005. [Google Scholar] [CrossRef]
- Vatzia, E.; Feest, K.; McNee, A.; Manjegowda, T.; Carr, B.V.; Paudyal, B.; Chrun, T.; Maze, E.A.; McCarron, A.; Morris, S.; et al. Immunization with matrix-, nucleoprotein and neuraminidase protects against H3N2 influenza challenge in pH1N1 pre-exposed pigs. NPJ Vaccines 2023, 8, 19. [Google Scholar] [CrossRef]
- Sipo, I.; Knauf, M.; Fechner, H.; Poller, W.; Planz, O.; Kurth, R.; Norley, S. Vaccine protection against lethal homologous and heterologous challenge using recombinant AAV vectors expressing codon-optimized genes from pandemic swine origin influenza virus (SOIV). Vaccine 2011, 29, 1690–1699. [Google Scholar] [CrossRef]
- Demminger, D.E.; Walz, L.; Dietert, K.; Hoffmann, H.; Planz, O.; Gruber, A.D.; von Messling, V.; Wolff, T. Adeno-associated virus-vectored influenza vaccine elicits neutralizing and Fcgamma receptor-activating antibodies. EMBO Mol. Med. 2020, 12, e10938. [Google Scholar] [CrossRef]
- Joshi, L.R.; Knudsen, D.; Pineyro, P.; Dhakal, S.; Renukaradhya, G.J.; Diel, D.G. Protective Efficacy of an Orf Virus-Vector Encoding the Hemagglutinin and the Nucleoprotein of Influenza A Virus in Swine. Front. Immunol. 2021, 12, 747574. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Liu, F.; Shen, Y.; Wang, S.; Xu, W.; Fang, F.; Sun, B.; Xie, Z.; Chen, Z. Cross-protection against influenza virus infection by intranasal administration of nucleoprotein-based vaccine with compound 48/80 adjuvant. Hum. Vaccines Immunother. 2015, 11, 397–406. [Google Scholar] [CrossRef]
- Guo, L.; Zheng, M.; Ding, Y.; Li, D.; Yang, Z.; Wang, H.; Chen, Q.; Sui, Z.; Fang, F.; Chen, Z. Protection against multiple influenza A virus subtypes by intranasal administration of recombinant nucleoprotein. Arch. Virol. 2010, 155, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.V.; Ebensen, T.; Schulze, K.; Cargnelutti, D.; Blazejewska, P.; Scodeller, E.A.; Guzman, C.A. Intranasal delivery of influenza rNP adjuvanted with c-di-AMP induces strong humoral and cellular immune responses and provides protection against virus challenge. PLoS ONE 2014, 9, e104824. [Google Scholar] [CrossRef] [PubMed]
- Cookenham, T.; Lanzer, K.G.; Gage, E.; Lorenzo, E.C.; Carter, D.; Coler, R.N.; Baldwin, S.L.; Haynes, L.; Reiley, W.W.; Blackman, M.A. Vaccination of aged mice with adjuvanted recombinant influenza nucleoprotein enhances protective immunity. Vaccine 2020, 38, 5256–5267. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Zhao, Y.; Chen, X. Potentiation of Recombinant NP and M1-Induced Cellular Immune Responses and Protection by Physical Radiofrequency Adjuvant. Vaccines 2021, 9, 1382. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X. CpG 1018 Is an Effective Adjuvant for Influenza Nucleoprotein. Vaccines 2023, 11, 649. [Google Scholar] [CrossRef]
- Yin, Y.; Li, B.; Zhou, L.; Luo, J.; Liu, X.; Wang, S.; Lu, Q.; Tan, W.; Chen, Z. Protein transduction domain-mediated influenza NP subunit vaccine generates a potent immune response and protection against influenza virus in mice. Emerg. Microbes Infect. 2020, 9, 1933–1942. [Google Scholar] [CrossRef]
- Tan, M.P.; Mohamed Alitheen, N.B.; Tan, W.S.; Yap, W.B. Expression of Influenza M2e-NP Recombinant Fusion Protein in Escherichia coli BL21 (DE3) and Its Binding to Antibodies. Vaccines 2022, 10, 2066. [Google Scholar] [CrossRef]
- Shahsavandi, S.; Ebrahimi, M.M.; Fotouhi, F.; Tebianian, M. A Combination of Recombinant HA1-and Nucleoprotein-Based Chitosan Nanoparticles Induces Early and Potent Immune Responses Against the H9N2 Influenza Virus. Viral Immunol. 2022, 35, 365–374. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; Dong, C.; Gonzalez, G.X.; Song, Y.; Zhu, W.; Kim, J.; Wei, L.; Wang, B.Z. Influenza NP core and HA or M2e shell double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nanomed. Nanotechnol. Biol. Med. 2022, 40, 102479. [Google Scholar] [CrossRef]
- Wei, J.; Li, Z.; Yang, Y.; Ma, G.; Su, Z.; Zhang, S. An Apoferritin-Hemagglutinin Conjugate Vaccine with Encapsulated Nucleoprotein Antigen Peptide from Influenza Virus Confers Enhanced Cross Protection. Bioconjugate Chem. 2020, 31, 1948–1959. [Google Scholar] [CrossRef]
- Del Campo, J.; Pizzorno, A.; Djebali, S.; Bouley, J.; Haller, M.; Perez-Vargas, J.; Lina, B.; Boivin, G.; Hamelin, M.E.; Nicolas, F.; et al. OVX836 a recombinant nucleoprotein vaccine inducing cellular responses and protective efficacy against multiple influenza A subtypes. NPJ Vaccines 2019, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Del Campo, J.; Bouley, J.; Chevandier, M.; Rousset, C.; Haller, M.; Indalecio, A.; Guyon-Gellin, D.; Le Vert, A.; Hill, F.; Djebali, S.; et al. OVX836 Heptameric Nucleoprotein Vaccine Generates Lung Tissue-Resident Memory CD8+ T-Cells for Cross-Protection Against Influenza. Front. Immunol. 2021, 12, 678483. [Google Scholar] [CrossRef] [PubMed]
- Withanage, K.; De Coster, I.; Cools, N.; Viviani, S.; Tourneur, J.; Chevandier, M.; Lambiel, M.; Willems, P.; Le Vert, A.; Nicolas, F.; et al. Phase 1 Randomized, Placebo-Controlled, Dose-Escalating Study to Evaluate OVX836, a Nucleoprotein-Based Influenza Vaccine: Intramuscular Results. J. Infect. Dis. 2022, 226, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Leroux-Roels, I.; Willems, P.; Waerlop, G.; Janssens, Y.; Tourneur, J.; De Boever, F.; Bruhwyler, J.; Alhatemi, A.; Jacobs, B.; Nicolas, F.; et al. Immunogenicity, safety, and preliminary efficacy evaluation of OVX836, a nucleoprotein-based universal influenza A vaccine candidate: A randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Infect. Dis. 2023, 23, 1360–1369. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.L.; Stack, A.; Misplon, J.A.; Lo, C.Y.; Mostowski, H.; Bennink, J.; Subbarao, K. Vaccination with DNA encoding internal proteins of influenza virus does not require CD8+ cytotoxic T lymphocytes: Either CD4+ or CD8+ T cells can promote survival and recovery after challenge. Int. Immunol. 2000, 12, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Tutykhina, I.; Esmagambetov, I.; Bagaev, A.; Pichugin, A.; Lysenko, A.; Shcherbinin, D.; Sedova, E.; Logunov, D.; Shmarov, M.; Ataullakhanov, R.; et al. Vaccination potential of B and T epitope-enriched NP and M2 against Influenza A viruses from different clades and hosts. PLoS ONE 2018, 13, e0191574. [Google Scholar] [CrossRef] [PubMed]
- Barefoot, B.E.; Sample, C.J.; Ramsburg, E.A. Recombinant vesicular stomatitis virus expressing influenza nucleoprotein induces CD8 T-cell responses that enhance antibody-mediated protection after lethal challenge with influenza virus. Clin. Vaccine Immunol. CVI 2009, 16, 488–498. [Google Scholar] [CrossRef]
- Heinen, P.P.; Rijsewijk, F.A.; de Boer-Luijtze, E.A.; Bianchi, A.T.J. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J. Gen. Virol. 2002, 83, 1851–1859. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, J.O.; Kim, J.Y.; Jung, H.E.; Lee, H.K.; Chang, J. Single mucosal vaccination targeting nucleoprotein provides broad protection against two lineages of influenza B virus. Antivir. Res. 2019, 163, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Langenmayer, M.C.; Luelf-Averhoff, A.T.; Marr, L.; Jany, S.; Freudenstein, A.; Adam-Neumair, S.; Tscherne, A.; Fux, R.; Rojas, J.J.; Blutke, A.; et al. Newly Designed Poxviral Promoters to Improve Immunogenicity and Efficacy of MVA-NP Candidate Vaccines against Lethal Influenza Virus Infection in Mice. Pathogens 2023, 12, 867. [Google Scholar] [CrossRef] [PubMed]
- McGee, M.C.; Huang, W. Evolutionary conservation and positive selection of influenza A nucleoprotein CTL epitopes for universal vaccination. J. Med. Virol. 2022, 94, 2578–2587. [Google Scholar] [CrossRef] [PubMed]
- van de Ven, K.; Lanfermeijer, J.; van Dijken, H.; Muramatsu, H.; Vilas Boas de Melo, C.; Lenz, S.; Peters, F.; Beattie, M.B.; Lin, P.J.C.; Ferreira, J.A.; et al. A universal influenza mRNA vaccine candidate boosts T cell responses and reduces zoonotic influenza virus disease in ferrets. Sci. Adv. 2022, 8, eadc9937. [Google Scholar] [CrossRef] [PubMed]
- Bouback, T.A.F.; Redwan, N.A. Approaches toward the development of DNA vaccine for influenza virus. Afr. J. Biotechnol. 2011, 10, 5209–5218. [Google Scholar] [CrossRef]
- Joe, P.T.; Christopoulou, I.; van Hoecke, L.; Schepens, B.; Ysenbaert, T.; Heirman, C.; Thielemans, K.; Saelens, X.; Aerts, J.L. Intranodal administration of mRNA encoding nucleoprotein provides cross-strain immunity against influenza in mice. J. Transl. Med. 2019, 17, 242. [Google Scholar] [CrossRef] [PubMed]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and immunogenicity of multimeric-001—A novel universal influenza vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef]
- Lowell, G.H.; Ziv, S.; Bruzil, S.; Babecoff, R.; Ben-Yedidia, T. Back to the future: Immunization with M-001 prior to trivalent influenza vaccine in 2011/12 enhanced protective immune responses against 2014/15 epidemic strain. Vaccine 2017, 35, 713–715. [Google Scholar] [CrossRef]
- Atsmon, J.; Caraco, Y.; Ziv-Sefer, S.; Shaikevich, D.; Abramov, E.; Volokhov, I.; Bruzil, S.; Haima, K.Y.; Gottlieb, T.; Ben-Yedidia, T. Priming by a novel universal influenza vaccine (Multimeric-001)-a gateway for improving immune response in the elderly population. Vaccine 2014, 32, 5816–5823. [Google Scholar] [CrossRef]
- Powell, T.J.; Peng, Y.; Berthoud, T.K.; Blais, M.E.; Lillie, P.J.; Hill, A.V.; Rowland-Jones, S.L.; McMichael, A.J.; Gilbert, S.C.; Dong, T. Examination of influenza specific T cell responses after influenza virus challenge in individuals vaccinated with MVA-NP+M1 vaccine. PLoS ONE 2013, 8, e62778. [Google Scholar] [CrossRef]
- Coughlan, L.; Sridhar, S.; Payne, R.; Edmans, M.; Milicic, A.; Venkatraman, N.; Lugonja, B.; Clifton, L.; Qi, C.; Folegatti, P.M.; et al. Heterologous Two-Dose Vaccination with Simian Adenovirus and Poxvirus Vectors Elicits Long-Lasting Cellular Immunity to Influenza Virus A in Healthy Adults. EBioMedicine 2018, 29, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Folegatti, P.M.; Bellamy, D.; Flaxman, A.; Mair, C.; Ellis, C.; Ramon, R.L.; Ramos Lopez, F.; Mitton, C.; Baker, M.; Poulton, I.; et al. Safety and Immunogenicity of the Heterosubtypic Influenza A Vaccine MVA-NP+M1 Manufactured on the AGE1.CR.pIX Avian Cell Line. Vaccines 2019, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.G.; Bussey, L.; Eagling-Vose, E.; Rutkowski, K.; Ellis, C.; Argent, C.; Griffin, P.; Kim, J.; Thackwray, S.; Shakib, S.; et al. Efficacy and safety of a universal influenza A vaccine (MVA-NP+M1) in adults when given after seasonal quadrivalent influenza vaccine immunisation (FLU009): A phase 2b, randomised, double-blind trial. Lancet Infect. Dis. 2022, 22, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Leroux-Roels, I.; Waerlop, G.; Tourneur, J.; De Boever, F.; Maes, C.; Bruhwyler, J.; Guyon-Gellin, D.; Moris, P.; Del Campo, J.; Willems, P.; et al. Randomized, Double-Blind, Reference-Controlled, Phase 2a Study Evaluating the Immunogenicity and Safety of OVX836, A Nucleoprotein-Based Influenza Vaccine. Front. Immunol. 2022, 13, 852904. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Bernstein, D.I.; Winokur, P.; Frey, S.E.; Angelo, L.S.; Bryant, C.; Ben-Yedidia, T.; Roberts, P.C.; El Sahly, H.M.; Keitel, W.A. Safety and immunogenicity of Multimeric-001 (M-001) followed by seasonal quadrivalent inactivated influenza vaccine in young adults—A randomized clinical trial. Vaccine 2023, 41, 2716–2722. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Robinson, S.; Fernandez, A.; Stoloff, G.A.; Mann, A.; Gilbert, A.; Balaratnam, G.; Wilkinson, T.; Lambkin-Williams, R.; Oxford, J.; et al. A Synthetic Influenza Virus Vaccine Induces a Cellular Immune Response That Correlates with Reduction in Symptomatology and Virus Shedding in a Randomized Phase Ib Live-Virus Challenge in Humans. Clin. Vaccine Immunol. CVI 2015, 22, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Robinson, S.; Stoloff, G.A.; Caparros-Wanderley, W. Synthetic Influenza vaccine (FLU-v) stimulates cell mediated immunity in a double-blind, randomised, placebo-controlled Phase I trial. Vaccine 2012, 30, 4655–4660. [Google Scholar] [CrossRef]
- Pleguezuelos, O.; Robinson, S.; Fernandez, A.; Stoloff, G.A.; Caparros-Wanderley, W. Meta-Analysis and Potential Role of Preexisting Heterosubtypic Cellular Immunity Based on Variations in Disease Severity Outcomes for Influenza Live Viral Challenges in Humans. Clin. Vaccine Immunol. CVI 2015, 22, 949–956. [Google Scholar] [CrossRef]
- Pleguezuelos, O.; James, E.; Fernandez, A.; Lopes, V.; Rosas, L.A.; Cervantes-Medina, A.; Cleath, J.; Edwards, K.; Neitzey, D.; Gu, W.; et al. Efficacy of FLU-v, a broad-spectrum influenza vaccine, in a randomized phase IIb human influenza challenge study. NPJ Vaccines 2020, 5, 22. [Google Scholar] [CrossRef]
- Pleguezuelos, O.; Dille, J.; de Groen, S.; Oftung, F.; Niesters, H.G.M.; Islam, M.A.; Naess, L.M.; Hungnes, O.; Aldarij, N.; Idema, D.L.; et al. Immunogenicity, Safety, and Efficacy of a Standalone Universal Influenza Vaccine, FLU-v, in Healthy Adults: A Randomized Clinical Trial. Ann. Intern. Med. 2020, 172, 453–462. [Google Scholar] [CrossRef]
- van Doorn, E.; Pleguezuelos, O.; Liu, H.; Fernandez, A.; Bannister, R.; Stoloff, G.; Oftung, F.; Norley, S.; Huckriede, A.; Frijlink, H.W.; et al. Evaluation of the immunogenicity and safety of different doses and formulations of a broad spectrum influenza vaccine (FLU-v) developed by SEEK: Study protocol for a single-center, randomized, double-blind and placebo-controlled clinical phase IIb trial. BMC Infect. Dis. 2017, 17, 241. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.N.; Bunce, C.J.; Horlock, C.; Watson, J.M.; Warrington, S.J.; Georges, B.; Brown, C.B. A novel peptide-based pan-influenza A vaccine: A double blind, randomised clinical trial of immunogenicity and safety. Vaccine 2015, 33, 396–402. [Google Scholar] [CrossRef] [PubMed]
- van de Sandt, C.E.; Kreijtz, J.H.; Rimmelzwaan, G.F. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 2012, 4, 1438–1476. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.T.; Heiny, A.T.; Miotto, O.; Salmon, J.; Marques, E.T.; Lemonnier, F.; August, J.T. Conservation and diversity of influenza A H1N1 HLA-restricted T cell epitope candidates for epitope-based vaccines. PLoS ONE 2010, 5, e8754. [Google Scholar] [CrossRef] [PubMed]
- Rimmelzwaan, G.F.; Kreijtz, J.H.; Bodewes, R.; Fouchier, R.A.; Osterhaus, A.D. Influenza virus CTL epitopes, remarkably conserved and remarkably variable. Vaccine 2009, 27, 6363–6365. [Google Scholar] [CrossRef] [PubMed]
- Voeten, J.T.; Bestebroer, T.M.; Nieuwkoop, N.J.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 2000, 74, 6800–6807. [Google Scholar] [CrossRef] [PubMed]
- Boon, A.C.; de Mutsert, G.; Graus, Y.M.; Fouchier, R.A.; Sintnicolaas, K.; Osterhaus, A.D.; Rimmelzwaan, G.F. Sequence variation in a newly identified HLA-B35-restricted epitope in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. J. Virol. 2002, 76, 2567–2572. [Google Scholar] [CrossRef]
- Rimmelzwaan, G.F.; Boon, A.C.; Voeten, J.T.; Berkhoff, E.G.; Fouchier, R.A.; Osterhaus, A.D. Sequence variation in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. Virus Res. 2004, 103, 97–100. [Google Scholar] [CrossRef]
- Korenkov, D.; Isakova-Sivak, I.; Rudenko, L. Basics of CD8 T-cell immune responses after influenza infection and vaccination with inactivated or live attenuated influenza vaccine. Expert Rev. Vaccines 2018, 17, 977–987. [Google Scholar] [CrossRef]
- Mohn, K.G.; Smith, I.; Sjursen, H.; Cox, R.J. Immune responses after live attenuated influenza vaccination. Hum. Vaccines Immunother. 2018, 14, 571–578. [Google Scholar] [CrossRef]
- Boon, A.C.; de Mutsert, G.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. The hypervariable immunodominant NP418-426 epitope from the influenza A virus nucleoprotein is recognized by cytotoxic T lymphocytes with high functional avidity. J. Virol. 2006, 80, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Isakova-Sivak, I.; Korenkov, D.; Smolonogina, T.; Tretiak, T.; Donina, S.; Rekstin, A.; Naykhin, A.; Shcherbik, S.; Pearce, N.; Chen, L.M.; et al. Comparative studies of infectivity, immunogenicity and cross-protective efficacy of live attenuated influenza vaccines containing nucleoprotein from cold-adapted or wild-type influenza virus in a mouse model. Virology 2017, 500, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Carragher, D.M.; Kaminski, D.A.; Moquin, A.; Hartson, L.; Randall, T.D. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J. Immunol. 2008, 181, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Vanderven, H.A.; Esterbauer, R.; Jegaskanda, S.; Tan, H.X.; Wheatley, A.K.; Kent, S.J. Poor protective potential of influenza nucleoprotein antibodies despite wide prevalence. Immunol. Cell Biol. 2022, 100, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Tomioka, Y.; Takakuwa, H.; Uechi, G.I.; Yabuta, T.; Ozaki, K.; Suyama, H.; Yamamoto, S.; Morimatsu, M.; Mai, L.Q.; et al. Cross-protective potential of anti-nucleoprotein human monoclonal antibodies against lethal influenza A virus infection. J. Gen. Virol. 2016, 97, 2104–2116. [Google Scholar] [CrossRef] [PubMed]
- van Wyke, K.L.; Hinshaw, V.S.; Bean, W.J., Jr.; Webster, R.G. Antigenic variation of influenza A virus nucleoprotein detected with monoclonal antibodies. J. Virol. 1980, 35, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi-Akiyama, T.; Yamashiro, T.; Mai, L.Q.; Narahara, K.; Miyamoto, A.; Shinagawa, S.; Mori, S.; Kitajima, H.; Kirikae, T. Discrimination of influenza A subtype by antibodies recognizing host-specific amino acids in the viral nucleoprotein. Influenza Other Respir. Viruses 2012, 6, 434–441. [Google Scholar] [CrossRef]
- Sautto, G.A.; Kirchenbaum, G.A.; Ecker, J.W.; Bebin-Blackwell, A.G.; Pierce, S.R.; Ross, T.M. Elicitation of Broadly Protective Antibodies following Infection with Influenza Viruses Expressing H1N1 Computationally Optimized Broadly Reactive Hemagglutinin Antigens. ImmunoHorizons 2018, 2, 226–237. [Google Scholar] [CrossRef]
- Wong, T.M.; Allen, J.D.; Bebin-Blackwell, A.G.; Carter, D.M.; Alefantis, T.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Computationally Optimized Broadly Reactive Hemagglutinin Elicits Hemagglutination Inhibition Antibodies against a Panel of H3N2 Influenza Virus Cocirculating Variants. J. Virol. 2017, 91, 01581-17. [Google Scholar] [CrossRef]
- Giles, B.M.; Crevar, C.J.; Carter, D.M.; Bissel, S.J.; Schultz-Cherry, S.; Wiley, C.A.; Ross, T.M. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J. Infect. Dis. 2012, 205, 1562–1570. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Ellis, D.; King, N.P. New Vaccine Design and Delivery Technologies. J. Infect. Dis. 2019, 219, S88–S96. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Wu, Y.; Kim, Y.B.; Oh, Y.K. Advances in vaccine delivery systems against viral infectious diseases. Drug Deliv. Transl. Res. 2021, 11, 1401–1419. [Google Scholar] [CrossRef] [PubMed]
- Kaurav, M.; Madan, J.; Sudheesh, M.S.; Pandey, R.S. Combined adjuvant-delivery system for new generation vaccine antigens: Alliance has its own advantage. Artif. Cells Nanomed. Biotechnol. 2018, 46, S818–S831. [Google Scholar] [CrossRef]
- Jin, H.; Subbarao, K. Live Attenuated Influenza Vaccine. In Influenza Pathogenesis and Control; Oldstone, M.B.A., Compans, R.W., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume II, pp. 181–204. [Google Scholar]
- Rudenko, L.; Yeolekar, L.; Kiseleva, I.; Isakova-Sivak, I. Development and approval of live attenuated influenza vaccines based on Russian master donor viruses: Process challenges and success stories. Vaccine 2016, 34, 5436–5441. [Google Scholar] [CrossRef] [PubMed]
- Isakova-Sivak, I.; Chen, L.M.; Matsuoka, Y.; Voeten, J.T.; Kiseleva, I.; Heldens, J.G.; den Bosch, H.; Klimov, A.; Rudenko, L.; Cox, N.J.; et al. Genetic bases of the temperature-sensitive phenotype of a master donor virus used in live attenuated influenza vaccines: A/Leningrad/134/17/57 (H2N2). Virology 2011, 412, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Kitame, F.; Kendal, A.P.; Maassab, H.F.; Naeve, C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology 1988, 167, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sobrido, L.; Peersen, O.; Nogales, A. Temperature Sensitive Mutations in Influenza a Viral Ribonucleoprotein Complex Responsible for the Attenuation of the Live Attenuated Influenza Vaccine. Viruses 2018, 10, 560. [Google Scholar] [CrossRef]
- Zhou, B.; Li, Y.; Speer, S.D.; Subba, A.; Lin, X.; Wentworth, D.E. Engineering temperature sensitive live attenuated influenza vaccines from emerging viruses. Vaccine 2012, 30, 3691–3702. [Google Scholar] [CrossRef]
- Nogales, A.; Rodriguez, L.; Chauche, C.; Huang, K.; Reilly, E.C.; Topham, D.J.; Murcia, P.R.; Parrish, C.R.; Martinez-Sobrido, L. Temperature-Sensitive Live-Attenuated Canine Influenza Virus H3N8 Vaccine. J. Virol. 2017, 91, 02211-16. [Google Scholar] [CrossRef]
- Solorzano, A.; Ye, J.; Perez, D.R. Alternative live-attenuated influenza vaccines based on modifications in the polymerase genes protect against epidemic and pandemic flu. J. Virol. 2010, 84, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.; Blanco-Lobo, P.; Reilly, E.C.; Maehigashi, T.; Nogales, A.; Smith, A.; Topham, D.J.; Dewhurst, S.; Kim, B.; Martinez-Sobrido, L. Comparative Study of the Temperature Sensitive, Cold Adapted and Attenuated Mutations Present in the Master Donor Viruses of the Two Commercial Human Live Attenuated Influenza Vaccines. Viruses 2019, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, A.J.; Santos, C.P.; Godbout, R.A.; Subbarao, K. The temperature-sensitive and attenuation phenotypes conferred by mutations in the influenza virus PB2, PB1, and NP genes are influenced by the species of origin of the PB2 gene in reassortant viruses derived from influenza A/California/07/2009 and A/WSN/33 viruses. J. Virol. 2014, 88, 12339–12347. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Steel, J.; Liu, W.C.; Lowen, A.C.; Rodriguez, L.; Chiem, K.; Cox, A.; Garcia-Sastre, A.; Albrecht, R.A.; Dewhurst, S.; et al. Mutation L319Q in the PB1 Polymerase Subunit Improves Attenuation of a Candidate Live-Attenuated Influenza A Virus Vaccine. Microbiol. Spectr. 2022, 10, e0007822. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Rodriguez, L.; El Ghouayel, M.; Nogales, A.; Chamberlain, J.M.; Sortino, K.; Reilly, E.; Feng, C.; Topham, D.J.; Martinez-Sobrido, L.; et al. A Live Attenuated Influenza Vaccine Elicits Enhanced Heterologous Protection When the Internal Genes of the Vaccine Are Matched to Those of the Challenge Virus. J. Virol. 2020, 94, 01065-19. [Google Scholar] [CrossRef] [PubMed]
- Isakova-Sivak, I.; Korenkov, D.; Rudenko, L. Reassortant viruses for influenza vaccines: Is it time to reconsider their genome structures? Expert Rev. Vaccines 2016, 15, 565–567. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Bennink, J.R.; Smith, G.L.; Moss, B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 1985, 82, 1785–1789. [Google Scholar] [CrossRef]
- Korenkov, D.A.; Laurie, K.L.; Reading, P.C.; Carolan, L.A.; Chan, K.F.; Isakova, S., II; Smolonogina, T.A.; Subbarao, K.; Barr, I.G.; Villanueva, J.; et al. Safety, immunogenicity and protection of A(H3N2) live attenuated influenza vaccines containing wild-type nucleoprotein in a ferret model. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018, 64, 95–104. [Google Scholar] [CrossRef]
- Prokopenko, P.; Matyushenko, V.; Rak, A.; Stepanova, E.; Chistyakova, A.; Goshina, A.; Kudryavtsev, I.; Rudenko, L.; Isakova-Sivak, I. Truncation of NS1 Protein Enhances T Cell-Mediated Cross-Protection of a Live Attenuated Influenza Vaccine Virus Expressing Wild-Type Nucleoprotein. Vaccines 2023, 11, 501. [Google Scholar] [CrossRef]
- Rekstin, A.; Isakova-Sivak, I.; Petukhova, G.; Korenkov, D.; Losev, I.; Smolonogina, T.; Tretiak, T.; Donina, S.; Shcherbik, S.; Bousse, T.; et al. Immunogenicity and Cross Protection in Mice Afforded by Pandemic H1N1 Live Attenuated Influenza Vaccine Containing Wild-Type Nucleoprotein. BioMed Res. Int. 2017, 2017, 9359276. [Google Scholar] [CrossRef]
- Isakova-Sivak, I.; Matyushenko, V.; Kotomina, T.; Kiseleva, I.; Krutikova, E.; Donina, S.; Rekstin, A.; Larionova, N.; Mezhenskaya, D.; Sivak, K.; et al. Sequential Immunization with Universal Live Attenuated Influenza Vaccine Candidates Protects Ferrets against a High-Dose Heterologous Virus Challenge. Vaccines 2019, 7, 61. [Google Scholar] [CrossRef]
- Korenkov, D.; Nguyen, T.H.O.; Isakova-Sivak, I.; Smolonogina, T.; Brown, L.E.; Kedzierska, K.; Rudenko, L. Live Attenuated Influenza Vaccines engineered to express the nucleoprotein of a recent isolate stimulate human influenza CD8(+) T cells more relevant to current infections. Hum. Vaccines Immunother. 2018, 14, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Bruder, D.; Srikiatkhachorn, A.; Enelow, R.I. Cellular immunity and lung injury in respiratory virus infection. Viral Immunol. 2006, 19, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.E.; Varga, S.M. The CD8 T Cell Response to Respiratory Virus Infections. Front. Immunol. 2018, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Mezhenskaya, D.; Isakova-Sivak, I.; Rudenko, L. M2e-based universal influenza vaccines: A historical overview and new approaches to development. J. Biomed. Sci. 2019, 26, 76. [Google Scholar] [CrossRef] [PubMed]
- Berkhoff, E.G.M.; Geelhoed-Mieras, M.M.; Fouchier, R.A.M.; Osterhaus, A.; Rimmelzwaan, G.F. Assessment of the extent of variation in influenza A virus cytotoxic T-lymphocyte epitopes by using virus-specific CD8+ T-cell clones. J. Gen. Virol. 2007, 88, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Amarasena, T.H.; Laurie, K.L.; Tan, H.X.; Butler, J.; Parsons, M.S.; Alcantara, S.; Petravic, J.; Davenport, M.P.; Hurt, A.C.; et al. Standard trivalent influenza virus protein vaccination does not prime antibody-dependent cellular cytotoxicity in macaques. J. Virol. 2013, 87, 13706–13718. [Google Scholar] [CrossRef] [PubMed]
- Rak, A.; Donina, S.; Zabrodskaya, Y.; Rudenko, L.; Isakova-Sivak, I. Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests. Viruses 2022, 14, 2041. [Google Scholar] [CrossRef]
- Rak, A.; Gorbunov, N.; Kostevich, V.; Sokolov, A.; Prokopenko, P.; Rudenko, L.; Isakova-Sivak, I. Assessment of Immunogenic and Antigenic Properties of Recombinant Nucleocapsid Proteins of Five SARS-CoV-2 Variants in a Mouse Model. Viruses 2023, 15, 230. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).