Identification of Food-Derived Electrophilic Chalcones as Nrf2 Activators Using Comprehensive Virtual Screening Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Machine Learning
2.3. Molecular Docking and Virtual Screening
2.4. Cell Culture
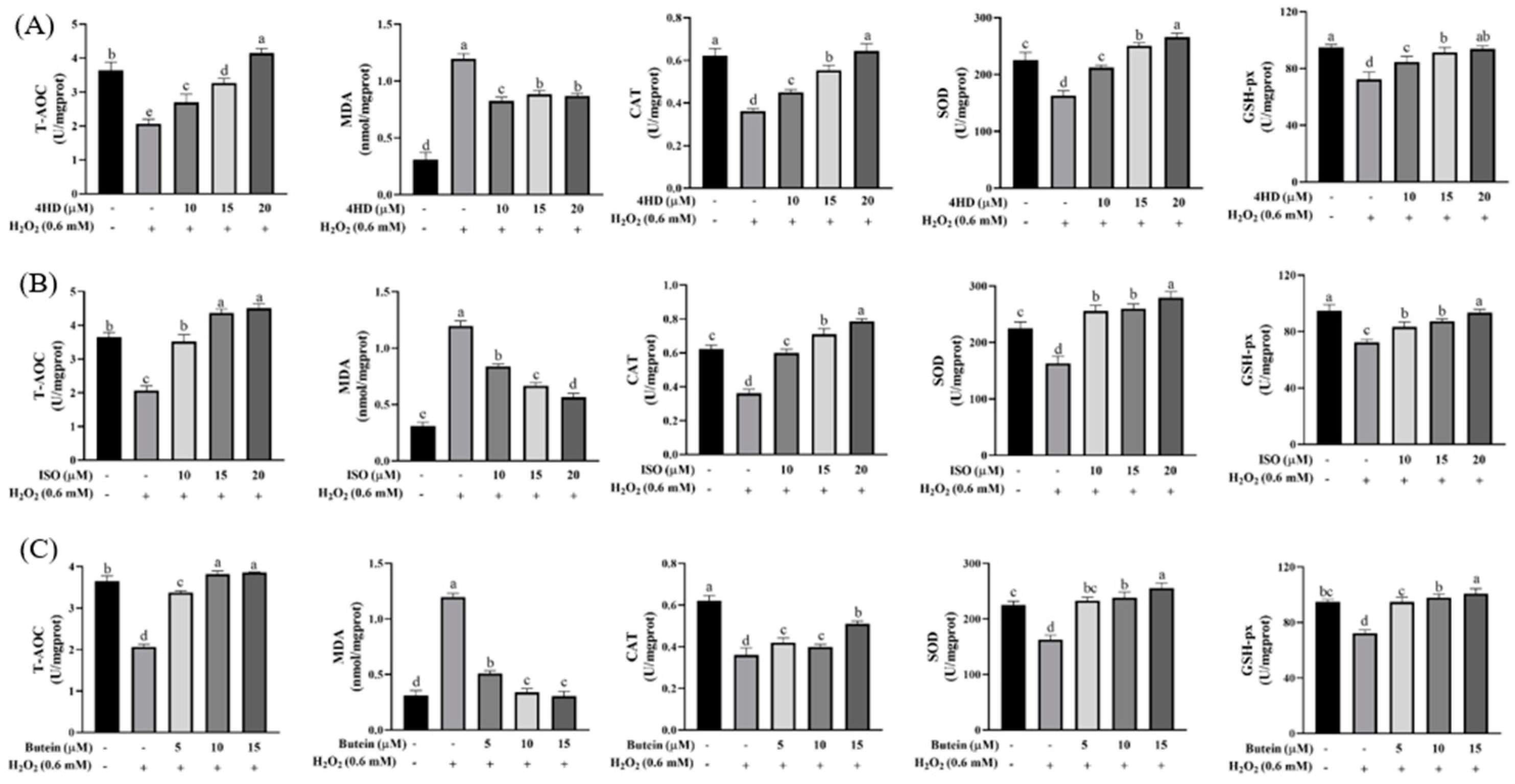
2.5. Determination of Antioxidant Biochemical Indexes
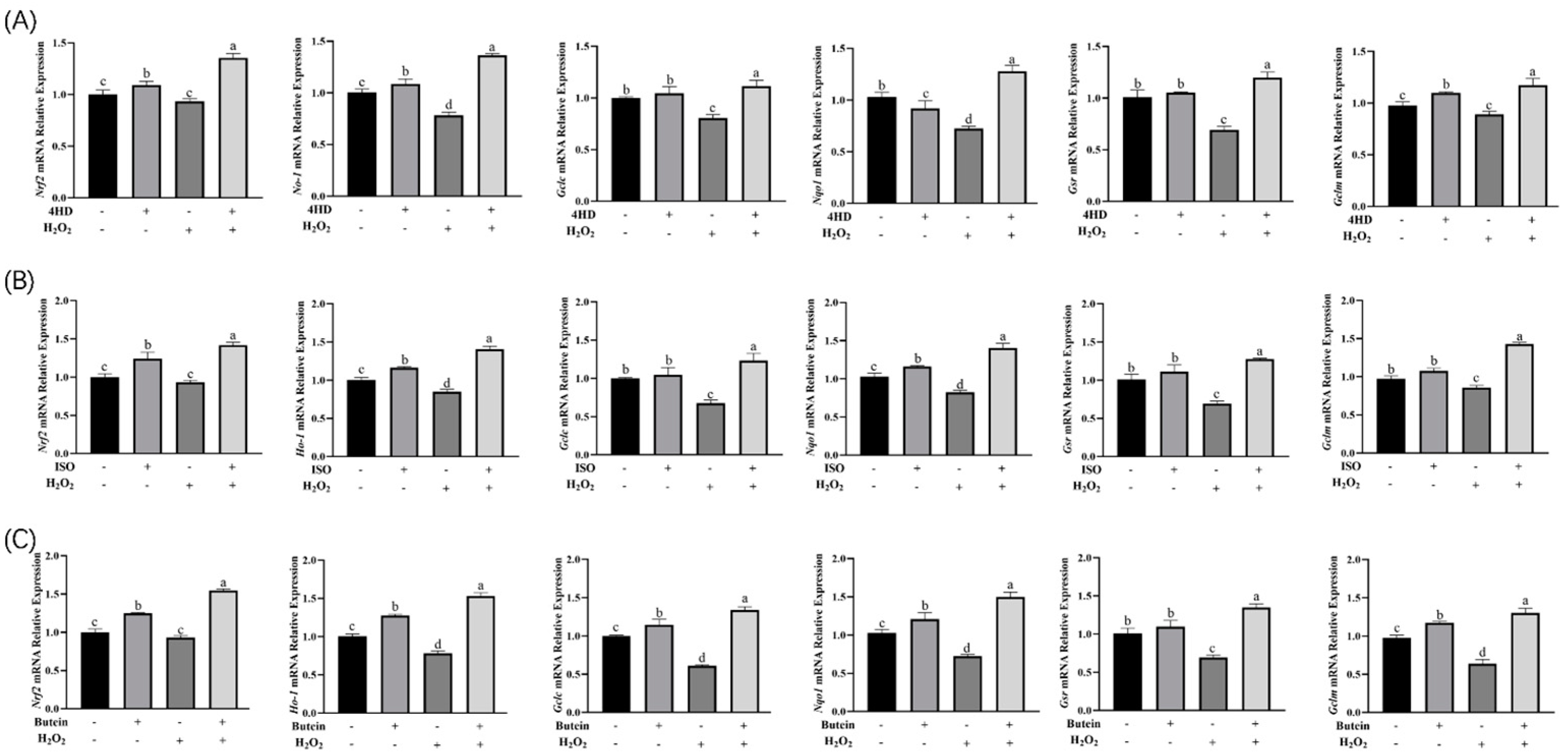
2.6. Real-Time Fluorescence Quantitative PCR
2.7. Statistical Analysis
3. Results
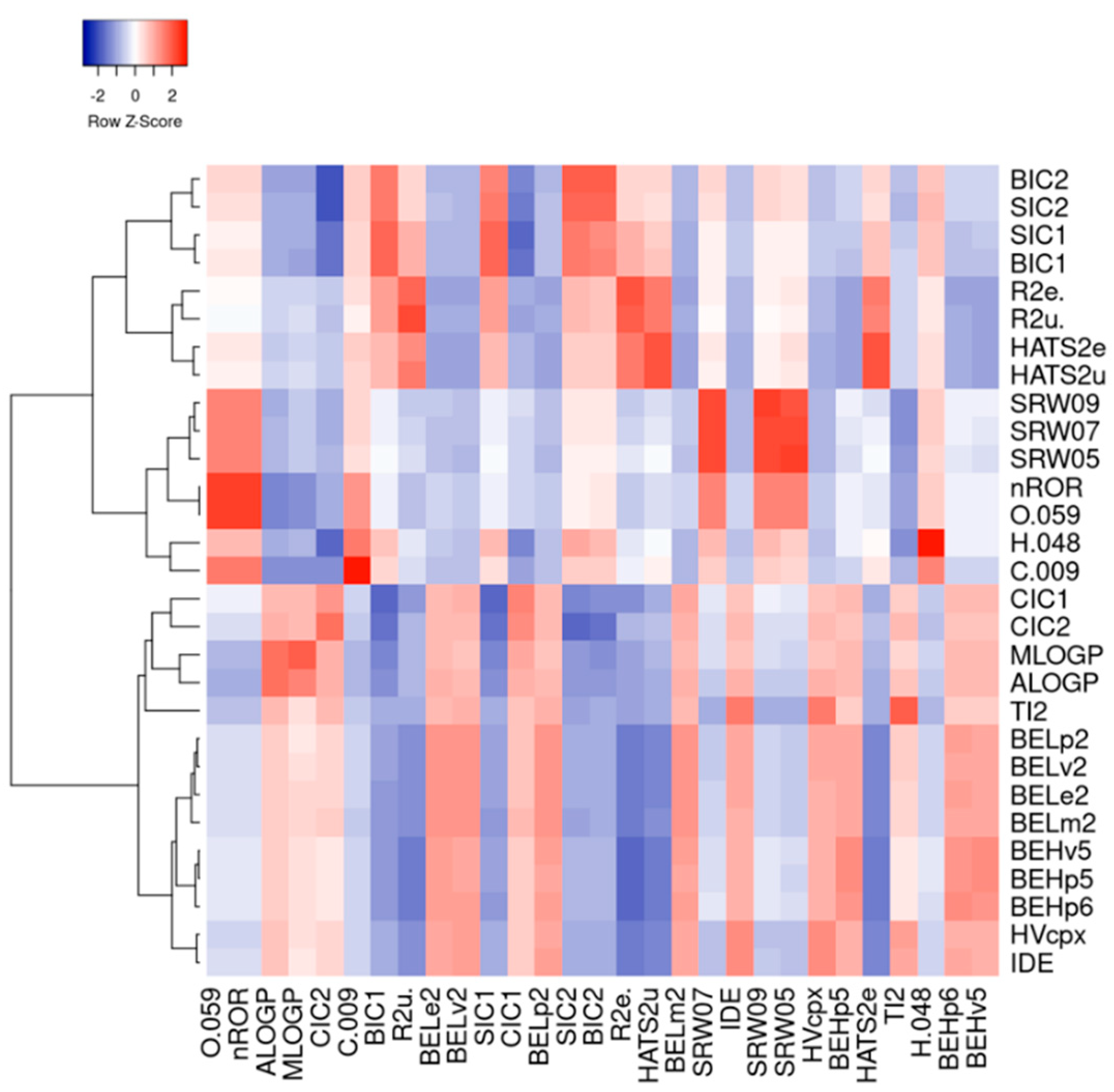
3.1. Machine Learning Analysis of Food-Derived Electrophilic Compounds
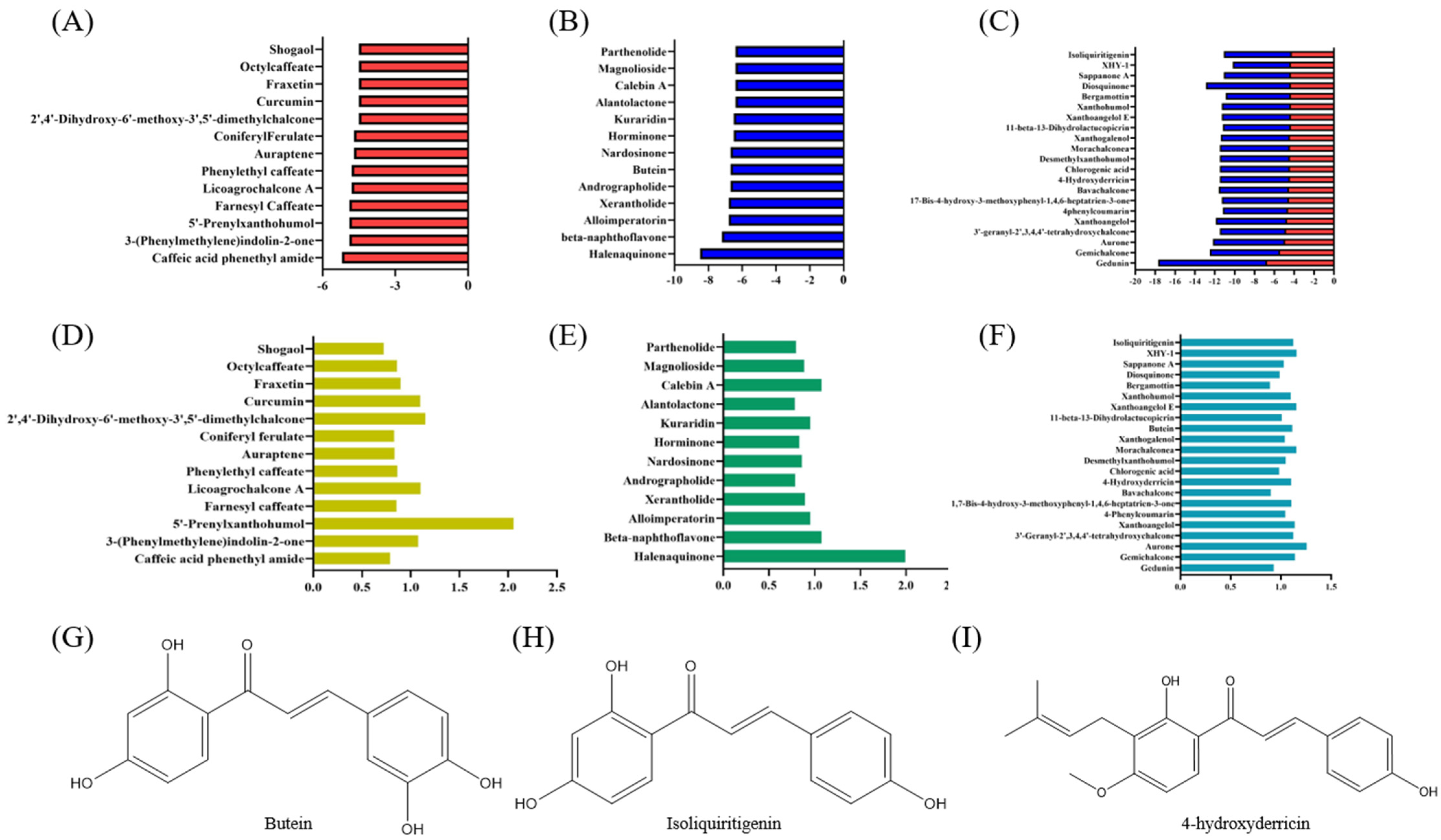
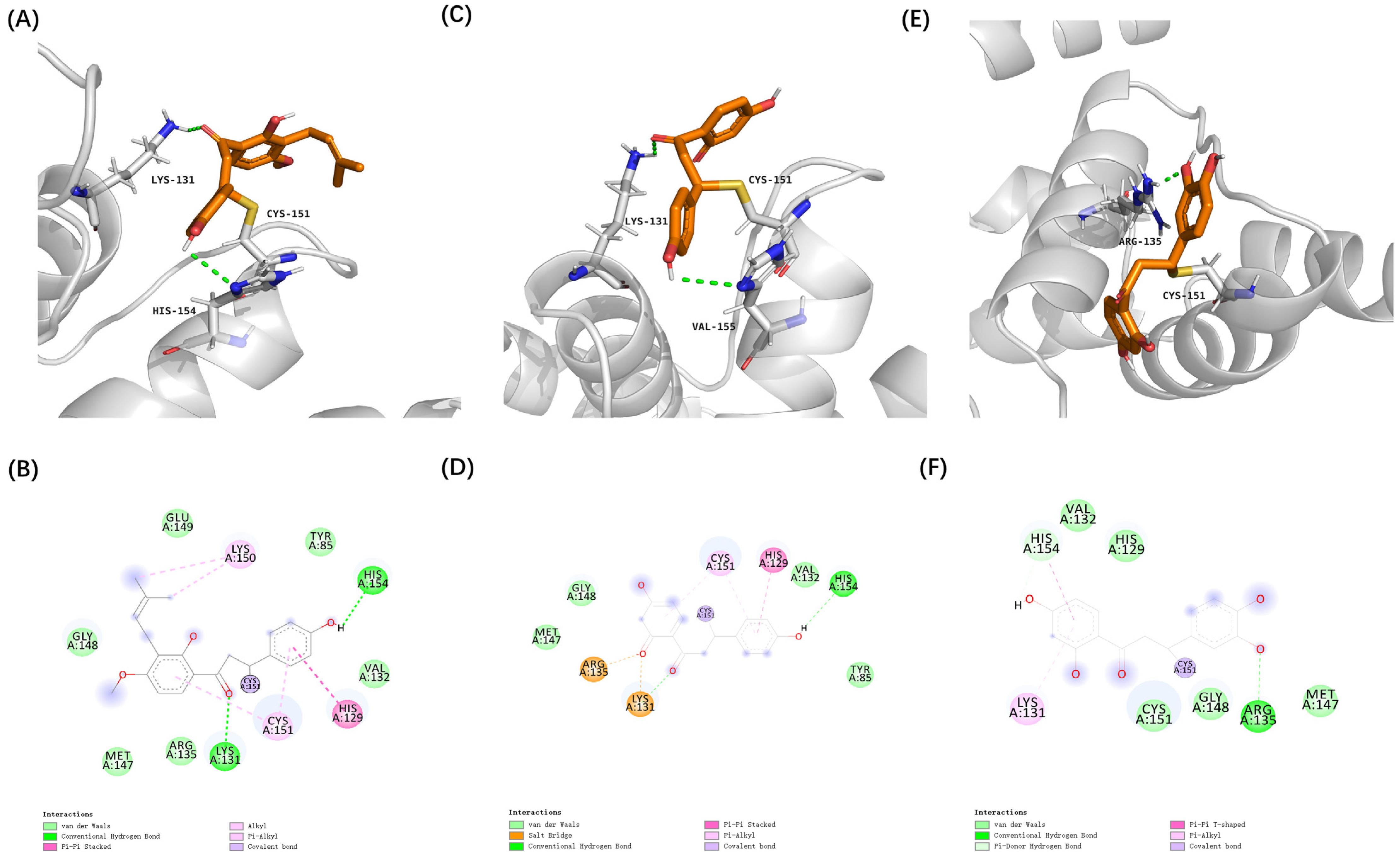
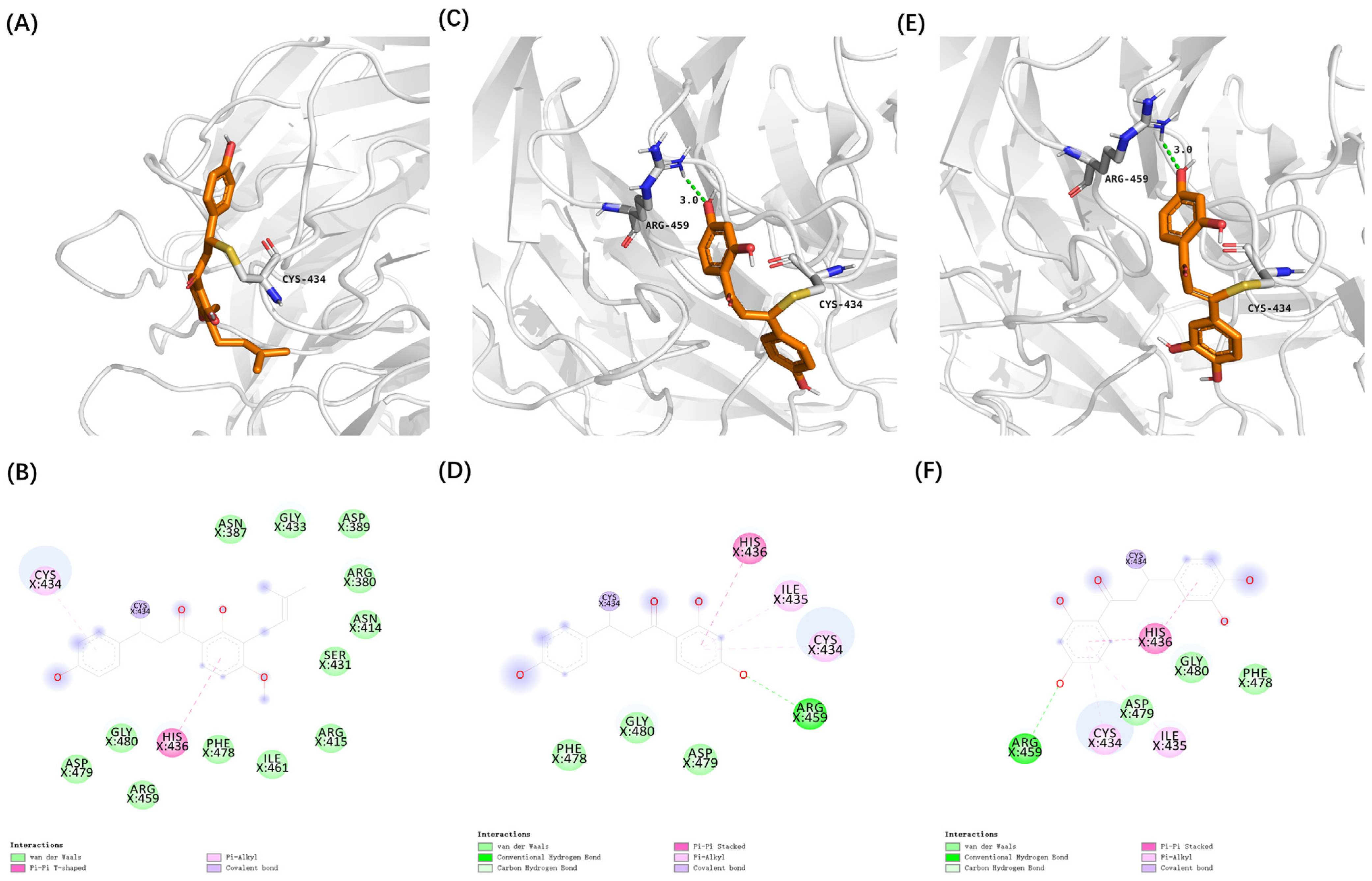
3.2. Virtual Screening of Cytoprotective Electrophilic Compounds
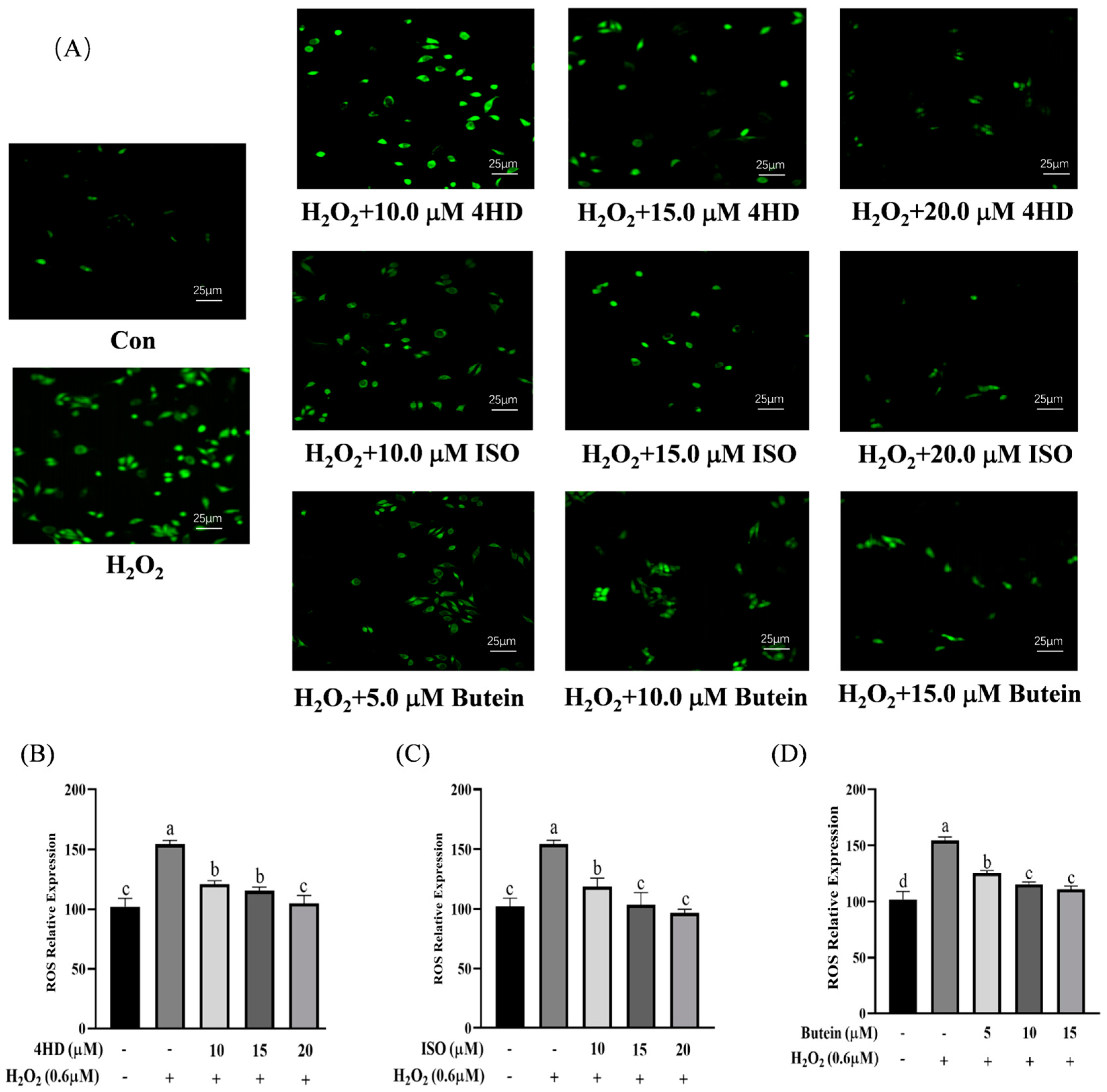
3.3. Verification of the Protective Effect of Chalcones on Oxidative-Damaged Cell Models
3.4. Expression Analysis of Nrf2 Signaling Pathway-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Islam, M.S. Molecular Regulations and Functions of the Transient Receptor Potential Channels of the Islets of Langerhans and Insulinoma Cells. Cells 2020, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Rezaie, T.; Seki, M.; Sunico, C.R.; Tabuchi, T.; Kitagawa, T.; Yanagitai, M.; Senzaki, M.; Kosegawa, C.; Taira, H.; et al. Dual neuroprotective pathways of a pro-electrophilic compound via HSF-1-activated heat-shock proteins and Nrf2-activated phase 2 antioxidant response enzymes. J. Neurochem. 2011, 119, 569–578. [Google Scholar] [CrossRef]
- Rahban, M.; Habibi-Rezaei, M.; Mazaheri, M.; Saso, L.; Moosavi-Movahedi, A.A. Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications. Antioxidants 2020, 9, 1228. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Chen, Q.M. Fresh Medium or L-Cystine as an Effective Nrf2 Inducer for Cytoprotection in Cell Culture. Cells 2023, 12, 291. [Google Scholar] [CrossRef] [PubMed]
- Muri, J.; Wolleb, H.; Broz, P.; Carreira, E.M.; Kopf, M. Electrophilic Nrf2 activators and itaconate inhibit inflammation at low dose and promote IL-1β production and inflammatory apoptosis at high dose. Redox Biol. 2020, 36, 101647. [Google Scholar] [CrossRef]
- Cheng, X.-R.; Yu, B.-T.; Song, J.; Ma, J.-H.; Chen, Y.-Y.; Zhang, C.-X.; Tu, P.-H.; Muskat, M.N.; Zhu, Z.-G. The Alleviation of Dextran Sulfate Sodium (DSS)-Induced Colitis Correlate with the logP Values of Food-Derived Electrophilic Compounds. Antioxidants 2022, 11, 2406. [Google Scholar] [CrossRef]
- Tian, X.; Fang, X.; Huang, J.Q.; Wang, L.J.; Mao, Y.B.; Chen, X.Y. A gossypol biosynthetic intermediate disturbs plant defence response. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180319. [Google Scholar] [CrossRef]
- Villa, S.M.; Heckman, J.; Bandyopadhyay, D. Medicinally Privileged Natural Chalcones: Abundance, Mechanisms of Action, and Clinical Trials. Int. J. Mol. Sci. 2024, 25, 9623. [Google Scholar] [CrossRef]
- Zhuang, C.; Zhang, W.; Sheng, C.; Zhang, W.; Xing, C.; Miao, Z. Chalcone: A Privileged Structure in Medicinal Chemistry. Chem. Rev. 2017, 117, 7762–7810. [Google Scholar] [CrossRef]
- Ducki, S. The development of chalcones as promising anticancer agents. IDrugs 2007, 10, 42–46. [Google Scholar]
- Chowdhary, S.; Preeti; Shekhar; Gupta, N.; Kumar, R.; Kumar, V. Advances in chalcone-based anticancer therapy: Mechanisms, preclinical advances, and future perspectives. Expert. Opin. Drug Discov. 2024, 19, 1417–1437. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.; Wang, R.; Fang, Z.; Sun, M.; Sun, X.; Wu, J.; Dang, Y.; Liu, J. Assessment of the Hypoglycemic and Hypolipidemic Activity of Flavonoid-Rich Extract from Angelica keiskei. Molecules 2022, 27, 6625. [Google Scholar] [CrossRef]
- Shang, A.; Liu, H.Y.; Luo, M.; Xia, Y.; Yang, X.; Li, H.Y.; Wu, D.T.; Sun, Q.; Geng, F.; Gan, R.Y. Sweet tea (Lithocarpus polystachyus rehd.) as a new natural source of bioactive dihydrochalcones with multiple health benefits. Crit. Rev. Food Sci. Nutr. 2022, 62, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.H.; Li, J.; Shen, Z.C.; Lin, X.F.; Chen, A.Q.; Wang, Y.F.; Gong, E.S.; Liu, D.; Zou, Q.; Wang, X.Y. Advances in health-promoting effects of natural polysaccharides: Regulation on Nrf2 antioxidant pathway. Front. Nutr. 2023, 10, 1102146. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef]
- Kemmerer, Z.A.; Ader, N.R.; Mulroy, S.S.; Eggler, A.L. Comparison of human Nrf2 antibodies: A tale of two proteins. Toxicol. Lett. 2015, 238, 83–89. [Google Scholar] [CrossRef]
- Park, C.L.; Kim, J.H.; Jeon, J.S.; Lee, J.H.; Zhang, K.; Guo, S.; Lee, D.H.; Gao, E.M.; Son, R.H.; Kim, Y.M.; et al. Protective Effect of Alpinia oxyphylla Fruit against tert-Butyl Hydroperoxide-Induced Toxicity in HepG2 Cells via Nrf2 Activation and Free Radical Scavenging and Its Active Molecules. Antioxidants 2022, 11, 1032. [Google Scholar] [CrossRef]
- Amoroso, R.; Maccallini, C.; Bellezza, I. Activators of Nrf2 to Counteract Neurodegenerative Diseases. Antioxidants 2023, 12, 778. [Google Scholar] [CrossRef]
- Dong, Y.; Kang, H.; Peng, R.; Liu, Z.; Liao, F.; Hu, S.A.; Ding, W.; Wang, P.; Yang, P.; Zhu, M.; et al. A clinical-stage Nrf2 activator suppresses osteoclast differentiation via the iron-ornithine axis. Cell Metab. 2024, 36, 1679–1695.e1676. [Google Scholar] [CrossRef]
- Ma, Q.; He, X. Molecular basis of electrophilic and oxidative defense: Promises and perils of Nrf2. Pharmacol. Rev. 2012, 64, 1055–1081. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Chu, X.Y.; Jiang, L.H.; Liu, M.Y.; Mei, Z.L.; Zhang, H.Y. Identification of Non-Electrophilic Nrf2 Activators from Approved Drugs. Molecules 2017, 22, 883. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Y.; Xu, L.L.; Lu, M.C.; Chen, Z.Y.; Yuan, Z.W.; Xu, X.L.; Guo, X.K.; Zhang, X.J.; Sun, H.P.; You, Q.D. Structure-Activity and Structure-Property Relationship and Exploratory in Vivo Evaluation of the Nanomolar Keap1-Nrf2 Protein-Protein Interaction Inhibitor. J. Med. Chem. 2015, 58, 6410–6421. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Dębski, K.J.; Dabrowski, M.; Czarnecka, A.M.; Szczylik, C. Gene set enrichment analysis and ingenuity pathway analysis of metastatic clear cell renal cell carcinoma cell line. Am. J. Physiol. Ren. Physiol. 2016, 311, F424–F436. [Google Scholar] [CrossRef]
- Hesse, J.; Malhan, D.; Yalçin, M.; Aboumanify, O.; Basti, A.; Relógio, A. An Optimal Time for Treatment-Predicting Circadian Time by Machine Learning and Mathematical Modelling. Cancers 2020, 12, 3103. [Google Scholar] [CrossRef]
- Xu, C.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef]
- Harigua-Souiai, E.; Oualha, R.; Souiai, O.; Abdeljaoued-Tej, I.; Guizani, I. Applied Machine Learning Toward Drug Discovery Enhancement: Leishmaniases as a Case Study. Bioinform. Biol. Insights 2022, 16, 11779322221090349. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef]
- Batey, R.T.; Gilbert, S.D.; Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 2004, 432, 411–415. [Google Scholar] [CrossRef]
- Varnek, A.; Baskin, I. Machine learning methods for property prediction in chemoinformatics: Quo Vadis? J. Chem. Inf. Model. 2012, 52, 1413–1437. [Google Scholar] [CrossRef]
- Korkmaz, S. Deep Learning-Based Imbalanced Data Classification for Drug Discovery. J. Chem. Inf. Model. 2020, 60, 4180–4190. [Google Scholar] [CrossRef]
- Altae-Tran, H.; Ramsundar, B.; Pappu, A.S.; Pande, V. Low Data Drug Discovery with One-Shot Learning. ACS Cent. Sci. 2017, 3, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Das, J.; Ghosh, J.; Roy, A.; Sil, P.C. Mangiferin exerts hepatoprotective activity against D-galactosamine induced acute toxicity and oxidative/nitrosative stress via Nrf2-NFκB pathways. Toxicol. Appl. Pharmacol. 2012, 260, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Elkhedir, A.; Yahya, A.; Mansour, M.; Korin, A.; Albahi, A.; Khalifa, I.; Maqsood, S.; Xu, X. Protective Effects of Capsaicinoid Glucoside from Fresh Hot Peppers Against Hydrogen peroxide-induced Oxidative Stress in HepG2 Cells Through-dependent Signaling Pathway. Plant Foods Hum. Nutr. 2024, 80, 25. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Yoo, J.H.; Lee, Y.S.; Park, E.J.; Lee, H.J. Ameliorative effects of black ginseng on nonalcoholic fatty liver disease in free fatty acid-induced HepG2 cells and high-fat/high-fructose diet-fed mice. J. Ginseng Res. 2020, 44, 350–361. [Google Scholar] [CrossRef]
- Matsagar, S.V.; Singh, R.K. Protective Effects of NRF2 Activator Sulforaphane in Polyinosinic:Polycytidylic Acid-Induced In Vitro and In Vivo Model. J. Biochem. Mol. Toxicol. 2024, 38, e70086. [Google Scholar] [CrossRef]
- Zou, B.; Xiao, G.; Xu, Y.; Wu, J.; Yu, Y.; Fu, M. Persimmon vinegar polyphenols protect against hydrogen peroxide-induced cellular oxidative stress via Nrf2 signalling pathway. Food Chem. 2018, 255, 23–30. [Google Scholar] [CrossRef]
- Erlank, H.; Elmann, A.; Kohen, R.; Kanner, J. Polyphenols activate Nrf2 in astrocytes via H2O2, semiquinones, and quinones. Free Radic. Biol. Med. 2011, 51, 2319–2327. [Google Scholar] [CrossRef]
- Satoh, T.; McKercher, S.R.; Lipton, S.A. Reprint of: Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic. Biol. Med. 2014, 66, 45–57. [Google Scholar] [CrossRef]
- Gersch, M.; Kreuzer, J.; Sieber, S.A. Electrophilic natural products and their biological targets. Nat. Prod. Rep. 2012, 29, 659–682. [Google Scholar] [CrossRef]
- Belka, M.; Gostyńska-Stawna, A.; Stawny, M.; Krajka-Kuźniak, V. Activation of Nrf2 and FXR via Natural Compounds in Liver Inflammatory Disease. Int. J. Mol. Sci. 2024, 25, 11213. [Google Scholar] [CrossRef]
- Sakanyan, V. Reactive Chemicals and Electrophilic Stress in Cancer: A Minireview. High Throughput 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Ciriano, I.; Bender, A. Improved Chemical Structure-Activity Modeling Through Data Augmentation. J. Chem. Inf. Model. 2015, 55, 2682–2692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Guttman, Y.; Kerem, Z. Dietary Inhibitors of CYP3A4 Are Revealed Using Virtual Screening by Using a New Deep-Learning Classifier. J. Agric. Food Chem. 2022, 70, 2752–2761. [Google Scholar] [CrossRef]
- Hilal, A.M.; Malibari, A.A.; Obayya, M.; Alzahrani, J.S.; Alamgeer, M.; Mohamed, A.; Motwakel, A.; Yaseen, I.; Hamza, M.A.; Zamani, A.S. Feature Subset Selection with Optimal Adaptive Neuro-Fuzzy Systems for Bioinformatics Gene Expression Classification. Comput. Intell. Neurosci. 2022, 2022, 1698137. [Google Scholar] [CrossRef]
- Ishii, T.; Warabi, E. Mechanism of Rapid Nuclear Factor-E2-Related Factor 2 (Nrf2) Activation via Membrane-Associated Estrogen Receptors: Roles of NADPH Oxidase 1, Neutral Sphingomyelinase 2 and Epidermal Growth Factor Receptor (EGFR). Antioxidants 2019, 8, 69. [Google Scholar] [CrossRef]
- Li, M.; Huang, W.; Jie, F.; Wang, M.; Zhong, Y.; Chen, Q.; Lu, B. Discovery of Keap1-Nrf2 small-molecule inhibitors from phytochemicals based on molecular docking. Food Chem. Toxicol. 2019, 133, 110758. [Google Scholar] [CrossRef]
- Cheng, Y.; Cheng, L.; Gao, X.; Chen, S.; Wu, P.; Wang, C.; Liu, Z. Covalent modification of Keap1 at Cys77 and Cys434 by pubescenoside a suppresses oxidative stress-induced NLRP3 inflammasome activation in myocardial ischemia-reperfusion injury. Theranostics 2021, 11, 861–877. [Google Scholar] [CrossRef]
- Wang, M.; Liu, C.Y.; Wang, T.; Yu, H.M.; Ouyang, S.H.; Wu, Y.P.; Gong, H.B.; Ma, X.H.; Jiao, G.L.; Fu, L.L.; et al. (+)-Clausenamide protects against drug-induced liver injury by inhibiting hepatocyte ferroptosis. Cell Death Dis. 2020, 11, 781. [Google Scholar] [CrossRef]
- Jackson, P.A.; Widen, J.C.; Harki, D.A.; Brummond, K.M. Covalent Modifiers: A Chemical Perspective on the Reactivity of α,β-Unsaturated Carbonyls with Thiols via Hetero-Michael Addition Reactions. J. Med. Chem. 2017, 60, 839–885. [Google Scholar] [CrossRef]
- Lee, C.; Park, C. Bacterial Responses to Glyoxal and Methylglyoxal: Reactive Electrophilic Species. Int. J. Mol. Sci. 2017, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.-L.; Zhang, Z.-W.; Lekkala, R.; Alsulami, H.; Rakesh, K.P. Chalcone hybrids as privileged scaffolds in antimalarial drug discovery: A key review. Eur. J. Med. Chem. 2020, 193, 112215. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell Biochem. 2020, 467, 1–12. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Li, W.; Ai, N.; Jin, H. Biliverdin/Bilirubin Redox Pair Protects Lens Epithelial Cells against Oxidative Stress in Age-Related Cataract by Regulating NF-κB/iNOS and Nrf2/HO-1 Pathways. Oxid. Med. Cell Longev. 2022, 2022, 7299182. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, P.S.; Soares, M.A.; Hameedi, S.G.; Kadle, R.L.; Mubasher, A.; Kowzun, M.; Ceradini, D.J. Dysregulation of Nrf2/Keap1 Redox Pathway in Diabetes Affects Multipotency of Stromal Cells. Diabetes 2019, 68, 141–155. [Google Scholar] [CrossRef]
- Rodrigo, R.; Retamal, C.; Schupper, D.; Vergara-Hernández, D.; Saha, S.; Profumo, E.; Buttari, B.; Saso, L. Antioxidant Cardioprotection against Reperfusion Injury: Potential Therapeutic Roles of Resveratrol and Quercetin. Molecules 2022, 27, 2564. [Google Scholar] [CrossRef]
- Schwarz, M.; Lossow, K.; Kopp, J.F.; Schwerdtle, T.; Kipp, A.P. Crosstalk of Nrf2 with the Trace Elements Selenium, Iron, Zinc, and Copper. Nutrients 2019, 11, 2112. [Google Scholar] [CrossRef]
- Francis, N.; Rao, S.; Blanchard, C.; Santhakumar, A. Black Sorghum Phenolic Extract Regulates Expression of Genes Associated with Oxidative Stress and Inflammation in Human Endothelial Cells. Molecules 2019, 24, 3321. [Google Scholar] [CrossRef]
- Liu, T.; Lv, Y.F.; Zhao, J.L.; You, Q.D.; Jiang, Z.Y. Regulation of Nrf2 by phosphorylation: Consequences for biological function and therapeutic implications. Free Radic. Biol. Med. 2021, 168, 129–141. [Google Scholar] [CrossRef]
- Matzinger, M.; Fischhuber, K.; Pölöske, D.; Mechtler, K.; Heiss, E.H. AMPK leads to phosphorylation of the transcription factor Nrf2, tuning transactivation of selected target genes. Redox Biol. 2020, 29, 101393. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zheng, X.; Ji, J.; Zhu, X.; Liu, X.; Liu, H.; Guo, L.; Ye, K.; Zhang, S.; Xu, Y.-j.; et al. The carotenoid torularhodin alleviates NAFLD by promoting Akkermanisa muniniphila-mediated adenosylcobalamin metabolism. Nat. Commun. 2025, 16, 3338. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Nrf2 | TCCAGTCAGAAACCAGTGGAT | GAATGTCTGCGCCAAAAGCTG |
| Nqo1 | GAAGAGCACTGATCGTACTGGC | GGATACTGAAAGTTCGCAGGG |
| Gclc | GGAGACCAGAGTATGGGAGTT | CCGGCGTTTTCGCATGTTG |
| Gclm | CATTTACAGCCTTACTGGGAGG | ATGCAGTCAAATCTGGTGGCA |
| HO-1 | AAGACTGCGTTCCTGCTCAAC | AAAGCCCTACAGCAACTGTCG |
| Gsr | CACGAGTGATCCCAAGCCC | CAATGTAACCTGCACCAACAATG |
| β-actin | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, B.; Tu, P.; Weng, J.; Zhang, Y.; Lin, Q.; Muskat, M.N.; Wang, J.; Tang, X.; Cheng, X. Identification of Food-Derived Electrophilic Chalcones as Nrf2 Activators Using Comprehensive Virtual Screening Techniques. Antioxidants 2025, 14, 546. https://doi.org/10.3390/antiox14050546
Bai B, Tu P, Weng J, Zhang Y, Lin Q, Muskat MN, Wang J, Tang X, Cheng X. Identification of Food-Derived Electrophilic Chalcones as Nrf2 Activators Using Comprehensive Virtual Screening Techniques. Antioxidants. 2025; 14(5):546. https://doi.org/10.3390/antiox14050546
Chicago/Turabian StyleBai, Bingyu, Piaohan Tu, Jiayi Weng, Yan Zhang, Quan Lin, Mitchell N. Muskat, Jie Wang, Xue Tang, and Xiangrong Cheng. 2025. "Identification of Food-Derived Electrophilic Chalcones as Nrf2 Activators Using Comprehensive Virtual Screening Techniques" Antioxidants 14, no. 5: 546. https://doi.org/10.3390/antiox14050546
APA StyleBai, B., Tu, P., Weng, J., Zhang, Y., Lin, Q., Muskat, M. N., Wang, J., Tang, X., & Cheng, X. (2025). Identification of Food-Derived Electrophilic Chalcones as Nrf2 Activators Using Comprehensive Virtual Screening Techniques. Antioxidants, 14(5), 546. https://doi.org/10.3390/antiox14050546







