Genome-Wide Identification and Characterization of Chinese Cabbage S1fa Transcription Factors and Their Roles in Response to Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of the S1fa Family Genes in Chinese Cabbage
2.2. Phylogenetic Analysis of the S1fa Genes in Chinese Cabbage
2.3. Cis-Element Analysis of S1fa
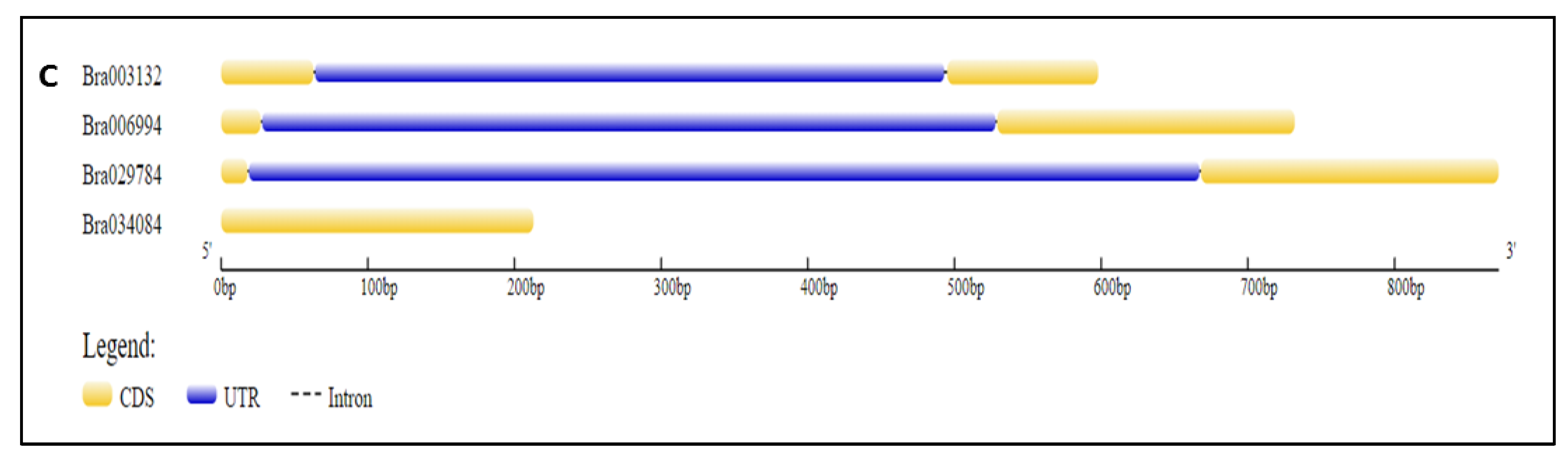
2.4. Structure and Motif Analysis of the S1fa Genes
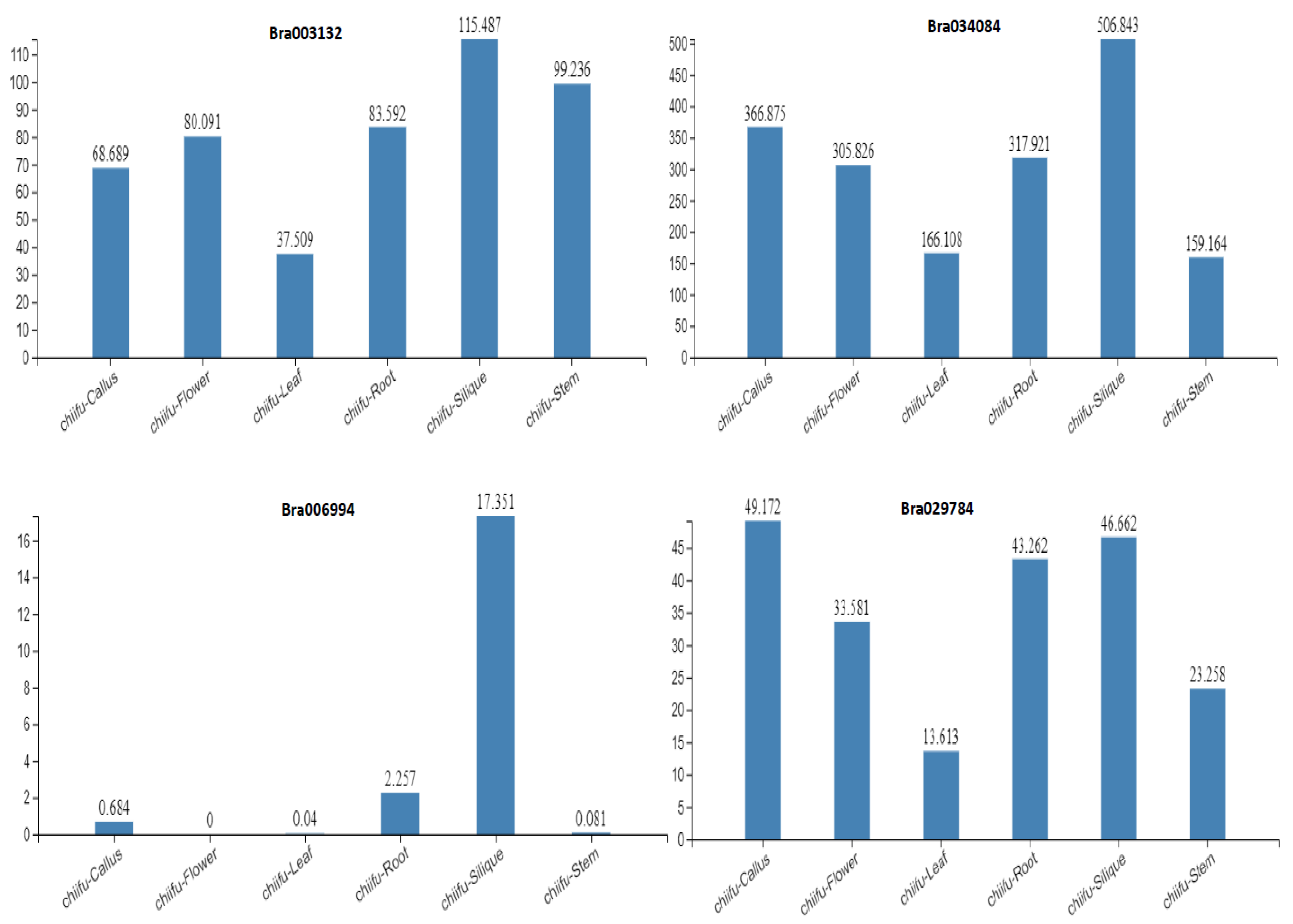
2.5. Expression Profiles of the S1fa Genes in Different Tissues
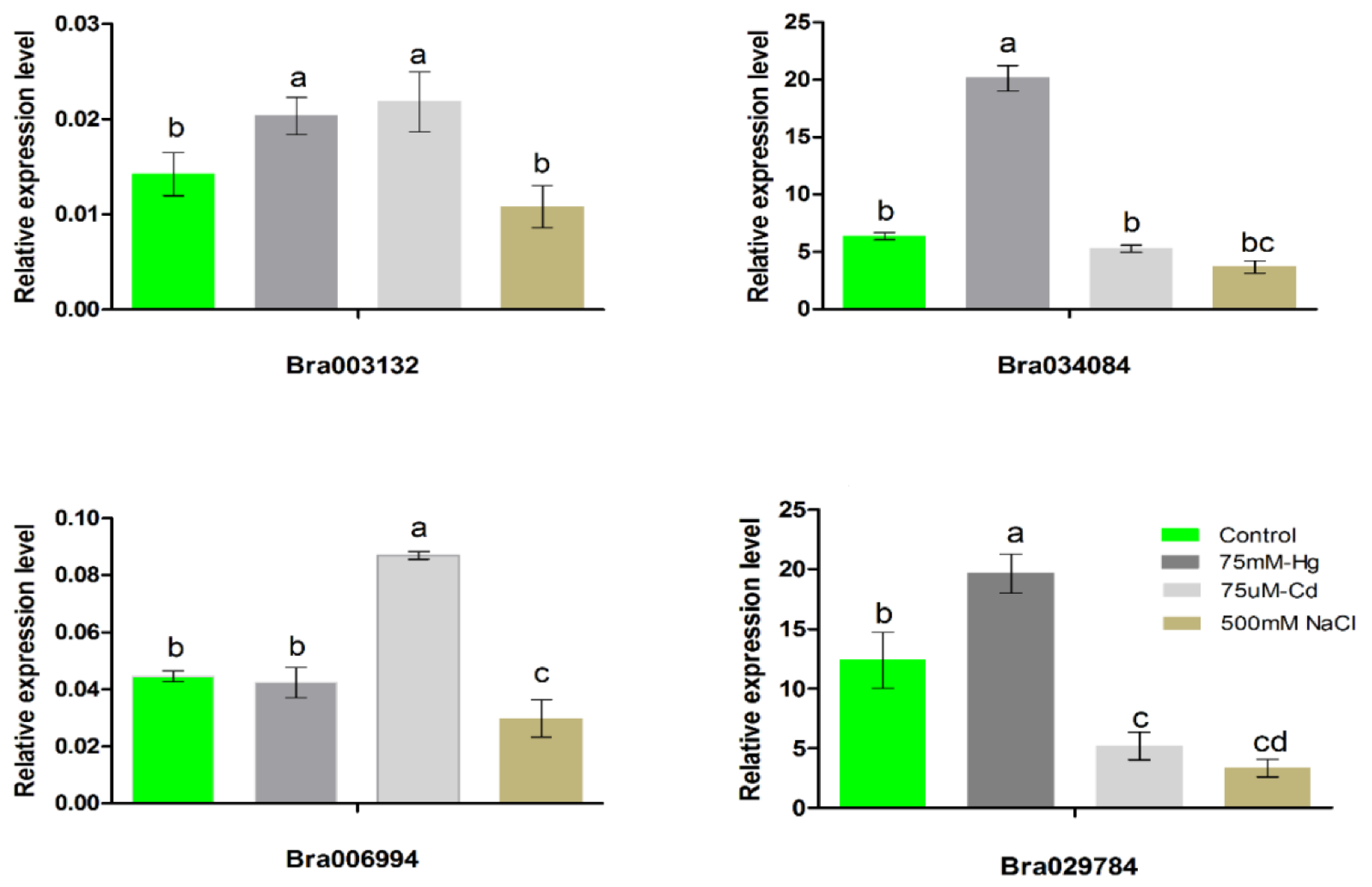
2.6. Expression Patterns of the S1fa Genes under Abiotic Stress
2.7. Prediction of miRNAs Targeting the S1fa Genes in Chinese Cabbage
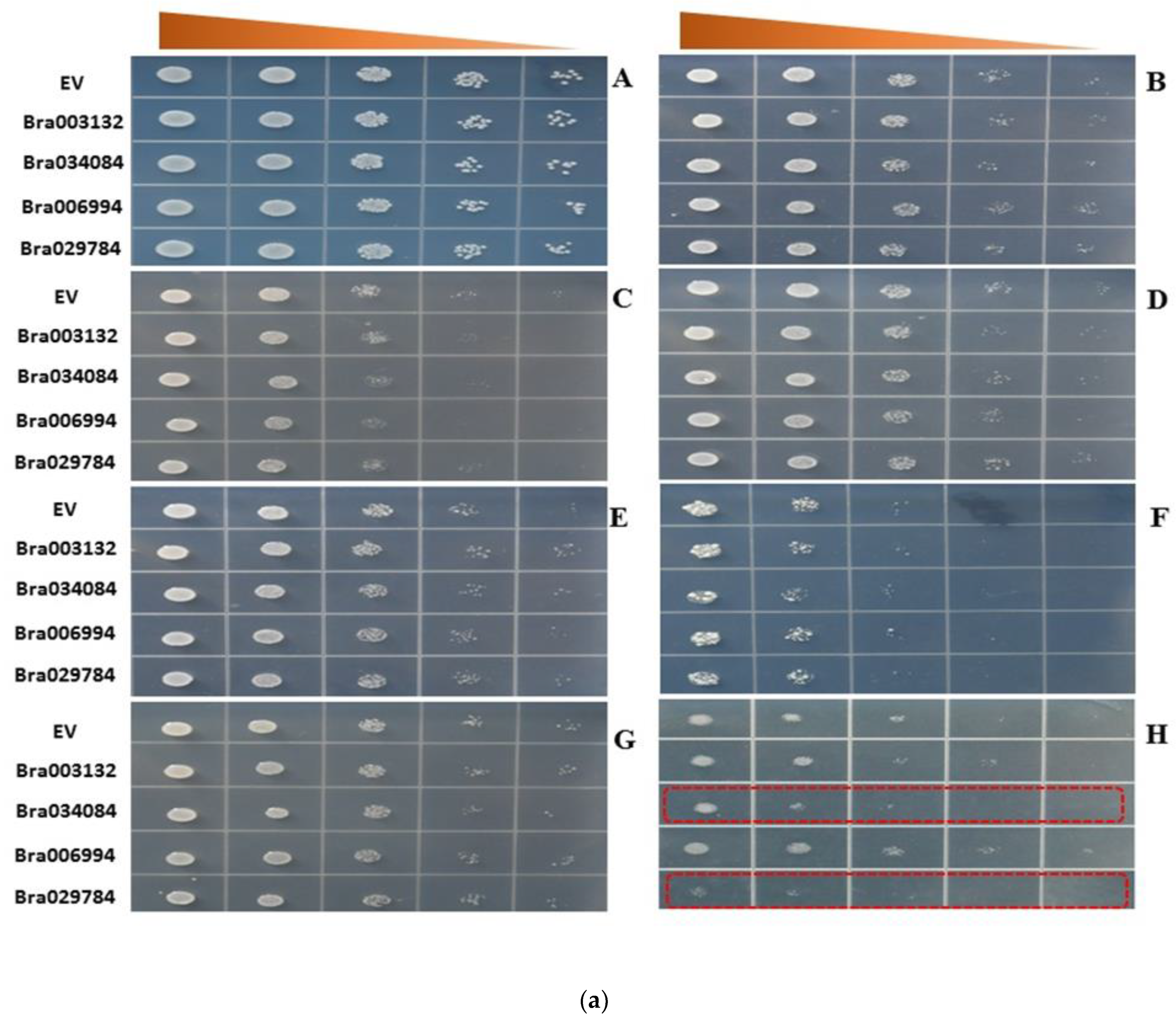
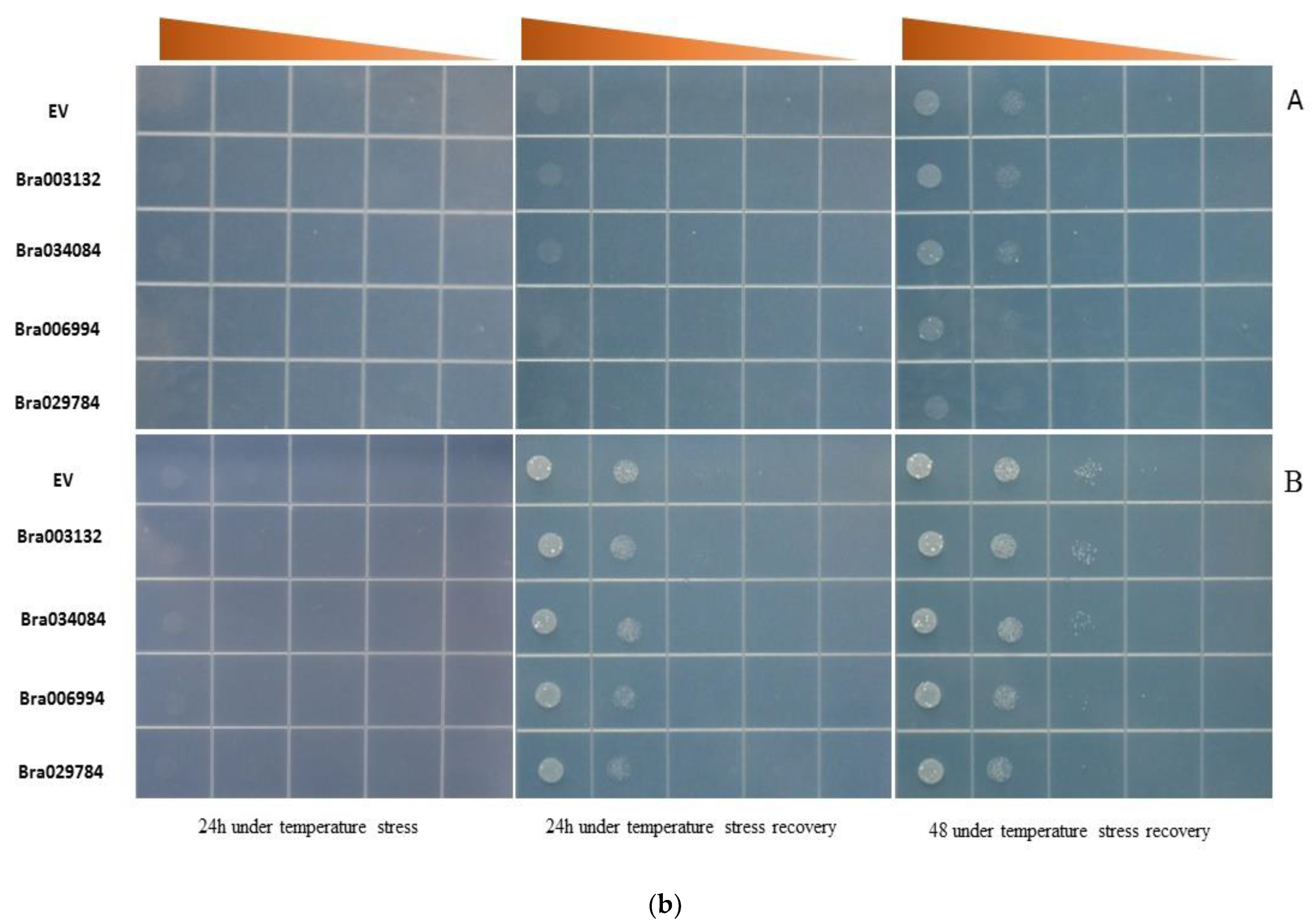
2.8. S1fa Overexpression in Response to Abiotic Stresses in Yeast
2.9. Growth Curve
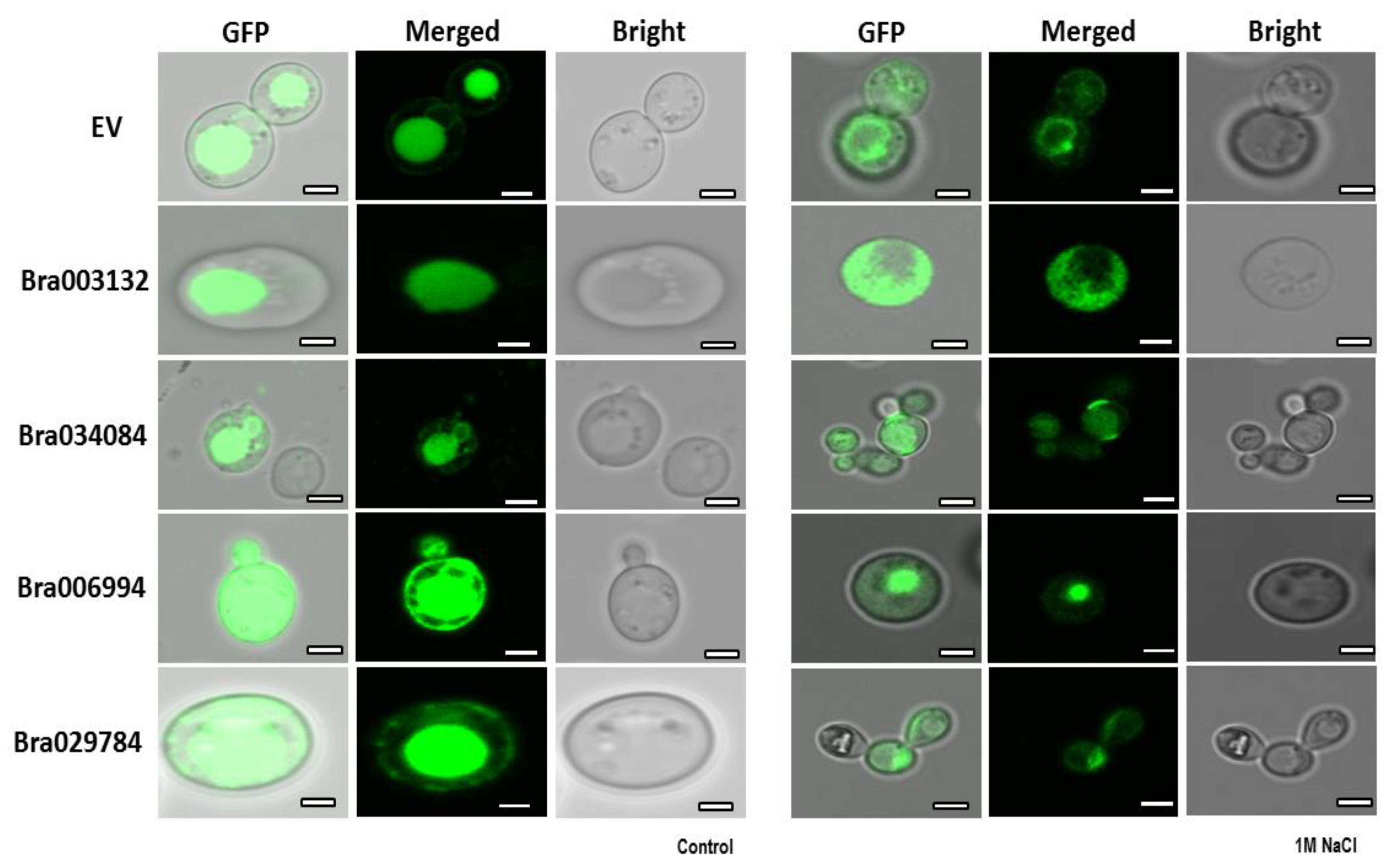
2.10. Subcellular Localization of the S1Fa Genes
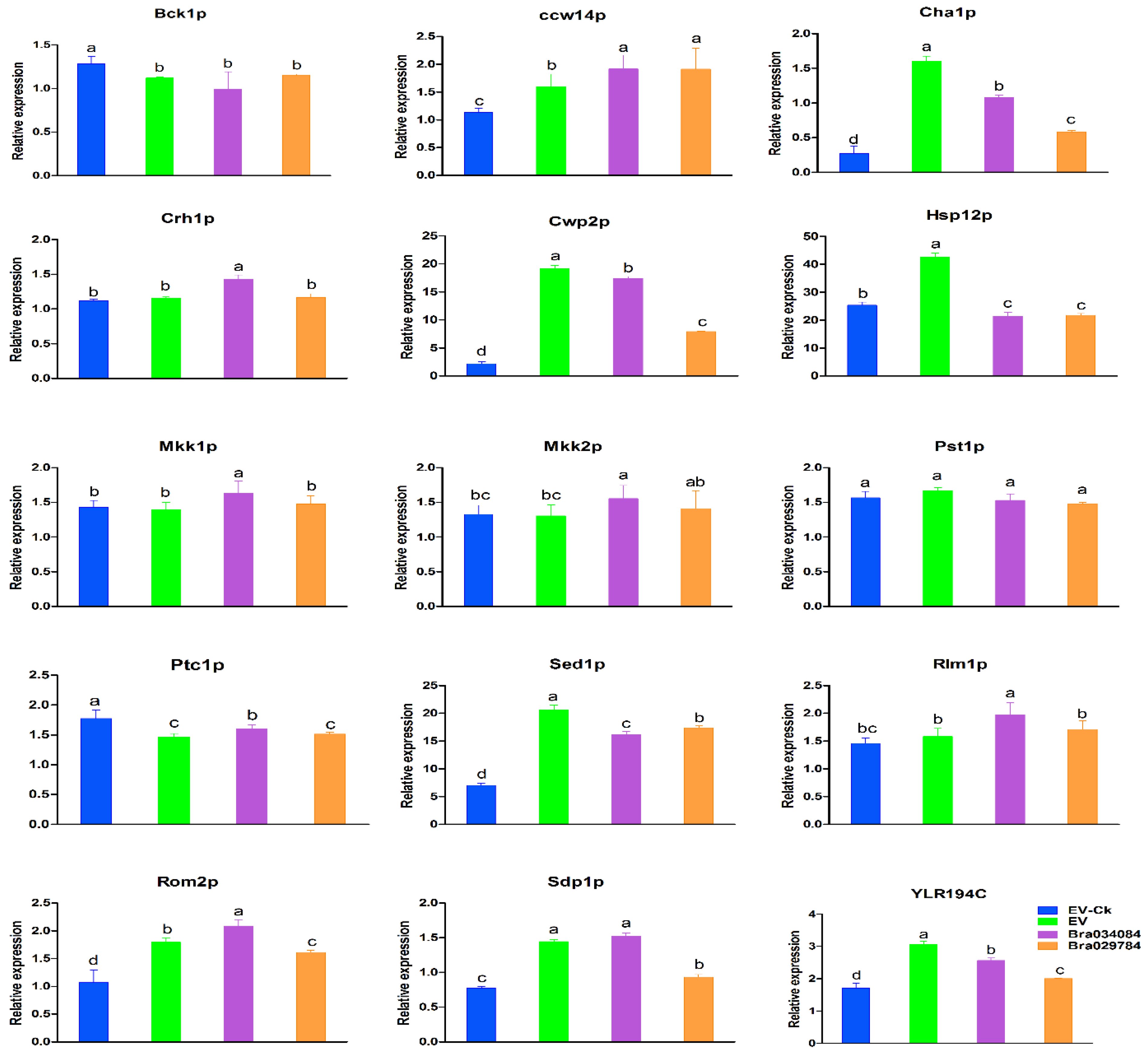
2.11. Responses of Cell Wall Biosynthesis Genes to NaCl Stresses
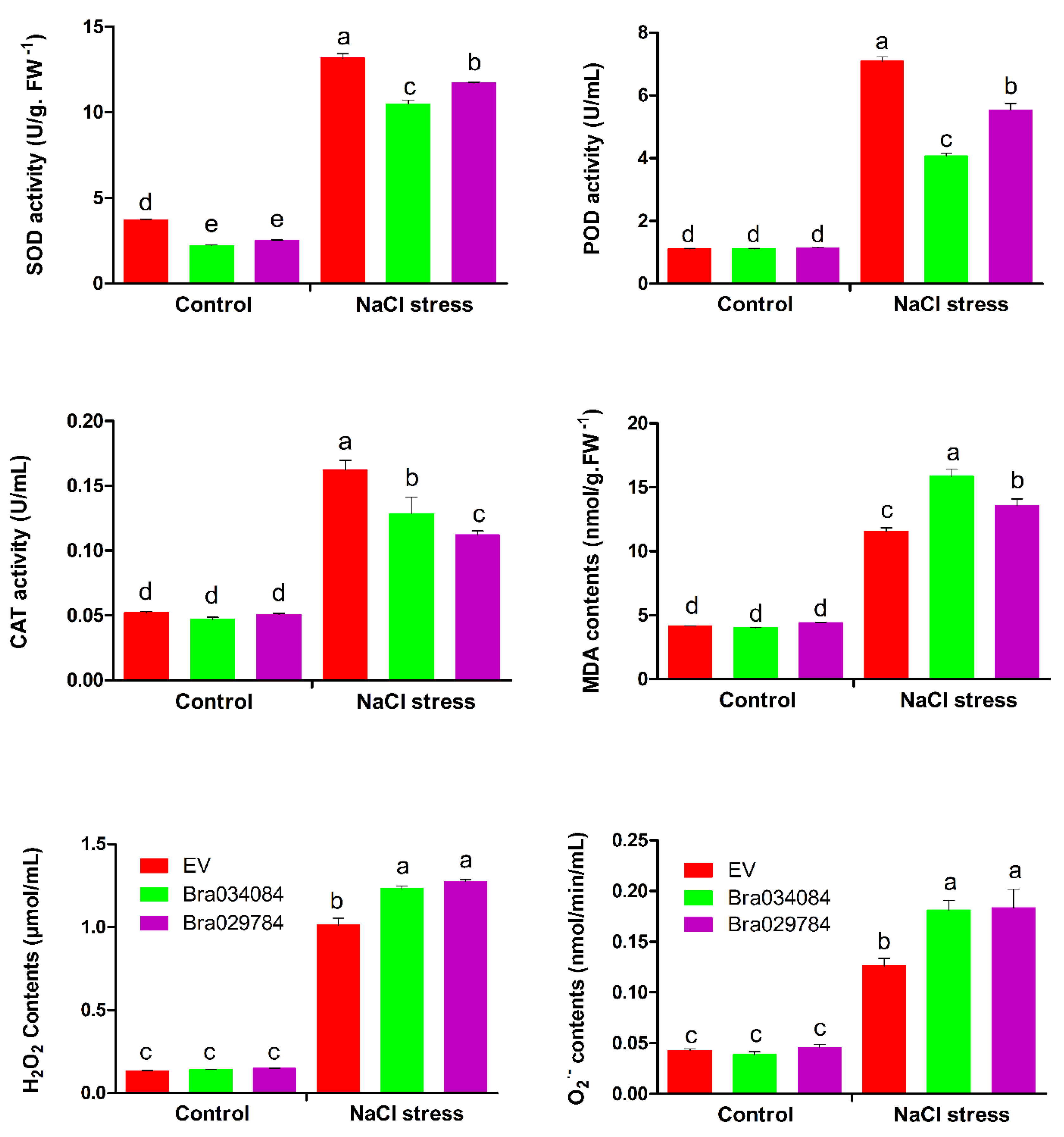
2.12. Antioxidant Enzyme Activities and ROS Accumulation under NaCl Stresses
3. Discussion
4. Materials and Method
4.1. Identification of the S1fa Genes in Chinese Cabbage
4.2. Phylogenetic Trees and Sequence Alignment
4.3. S1fa Structure and Conserved Motif
4.4. S1fa Gene Promoter Analysis and miRNA Prediction
4.5. Total RNA Extraction and qRT-PCR Analysis
4.6. Yeast Constructs
4.7. Tolerance Assay and Growth Curve
4.8. Determination of Antioxidant Enzyme Activities and ROS Contents
5. Statistical Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mabuchi, R.; Tanaka, M.; Nakanishi, C.; Takatani, N.; Tanimoto, S. Analysis of Primary Metabolites in Cabbage (Brassica oleracea var. capitata) Varieties Correlated with Antioxidant Activity and Taste Attributes by Metabolic Profiling. Molecules 2019, 24, 4282. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Li, N.; Ty, S.; Xh, L.; Brestic, M.; Shao, H.; Li, J.; Rki, S. Accumulation capacity of ions in cabbage (Brassica oleracea L.) supplied with sea water. Plant Soil Environ. 2016, 62, 314–320. [Google Scholar]
- Park, S.; Valan Arasu, M.; Lee, M.K.; Chun, J.H.; Seo, J.M.; Lee, S.W.; Al-Dhabi, N.A.; Kim, S.J. Quantification of glucosinolates, anthocyanins, free amino acids, and vitamin C in inbred lines of cabbage (Brassica oleracea L.). Food Chem. 2014, 145, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, Y.; Wang, X.; Lv, J.; Tang, Z.; Hu, L.; Luo, S.; Wang, R.; Ali, B.; Yu, J. Physiological Mechanism of Exogenous 5-Aminolevulinic Acid Improved the Tolerance of Chinese Cabbage (Brassica pekinensis L.) to Cadmium Stress. Front. Plant Sci. 2022, 13, 845396. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Hu, J.; Gao, C.; Chen, G.; Wang, B.; Lin, C.; Song, L.; Ding, Y.; Zhou, G. Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp. chinensis). Sci. Rep. 2019, 9, 5002. [Google Scholar] [CrossRef]
- Anwar, A.; Kim, J.K. Transgenic Breeding Approaches for Improving Abiotic Stress Tolerance: Recent Progress and Future Perspectives. Int. J. Mol. Sci. 2020, 21, 2695. [Google Scholar] [CrossRef]
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 2018, 51, 46. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI Pathway: Regulation of the Transcriptional Adaptive Response to Cell Wall Stress in Yeast. J. Fungi 2017, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Lippold, F.; Sanchez, D.H.; Musialak, M.; Schlereth, A.; Scheible, W.R.; Hincha, D.K.; Udvardi, M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009, 149, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Schippers, J.H.; Mieulet, D.; Obata, T.; Fernie, A.R.; Guiderdoni, E.; Mueller-Roeber, B. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J. 2013, 76, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Zagorchev, L.; Kamenova, P.; Odjakova, M. The Role of Plant Cell Wall Proteins in Response to Salt Stress. Sci. World J. 2014, 2014, 764089. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Niu, Y.; Dong, H.; Jia, Y.; Wang, Y. Characterization of the Function of Two S1Fa-Like Family Genes From Populus trichocarpa. Front. Plant Sci. 2021, 12, 753099. [Google Scholar] [CrossRef]
- Kim, S.-I.; Lee, K.H.; Kwak, J.S.; Kwon, D.H.; Song, J.T.; Seo, H.S. Overexpression of Rice OsS1Fa1 Gene Confers Drought Tolerance in Arabidopsis. Plants 2021, 10, 2181. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.X.; Li, Y.F.; Rocipon, M.; Mache, R. Sequence-specific interaction between S1F, a spinach nuclear factor, and a negative cis-element conserved in plastid-related genes. J. Biol. Chem. 1992, 267, 23515–23519. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W.; Yao, N.; Feng, Y.; Chai, R.; Yang, G.; et al. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef] [PubMed]
- Bo, W.; Zhaohui, Z.; Huanhuan, Z.; Xia, W.; Binglin, L.; Lijia, Y.; Xiangyan, H.; Deshui, Y.; Xuelian, Z.; Chunguo, W.; et al. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice. Rice Sci. 2019, 26, 98–108. [Google Scholar] [CrossRef]
- Ohnishi, T.; Sugahara, S.; Yamada, T.; Kikuchi, K.; Yoshiba, Y.; Hirano, H.Y.; Tsutsumi, N. OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 2005, 80, 135–139. [Google Scholar] [CrossRef]
- Wei, Z.-Z.; Hu, K.-D.; Zhao, D.-L.; Tang, J.; Huang, Z.-Q.; Jin, P.; Li, Y.-H.; Han, Z.; Hu, L.-Y.; Yao, G.-F.; et al. MYB44 competitively inhibits the formation of the MYB340-bHLH2-NAC56 complex to regulate anthocyanin biosynthesis in purple-fleshed sweet potato. BMC Plant Biol. 2020, 20, 258. [Google Scholar] [CrossRef]
- Quan, X.; Liu, J.; Zhang, N.; Xie, C.; Li, H.; Xia, X.; He, W.; Qin, Y. Genome-Wide Association Study Uncover the Genetic Architecture of Salt Tolerance-Related Traits in Common Wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 663941. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Zhou, R.; Dossa, K.; Yu, J.; Li, D.; Liu, A.; Mmadi, M.A.; Zhang, X.; You, J. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS ONE 2018, 13, e0200850. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.X.; Li, H.; Teng, R.M.; Wang, Y.; Zhuang, J. Genome-Wide Identification and Expression Analysis of Calcineurin B-Like Protein and Calcineurin B-Like Protein-Interacting Protein Kinase Family Genes in Tea Plant. DNA Cell Biol. 2019, 38, 824–839. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Miao, L.; Huang, B.; Gao, L.; He, C.; Yan, Y.; Wang, J.; Yu, X.; Li, Y. Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones. Int J Mol Sci 2019, 20, 5048. [Google Scholar] [CrossRef]
- Liu, Q.-L.; Xu, K.-D.; Pan, Y.-Z.; Jiang, B.-B.; Liu, G.-L.; Jia, Y.; Zhang, H.-Q. Functional Analysis of a Novel Chrysanthemum WRKY Transcription Factor Gene Involved in Salt Tolerance. Plant Mol. Biol. Rep. 2014, 32, 282–289. [Google Scholar] [CrossRef]
- Marand, A.P.; Schmitz, R.J. Single-cell analysis of cis-regulatory elements. Curr. Opin. Plant Biol. 2022, 65, 102094. [Google Scholar] [CrossRef] [PubMed]
- Davoudi, M.; Chen, J.; Lou, Q. Genome-Wide Identification and Expression Analysis of Heat Shock Protein 70 (HSP70) Gene Family in Pumpkin (Cucurbita moschata) Rootstock under Drought Stress Suggested the Potential Role of these Chaperones in Stress Tolerance. Int. J. Mol. Sci. 2022, 23, 1918. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Chen, K.; Deng, J.; Zhang, L.; Wang, W.; Kong, J.; Klosterman, S.J.; Zhang, X.; Aierxi, A.; Zhu, L. miR398b negatively regulates cotton immune responses to Verticillium dahliae via multiple targets. Crop J. 2022, 10, 1026–1036. [Google Scholar] [CrossRef]
- Cui, C.; Wang, J.J.; Zhao, J.H.; Fang, Y.Y.; He, X.F.; Guo, H.S.; Duan, C.G. A Brassica miRNA Regulates Plant Growth and Immunity through Distinct Modes of Action. Mol. Plant 2020, 13, 231–245. [Google Scholar] [CrossRef]
- Collin, A.; Daszkowska-Golec, A.; Szarejko, I. Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants. Cells 2021, 10, 1996. [Google Scholar] [CrossRef]
- Nath, M.; Bhatt, D.; Jain, A.; Saxena, S.C.; Saifi, S.K.; Yadav, S.; Negi, M.; Prasad, R.; Tuteja, N. Salt stress triggers augmented levels of Na+, Ca2+ and ROS and alter stress-responsive gene expression in roots of CBL9 and CIPK23 knockout mutants of Arabidopsis thaliana. Environ. Exp. Bot. 2019, 161, 265–276. [Google Scholar] [CrossRef]
- Zhang, H.-f.; Liu, S.-y.; Ma, J.-h.; Wang, X.-k.; Haq, S.U.; Meng, Y.-c.; Zhang, Y.-m.; Chen, R.-g. CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 26. [Google Scholar] [CrossRef]
- Gerik, K.J.; Donlin, M.J.; Soto, C.E.; Banks, A.M.; Banks, I.R.; Maligie, M.A.; Selitrennikoff, C.P.; Lodge, J.K. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 2005, 58, 393–408. [Google Scholar] [CrossRef]
- He, Y.; Chen, Y.; Song, W.; Zhu, L.; Dong, Z.; Ow, D.W. A Pap1–Oxs1 signaling pathway for disulfide stress in Schizosaccharomyces pombe. Nucleic Acids Res. 2017, 45, 106–114. [Google Scholar] [CrossRef]
- Techo, T.; Charoenpuntaweesin, S.; Auesukaree, C. Involvement of the Cell Wall Integrity Pathway of Saccharomyces cerevisiae in Protection against Cadmium and Arsenate Stresses. Appl. Environ. Microbiol. 2020, 86, e01339-20. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Zhou, D.; Song, J.; Gao, J. Nitrogen Uptake and Distribution in Different Chinese Cabbage Genotypes under Low Nitrogen Stress. Int. J. Mol. Sci. 2022, 23, 1573. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Ren, Z.; Zhou, Y.; Zhao, J.; Zhang, F.; Feng, J.; Liu, W.; Ma, X. Genome-Wide Identification of the Gossypium hirsutum NHX Genes Reveals that the Endosomal-Type GhNHX4A is Critical for the Salt Tolerance of Cotton. Int. J. Mol. Sci. 2020, 21, 7712. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Shi, L.; Li, X.; Zheng, H.; Gao, J.; Wang, M.; He, L.; Zhang, W. OXS2 is Required for Salt Tolerance Mainly through Associating with Salt Inducible Genes, CA1 and Araport11, in Arabidopsis. Sci. Rep. 2019, 9, 20341. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Jing, Y.; Shen, J.; Li, X.; Liu, H.; Geng, Z.; Wang, M.; Li, Y.; Chen, D.; Gao, J.; et al. Mitochondrial Pyruvate Carriers Prevent Cadmium Toxicity by Sustaining the TCA Cycle and Glutathione Synthesis. Plant Physiol. 2019, 180, 198–211. [Google Scholar] [CrossRef] [Green Version]
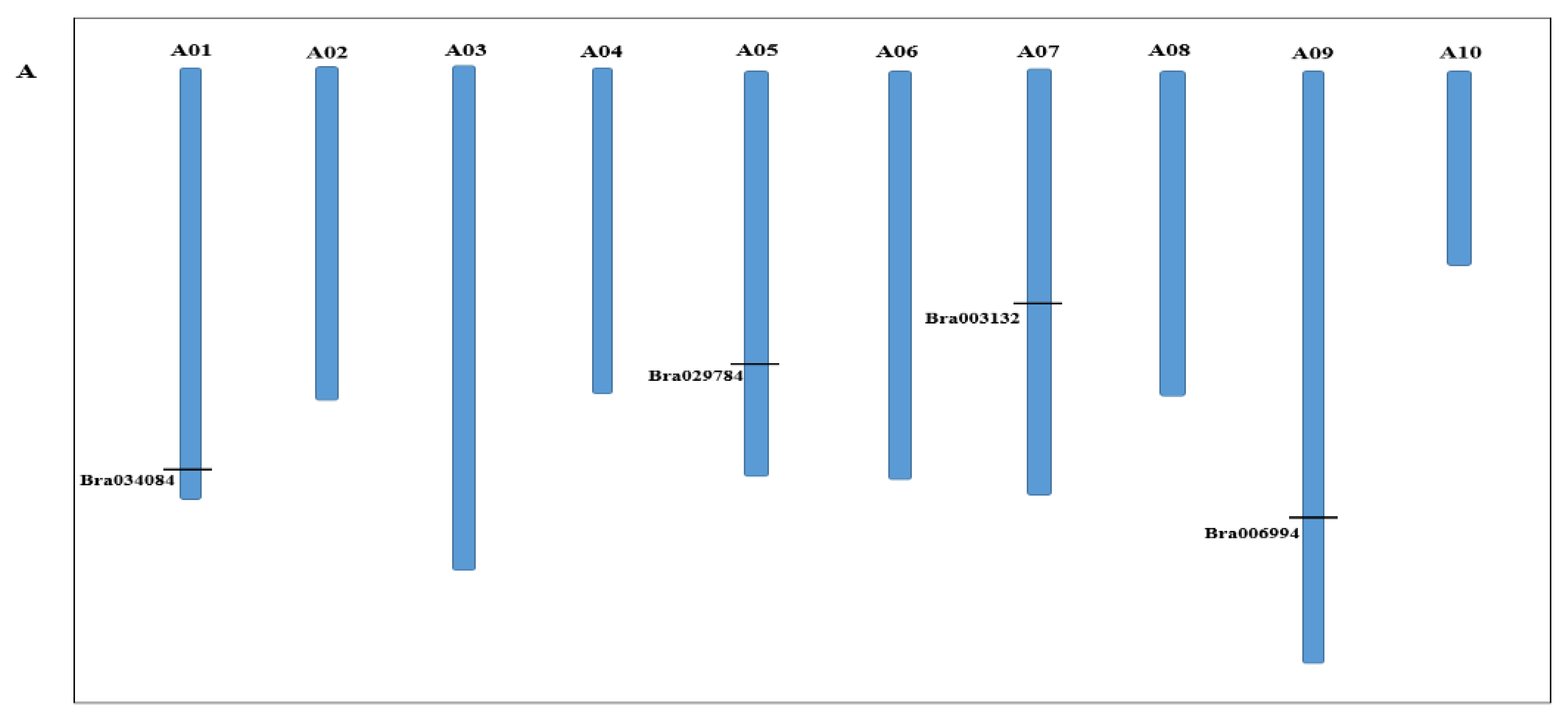

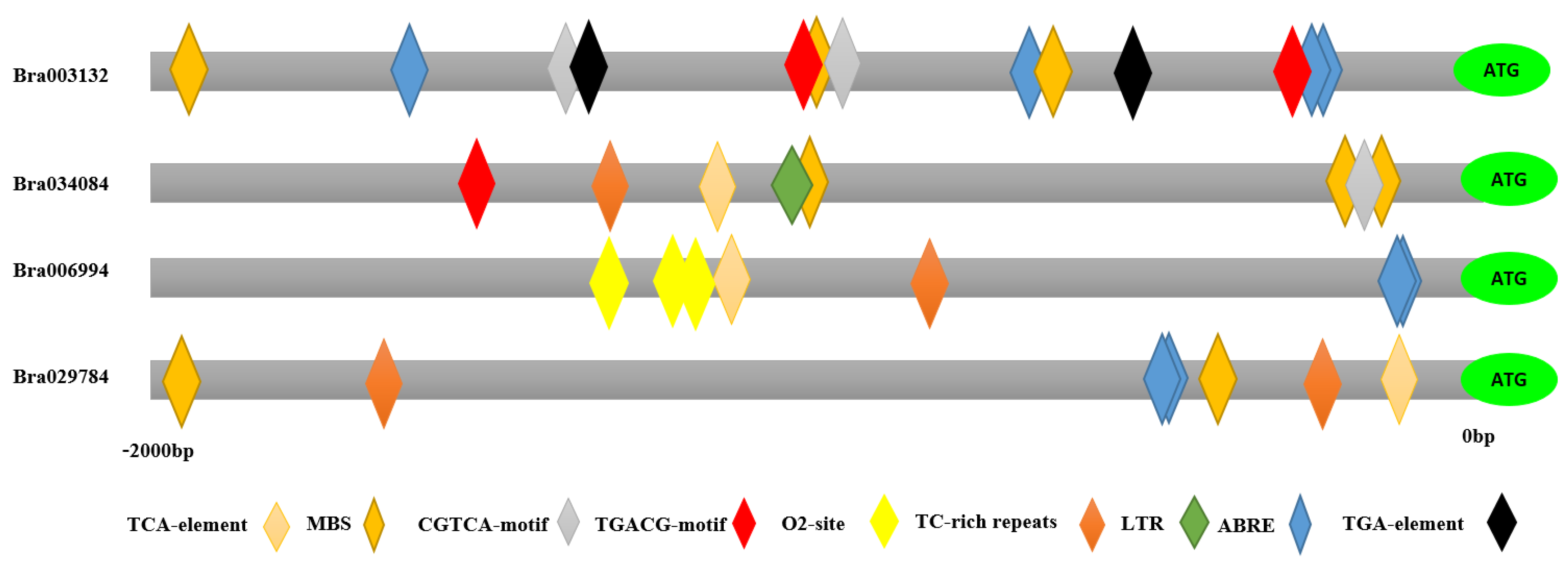
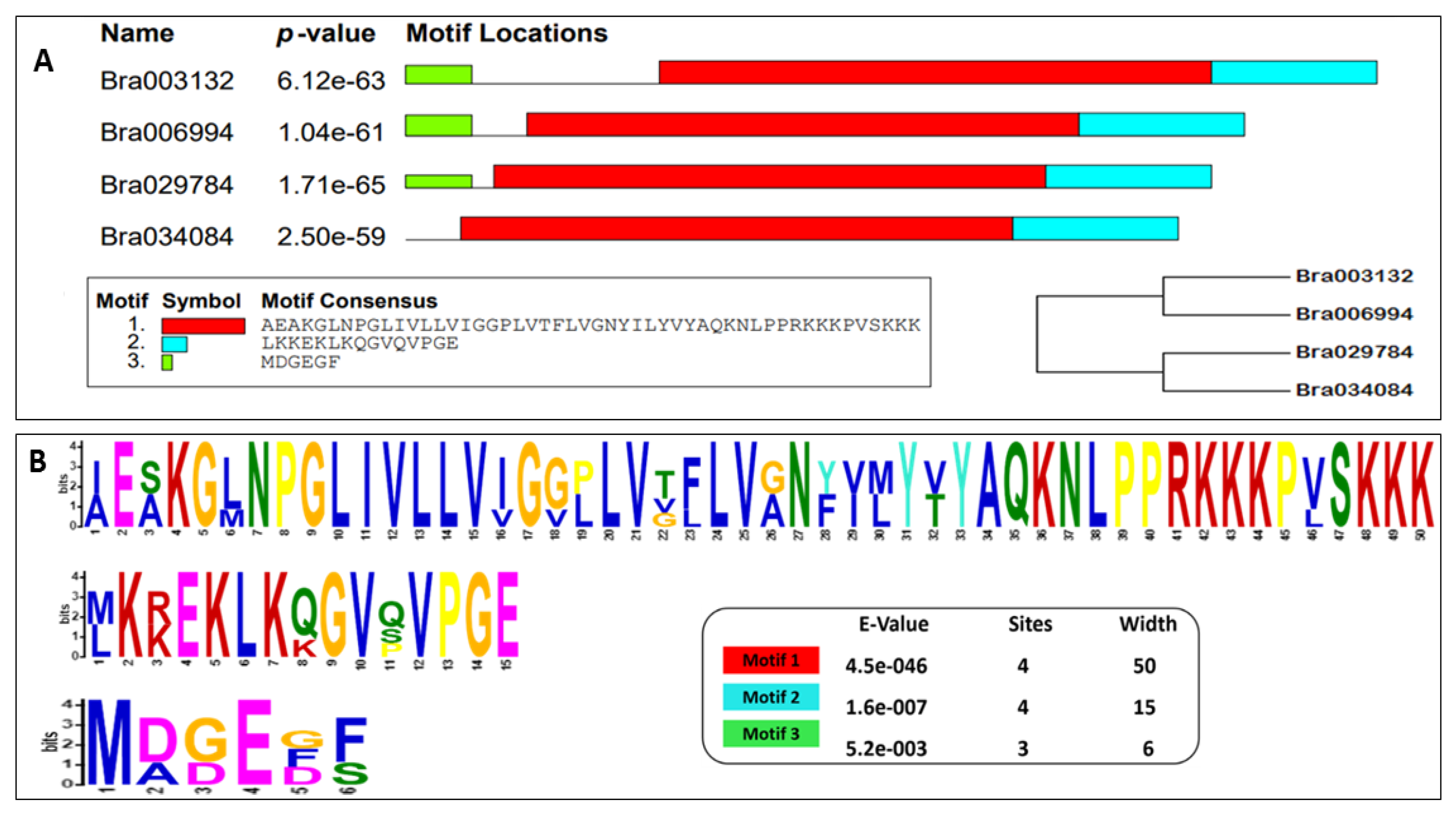

| Gene ID | Protein Length (aa) | Molecular Weight (KD) | Chromosome Location | pl | Strand Direction | Subcellular Location |
|---|---|---|---|---|---|---|
| Bra003132 | 88 | 9.3 | A07: 14791441..14792038 | 10.06 | - | Nuclear |
| Bra034084 | 70 | 7.8 | A01: 25080494..25080706 | 10.38 | + | Nuclear |
| Bra006994 | 76 | 8.3 | A09: 28232711..28233442 | 10.05 | - | Nuclear |
| Bra029784 | 73 | 8.1 | A05: 23013673..23014543 | 10.06 | + | Nuclear |
| miRNA | Target S1fa | Target Site | miRNA Fragment | Alignment | Target Fragment | Inhibition |
|---|---|---|---|---|---|---|
| ath-miR5661 | Bra003132 | 146–166 | AGAGGUACAUCAUGUAGUCUG | :::::.:::::::: : : | CCAACUACGUGAUGUACGUGU | Cleavage |
| ptc-miR397c | Bra003132 | 114–134 | UCAAUGAGUGGAGCUUUGAUG | ..::..:: :::.::..::: | UGUCGGAGGUCCGCUUGUUGU | Cleavage |
| mtr-miR2641 | Bra003132 | 71–90 | GUUUGAUCCUUUACGUUUAU | :.:: :::::::..::: | CUGAAGCCAAAGGAUUGAAC | Cleavage |
| hvu-miR6214 | Bra003132 | 99–118 | CGACGACGACGAGCACGACA | ::::::: :.::.:::: | AAUCGUGCUGCUUGUUGUCG | Translation |
| hme-miR-278 | Bra003132 | 3–23 | UCGGUGGGAUCUUCGUCCGUUU | ..:.::: ::::.:..:.:::. | GGAUGGA-GAAGGUUUCGCCGG | Cleavage |
| ptc-miR397c | Bra006994 | 78–98 | UCAAUGAGUGGAGCUUUGAUG | .:::..::.::::::..:::. | UAUCGGAGUUCCACUUGUUGG | Cleavage |
| zma-miR399e-5p | Bra006994 | 148–168 | GGGCUUCUCUUUCUUGGCAGG | ::: : :.:::. :::::::: | CCUCCAAGGAAGAAGAAGCCC | Cleavage |
| aly-miR160c-3p | Bra006994 | 91–110 | GCGUACAAGGAGCCAAGCAUG | : ::.: :::::::::: ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| ath-miR5661 | Bra006994 | 110–130 | AGAGGUACAUCAUGUAGUCUG | :::::.:::::::: : : | CCAACUACGUGAUGUACGUGU | Cleavage |
| osa-miR160a-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| osa-miR160b-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| zma-miR160b-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| zma-miR160g-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| bdi-miR160b-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| bdi-miR160c-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| ata-miR160c-3p | Bra006994 | 91–110 | GCGUGCAAGGAGCCAAGCAUG | : ::.: :::::::::. ::: | CUUGUUGGGCUCCUUGU-CGC | Cleavage |
| aly-miR838-3p | Bra029784 | 161–181 | UUUUCUUCUUCUUCUUGCACA | : : ::::::::::: :::.: | UUUCCAAGAAGAAGAUGAAGA | Cleavage |
| zma-miR399e-5p | Bra029784 | 139–159 | GGGCUUCUCUUUCUUGGCAGG | ::: : :.:::. :::::::: | CCUCCGAGGAAGAAGAAGCCC | Cleavage |
| ath-miR838 | Bra029784 | 161–181 | UUUUCUUCUACUUCUUGCACA | : : ::::::: ::: :::.: | UUUCCAAGAAGAAGAUGAAGA | Translation |
| osa-miR3982-3p | Bra029784 | 86–106 | AGUUGCCUACAUGGAGCGCCA | :: ::.::: :::::: :::: | UGACGUUCCUUGUAGGAAACU | Cleavage |
| bdi-miR398b | Bra029784 | 75–96 | CAGGAGUGUCACUGAGAACACA | : ::: .::::::..:::: | AGGGUUGCUAGUGACGUUCCUU | Cleavage |
| osa-miR2095-3p | Bra029784 | 58–77 | CUUCCAUUUAUGAUAAGUAU | .: ::: ::.:.: ::::.: | GUCCUUCUCGUGAUUGGAGG | Cleavage |
| aly-miR4248a | Bra029784 | 158–178 | ACAUUUUAUUUUUGGCAAUCA | .:: ::::.:: ::.::: | CCGUUUCCAAGAAGAAGAUGA | Cleavage |
| aly-miR4248b | Bra029784 | 158–178 | ACAUUUUAUUUUUGGCAAUCA | .:: ::::.:: ::.::: | CCGUUUCCAAGAAGAAGAUGA | Cleavage |
| aly-miR4248c | Bra029784 | 158–178 | ACAUUUUAUUUUUGGCAAUCA | .:: ::::.:: ::.::: | CCGUUUCCAAGAAGAAGAUGA | Cleavage |
| gma-miR4363 | Bra029784 | 52–73 | CGAUUACCAGAAGGCUUAUUAG | ::.:: . :::::: ::.::.: | CUGAUCGUCCUUCUCGUGAUUG | Cleavage |
| bdi-miR7757-3p.1 | Bra029784 | 192–212 | GGUAGUUGAAUGUUUUGUUUA | ::.:::..:.::::: : :: | GAAGCAAGGCGUUCAAGUUCC | Cleavage |
| aly-miR838-3p | Bra034084 | 152–172 | UUUUCUUCUUCUUCUUGCACA | : : ::::::::::: :::.: | UUUCCAAGAAGAAGAUGAAGA | Cleavage |
| zma-miR399e-5p | Bra034084 | 130–150 | GGGCUUCUCUUUCUUGGCAGG | ::: : :.:::. :::::::: | CCUCCGAGGAAGAAGAAGCCC | Cleavage |
| ath-miR838 | Bra034084 | 152–172 | UUUUCUUCUACUUCUUGCACA | : : ::::::: ::: :::.: | UUUCCAAGAAGAAGAUGAAGA | Translation |
| mtr-miR2673a | Bra034084 | 159–180 | CCUCUUCCUCUUCCUCUUCCAC | ::::: ::::: :.::: : | GAAGAAGAUGAAGAAGGAGAAG | Cleavage |
| mtr-miR2673b | Bra034084 | 159–180 | CCUCUUCCUCUUCCUCUUCCAC | ::::: ::::: :.::: : | GAAGAAGAUGAAGAAGGAGAAG | Cleavage |
| gma-miR4363 | Bra034084 | 43–64 | CGAUUACCAGAAGGCUUAUUAG | ::.:: . :::::: ::.:::: | CUGAUCGUCCUUCUUGUGAUCG | Cleavage |
| bdi-miR398b | Bra034084 | 66–87 | CAGGAGUGUCACUGAGAACACA | : ::: .::::::..:::: | AGGGUUGCUAGUGACGUUCCUU | Cleavage |
| osa-miR2055 | Bra034084 | 144–163 | UUUCCUUGGGAAGGUGGUUUC | :::.:: ::::.::::: :.: | GAAGCC-CCUUUCCAAGAAGA | Cleavage |
| osa-miR3982-3p | Bra034084 | 77–97 | AGUUGCCUACAUGGAGCGCCA | :: ::.::: :::.:: :::: | UGACGUUCCUUGUGGGAAACU | Cleavage |
| cca-miR6116-3p | Bra034084 | 49–69 | UCAUUUGAUCACAAGCAUGAG | :: : :::::::::..: :. | GUCCUUCUUGUGAUCGGAGGG | Cleavage |
| stu-miR8050-3p | Bra034084 | 182–202 | UGACUUGAGAUUCCUACUUGG | ::: :::.: .::::::. | UGAAGAAGGGAGUUCAAGUUC | Translation |
| gma-miR9752 | Bra034084 | 162–182 | UGCUUCUUCUUUUCCCUGUUU | .:: : : :.:::.:::::: | GAAGAUGAAGAAGGAGAAGCU | Cleavage |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwar, A.; Zhang, S.; Wang, L.-X.; Wang, F.; He, L.; Gao, J. Genome-Wide Identification and Characterization of Chinese Cabbage S1fa Transcription Factors and Their Roles in Response to Salt Stress. Antioxidants 2022, 11, 1782. https://doi.org/10.3390/antiox11091782
Anwar A, Zhang S, Wang L-X, Wang F, He L, Gao J. Genome-Wide Identification and Characterization of Chinese Cabbage S1fa Transcription Factors and Their Roles in Response to Salt Stress. Antioxidants. 2022; 11(9):1782. https://doi.org/10.3390/antiox11091782
Chicago/Turabian StyleAnwar, Ali, Shu Zhang, Li-Xia Wang, Fengde Wang, Lilong He, and Jianwei Gao. 2022. "Genome-Wide Identification and Characterization of Chinese Cabbage S1fa Transcription Factors and Their Roles in Response to Salt Stress" Antioxidants 11, no. 9: 1782. https://doi.org/10.3390/antiox11091782
APA StyleAnwar, A., Zhang, S., Wang, L.-X., Wang, F., He, L., & Gao, J. (2022). Genome-Wide Identification and Characterization of Chinese Cabbage S1fa Transcription Factors and Their Roles in Response to Salt Stress. Antioxidants, 11(9), 1782. https://doi.org/10.3390/antiox11091782








