Abstract
The interactions of nanoparticles with living organisms are driven by an interface called the protein corona. This interface is formed when nanoparticles are introduced in biological milieu and proteins are adsorbed at nanoparticles’ surfaces. Understanding the factors that are responsible for the formation and the composition of the protein corona could reveal mechanistic insights that are involved in the interaction of nanoparticles with biological structures. Multiple studies have qualitatively described the protein corona, but just a few have proposed quantification methods, especially for gold nanoparticles. Using bovine serum albumin conjugated with fluorescein-5-isothiocyanate as a model protein, we developed a fluorescent-based quantification method for gold nanoparticles’ protein coronas. The impact of nanoparticle size and surface chemistry was studied, and our research emphasizes that size and surface chemistry are determinant factors: Bigger nanoparticles and amino-modified surface chemistry are responsible for higher protein adsorption compared to smaller ones and carboxyl- or methoxy-modified surface chemistry. The proposed method can be used to complete the full picture of the interactions of nanoparticles with biological milieu and to describe the parameters which govern these interactions for the better development of nanomedicines.
1. Introduction
Nanomaterials have emerged as promising agents in biomedicine, with applications of nanoparticles ranging from photodynamic therapy to drug delivery or biological sensing [1]. Based on their dimensionality, nanomaterials have been classified as zero-dimensional nanoparticles, one-dimensional nanomaterials such as nanorods or nanotubes, two-dimensional nanomaterials such as nanosheets or nanofilms, and three-dimensional nanoparticles [2].
Among the broad types of nanoparticles, gold nanoparticles have drawn special attention due to their unique properties, such as controllable size, optical and electronic characteristics, and large surface-to-volume ratio [3,4]. Moreover, their high surface energy allows them to interact with various biomolecules from biological fluids. The resulting nanoparticles, coated with various proteins (known as protein corona) gain a new biological identity, as the protein layer can significantly affect their biodistribution and functionality [5,6].
A protein corona is actually made up of two component layers, the “hard” corona and the “soft” corona. The “hard” corona is represented by strongly-adsorbed proteins at the nanomaterial surface that usually remains irreversibly bounded. In contrast, the “soft” corona consists of proteins with a weaker binding affinity, so it exhibits a dynamic exchange [7,8]. Because of these rearrangements, the surface of a protein corona changes over time, which is reflected in the biological performance of the nanoparticle.
The screening of the nanoparticle surface by the corona has proved applicable in active targeting via specific ligands. However, it has been shown that their binding capacity is lost when these functionalized particles are placed in a biological environment [9,10]. Apparently, this loss of targeting specificity is caused by their interaction with other proteins in the medium and the formation of the protein corona. As a result, when placed in a biological medium, nanoparticles can easily fail to recognize and target relevant pathways. As nanoparticles continue to enter cells, the failure to target the intended pathways may lead to the accumulation in targeted and even in the non-targeted cells [11]. Thus, it is critical to establish analytical methods for the in-depth characterization of the protein corona and its interaction with other proteins.
Thus far, the nanoparticle–protein corona complexes have been mainly characterized qualitatively by assessing the factors which control protein corona formation (nanoparticle size, surface charge, morphology, pre-coating, abundance, and association/dissociation rate of proteins) or the conformational changes of proteins. The methods used for the primary characterization of protein corona have been UV-visible spectroscopy, Fourier transformed infrared spectroscopy, dynamic light scattering (DLS), circular dichroism spectroscopy, and isothermal titration calorimetry [12,13]. All these methods have provided valuable qualitative information about the protein corona, but the available methods for the direct quantification of gold nanoparticles’ (AuNPs) protein coronas are scarce, and research on this topic is still emerging. The relative quantification of different proteins forming AuNPs’ protein coronas was achieved by Konduru et al. by using gel-based densitometry [14]. Furthermore, DLS was employed by Cui et al. for quantification, but the method lacks sensitivity for low amounts of protein [15]. For polymeric nanoparticles, the total quantification of proteins can be relatively easily achieved by using spectroscopic UV-Vis assays (i.e., by Bradford) since there are no interferences. In contrast, the quantification of AuNPs’ protein coronas can be tedious since their absorption peak is in the same range as colorimetric protein assays [16] and no standard method has been established.
The present study aims to present a novel approach for protein corona quantification using a fluorescent-labeled protein model (BSA) and to evaluate the impact of gold nanoparticle size and surface chemistry on protein corona formation. The assessment of the exact amount of coated protein can provide essential information for understanding the intricate interactions of the protein corona with target molecules and other proteins. These interactions are of great interest as applications of nanoparticles in medicine, drug delivery, protein and peptide delivery are continuously expanding.
2. Materials and Methods
2.1. Gold Nanoparticles (AuNPs) Synthesis
Chloroauric acid trihydrate (HAuCl4⋅3H2O), sodium citrate trihydrate, sodium carbonate, sodium hydrogen carbonate, hetero-bifunctional poly(ethylene glycol)s (HS-PEG5kDa-X, X: COOH, NH2 or OCH3 and HS-PEG20kDa-COOH), bovine serum albumin (BSA) and FITC (fluorescein-5-isothiocyanate isomer 1) were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used without further purification. KCN was purchased from Chemapol. All glassware used in the syntheses were thoroughly cleaned in aqua regia (HCl: HNO3 = 3:1 v/v), rinsed in ultrapure water, and oven-dried prior to use.
The synthesis of spherical citrate-capped gold nanoparticles was employed, adopting a seeded growth approach developed by Puntes et al. [17]. Briefly, 150 mL of 2.2 mM sodium citrate was heated to boiling in a 250 mL three-neck round-bottomed flask. When the solution started to boil, 1 mL of HAuCl4 25 mM was added under vigorous stirring. After 10 min, the color of the solution turned soft pink, indicating the formation of AuNP seeds. For the growing steps, the temperature of the solution was decreased to 90 °C, and 1 mL of sodium citrate 60 mM and 1 mL of HAuCl4 25 mM were successively added. This process was repeated every 30 min for a total of 3 times, thus resulting in a new generation of nanoparticles. When the reaction was finished, the sample was diluted by extracting 55 mL of the sample and adding 53 mL of ultrapure water and 2 mL of 60 mM sodium citrate. The extracted sample was characterized, and the remaining one was used as seeds for the next generation. For this study, 2nd generation (24 nm) and 6th generation (56 nm) AuNPs were used.
The PEGylation of AuNPs was performed as described below. HS-PEG-X (X: COOH, NH2, OCH3) with MW = 5 kDa and HS-PEG-COOH with MW = 20 kDa were dissolved in water at a concentration of 5 mg/mL and added to 10 mL of AuNP solution to a ratio of 10 PEG molecules per nm2. The solutions were stirred overnight at room temperature, and the excess PEG molecules were washed by centrifugation (three times) [18].
2.2. Characterization of AuNP
UV-Vis spectroscopy:
For all AuNP suspensions, UV-visible spectra were recorded on a Specord 200 PLUS UV-visible spectrophotometer (Analytik Jena, Jena, Germany) by measuring in the 300–800 nm range using 1 cm quartz cuvettes. The appreciation of the size and the concentration of the colloids as determined using the equations developed by Haiss et al. [19].
Dynamic light scattering (DLS) and electrophoretic light scattering (ELS):
The hydrodynamic diameter of the nanoparticles was measured at 25 °C using a Malvern Zetasizer Nano ZS-90 (Malvern, UK) operating at a light source wavelength of 532 nm and a fixed scattering angle of 90°. AuNPs were diluted with water in a 1 cm optical path cell, and a DLS analysis was performed.
The ζ potential of AuNPs was measured at 25 °C by using the Malvern Zetasizer Nano ZS-90 by electrophoretic light scattering. ζ potential values are an average of three independent measurements.
2.3. BSA-FITC Conjugation
For protein corona studies, a model protein—bovine serum albumin (BSA)—was used. The choice of the protein model was based on the fact that serum albumin is the most abundant protein in the serum/plasma, and multiple mass spectrometry based studies have demonstrated that, usually, for metallic and polymeric nanoparticles, it is the protein that contributes the most to the protein corona formation [20,21]. BSA was labeled with FITC using the protocol described by Polo et al. [22]. FITC was dissolved in dimethyl sulfoxide (DMSO) (5 mg/mL) and was added to 1 mL of BSA 10 mg/mL dissolved in carbonate buffer (50 mM, pH = 9.2) at a ratio of 25 dye molecules per 1 BSA molecule. The solution was stirred for 4 h at room temperature, and the excess dye was removed using a size exclusion column (PD-10 desalting column, molecular weight cut off (MWCO) = 5000 Da) resulting in a purified fluorescently-labeled protein (BSA-FITC).
The concentration of the FITC-labeled protein was determined using Quick Start™ Bradford Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA) using a calibration curve with BSA as standard in a working range of 1–12 µg/mL. The samples absorbance was read at 595 nm using a CLARIOstar Plus microplate reader (BMG Labtech, Ortenberg, Germany). The fluorescence emission spectra and fluorescence emission intensity dependence of BSA-FITC vs. concentration ere determined using a CLARIOstar Plus microplate reader.
For the calibration curve, the fluorescence intensity of 12 serial dilutions of BSA-FITC (0.125–120 ng) in phosphate-buffered saline (pH = 7.4) mixed with KCN (final concentration 50 mM) was measured. The effect of AuNP dissolution on the recovery was evaluated by adding three known amounts (1, 50 or 100 ng) of BSA-FITC to a solution containing 1 × 1010 AuNPs/mL. BSA-FITC % recovery was calculated as the % of BSA-FITC recovered from the added amount after AuNP dissolution.
2.4. Colloidal Stability in Biological Media
In vitro stability was determined using the same conditions as for protein corona formation. Briefly, a volume of AuNPs corresponding to 1 × 1010 NPs was added to BSA-FITC (ratio 1:2500) at room temperature. As a control, ultrapure water was used. The UV-Vis spectrum of AuNPs was monitored for 12 h.
2.5. Protein Corona Formation and Quantification
AuNPs (final concentration 1 × 1010 NP/mL) were incubated with BSA-FITC at room temperature and protected from light for 1 h under continuous shaking (450 rpm). After 1 h, the nanoparticles covered with protein corona were washed three times by centrifuging and re-dispersed in 1 mL phosphate-buffered saline (pH = 7.4). In the last step, the NPs were re-dispersed in 50 µL buffer (Figure 1).
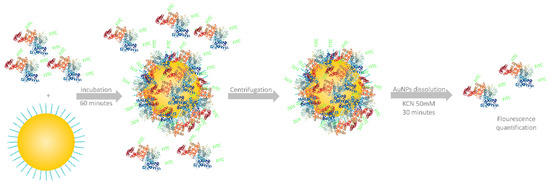
Figure 1.
A general overview of the workflow employed for protein corona quantification.
The fluorescence quenching effect of AuNPs was studied by adding 50 ng of BSA-FITC to solutions containing 1 × 1010 AuNPs.
For protein corona quantification, 10 uL of AuNP solution was mixed with 40 µL of buffer and 50 µL KCN 100 mM. After 30 min, the complete dissolution of AuNPs was achieved, and the protein content was quantified by fluorescence intensity measurement (λexcitation = 480 nm and λemission = 530 nm). As an additional method to confirm the AuNP number, a UV-Vis spectrophotometric determination was performed by diluting 10 µL from the same sample.
3. Results and Discussion
3.1. Gold Nanoparticle Synthesis and Characterization
Citrate-stabilized AuNPs of two different sizes (24 and 56 nm) were synthesized using an efficient seeded growth approach. Nanoparticles were characterized by the means of UV-Vis spectroscopy, dynamic light scattering (DLS), and electrophoretic light scattering (ELS) to determine their size, concentration, hydrodynamic diameter and zeta-potential. The UV-Vis absorbance spectra for citrate-capped AuNPs displayed a characteristic peak at 523.5 nm for the smaller particles and a peak at 534.5 nm for the bigger ones. The hydrodynamic diameter measurements were in accordance with the result of UV-Vis measurements, indicating average sizes of 30.4 and 64.2 nm. The zeta potentials of the AuNPs were also determined.
After characterization, AuNPs with different surface chemistries were produced by a ligand exchange process using different bifunctional thiolated-PEGs, resulting in four types of AuNPs for each size taken into study. PEGylation was successfully achieved using Au-SH chemical bonding. PEG attachments to the nanoparticles were confirmed by the slight redshift of approximately 3 nm in the plasmon absorption bands as a result of dielectric constant change at the nanoparticles’ surface (Figure 2) [23,24]. The characteristics of the PEGylated nanoparticles are presented in Table 1.
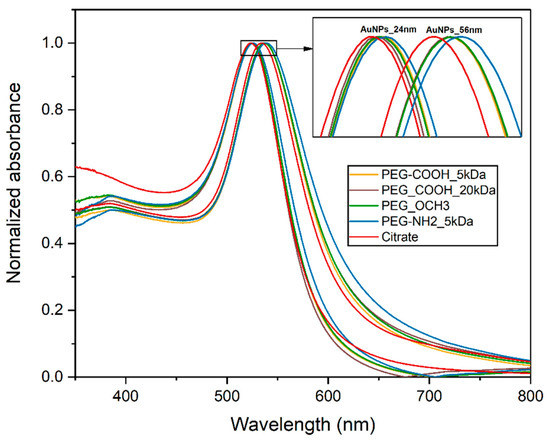
Figure 2.
UV-Vis spectra of gold nanoparticles (AuNPs).

Table 1.
AuNPs’ characterization.
It is well established that AuNPs’ interaction with biological media is strongly conditioned by their surface chemistry, PEGylation being one of the most common methods for improving the stability and the biocompatibility of AuNPs [25,26].
Furthermore, we aimed to assess the impact of size, different functional groups (-COOH, OCH3 and NH2, all with MW = 5 kDa) and the ligand molecular weight impact (-COOH 5 kDa vs. -COOH 20 kDa) on the protein corona quantity.
3.2. Stability Studies
The stability of AuNPs in biological media is crucial, since the interaction of the nanoparticles with proteins is strongly determined by their size, shape or surface chemistry. The presence of proteins may diminish or increase colloidal stability depending on different factors (protein concentration, ionic strength, etc.). Citrate capped-AuNPs are known to form aggregates in biological media, PEGylation being one of the most successful methods to increase the colloidal stability of AuNPs.
Prior to protein corona quantification, the stability of AuNPs was tested in the same conditions used for the quantification experiments. After 12 h, independent of surface chemistry, the dispersions of our AuNPs in protein-rich media have not yet displayed any modifications of the UV-Vis spectra (Figure 3), suggesting that the synthesized AuNPs were stable in the tested conditions.
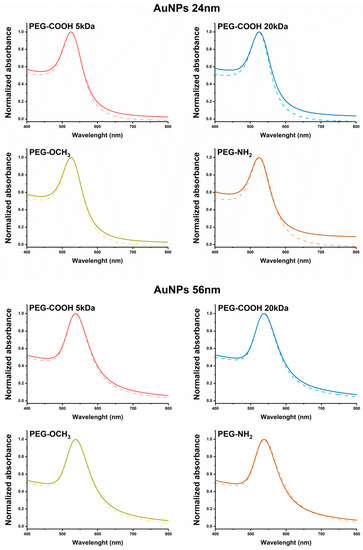
Figure 3.
UV-Vis spectra of 24 nm AuNPs (top) and 56 nm AuNPs (bottom) before (dotted line) and after (full line) incubation with bovine serum albumin-fluorescein-5-isothiocyanate isomer 1 (BSA-FITC) for 12 h.
3.3. Protein Corona Quantification
AuNPs are known to exhibit a fluorescence quenching effect due to surface plasmon resonance band which absorbs close to the wavelength emission band of the FITC molecule (Figure 4). For the fluorescence intensity quantification, a complete dissolution of the gold nanoparticles was mandatory. This was accomplished by adding CN− in excess, which led to the formation of complex AuCN2−, a colorless non-fluorescent ion. The addition of 50 mM KCN had no impact on the fluorescence of the FITC-BSA, as Figure 4 shows; however, as a precautionary measure, KCN was also added to the dilutions used for the calibration curve in order to elude any effect of the cyanide ions on the fluorescence.
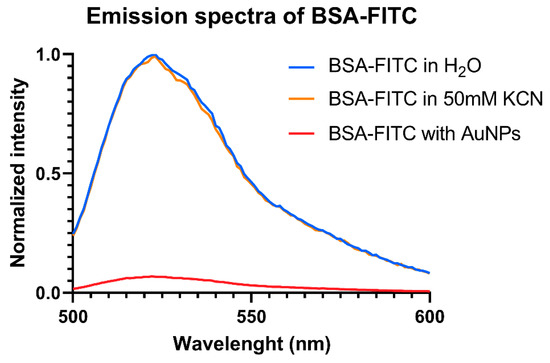
Figure 4.
Normalized fluorescence emission spectra of BSA-FITC.
The calibration curve used for protein corona quantification is presented in Figure 5. A linear plot was obtained over the range 0.125–120 ng, with a correlation coefficient of R2 = 0.9996. An amount of 1, 50 and 100 ng of FITC-labeled protein was used to estimate the BSA-FITC % recovery. The experimental data are presented in Table 2.
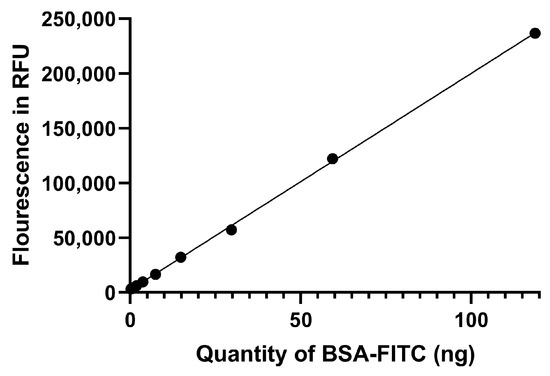
Figure 5.
The calibration curve obtained after plotting the fluorescent intensities against the amount of BSA-FITC.

Table 2.
BSA-FITC recovery data.
Figure 6 shows the total amount of proteins adsorbed on each type of nanoparticles. As expected, the amount of protein adsorbed on the 56 nm AuNPs was higher compared to smaller particles due to a higher surface area (9850 nm2/NP vs. 1820 nm2/NP) for all types of surface chemistries. As described by Piella et al., particle size plays a crucial role into formation of protein corona, with bigger particles forming a thicker layer of proteins due to different forming kinetics compared to the smaller ones [27]. Another main factor that controls protein affinity for nanoparticles’ surfaces is represented by surface chemistry. Several studies have clearly established that surface chemistry quantitatively and qualitatively influences the protein corona (by modulating the type of proteins that are adsorbed on the surface of the nanoparticles from a mixture of proteins) [10,28,29].
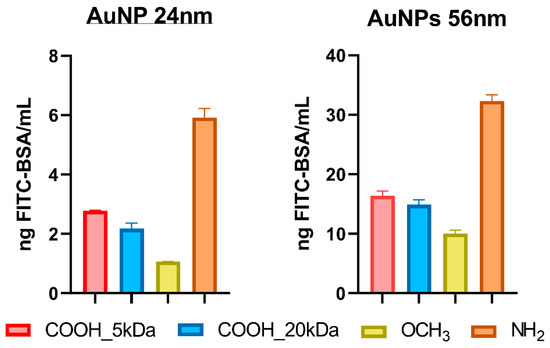
Figure 6.
The amount of fluorescently-labeled BSA adsorbed onto the AuNPs (24 and 56 nm). (concentration of AuNPs: 1 × 1010 NP/mL). Results are given as mean +/− standard deviation of three independent replicates.
Our results indicate that the highest amount of proteins was adsorbed by the positively charged, PEG-NH2 coated AuNPs. In contrast, the neutrally charged PEG-OCH3 AuNPs absorbed the least, while the negative charged PEG-COOH-coated AuNPs adsorbed an intermediate amount of proteins. The results can be explained by considering the intrinsic properties of BSA, which has an isoelectric point of 4.7 [30] and a surface charge of the nanoparticles. At pH = 7.4, the protein was slightly negatively charged, which was translated into a higher affinity for the positively charged NH2 modified surface, as the zeta-potential measurements indicated.
We also examined the influence of the molecular weight of the PEG ligand bearing the same functional group on the protein corona. Our results indicated that the PEG chain length had no influence on the quantity of protein attached to the surface of AuNPs. No significant differences were observed between PEG-COOH 5 kDa and PEG-COOH 20 kDa. The results are in agreement with previous studies, which established that above the molecular weight of 5 kDa, no significant decrease in protein corona content can be noticed [31].
4. Conclusions
A novel approach for the total quantification of the protein corona of AuNPs was developed using a model protein (BSA) labeled with FITC. The impact of size and the surface chemistry of AuNPs on the amount of adsorbed protein was studied. Our research confirmed that the PEGylation of the nanoparticles does not completely inhibit protein adsorption, and the major factors that impact protein adsorption are the size of the nanoparticles and the surface chemistry. The developed method could be useful in studying gold nanoparticle–protein interactions to decipher the subtle mechanism that governs those interactions.
Author Contributions
Conceptualization R.N., C.A.I. and F.L.; methodology R.N.; formal analysis R.N., A.U. and M.I.; investigation R.N. and M.I.; Writing—Original Draft preparation R.N. and A.U.; Writing—Review and Editing R.N., A.U., M.I., C.A.I. and F.L.; supervision C.A.I. and F.L.
Funding
This research was funded by “Iuliu Hațieganu” University of Medicine and Pharmacy, Cluj-Napoca, Romania, grant number 7690/79/15.04.2016 and 5200/68/01.03.2017 awarded to Raul Nicoară.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Versiani, A.F.; Andrade, L.M.; Martins, E.M.N.; Scalzo, S.; Geraldo, J.M.; Chaves, C.R.; Ferreira, D.C.; Ladeira, M.; Guatimosim, S.; Ladeira, L.O. Gold nanoparticles and their applications in biomedicine. Future Virol. 2016, 11, 293–309. [Google Scholar] [CrossRef]
- Sudha, P.N.; Sangeetha, K.; Vijayalakshmi, K.; Barhoum, A. Nanomaterials History, Classification, Unique Properties, Production and Market. In Emerging Applications of Nanoparticles and Architectural Nanostructures: Current Prospects and Future Trends; Elsevier: Amsterdam, The Netherlands, 2018; pp. 341–384. [Google Scholar]
- Elahi, N.; Kamali, M.; Baghersad, M.H. Recent Biomedical Applications of Gold Nanoparticles: A Review. Talanta 2018, 184, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Dreaden, E.C.; Alkilany, A.M.; Huang, X.; Murphy, C.J.; El-Sayed, M.A. The golden age: Gold nanoparticles for biomedicine. Chem. Soc. Rev. 2012, 41, 2740–2779. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Milani, S.; Baldelli Bombelli, F.; Pitek, A.S.; Dawson, K.A.; Rädler, J. Reversible versus Irreversible Binding of Transferrin to Polystyrene Nanoparticles: Soft and Hard Corona. ACS Nano 2012, 6, 2532–2541. [Google Scholar] [CrossRef] [PubMed]
- Cagliani, R.; Gatto, F.; Bardi, G. Protein adsorption: A feasible method for nanoparticle functionalization? Materials 2019, 12, 1991. [Google Scholar] [CrossRef]
- Mirshafiee, V.; Mahmoudi, M.; Lou, K.; Cheng, J.; Kraft, M.L. Protein corona significantly reduces active targeting yield. Chem. Commun. 2013, 49, 2557. [Google Scholar] [CrossRef]
- Kurtz-Chalot, A.; Villiers, C.; Pourchez, J.; Boudard, D.; Martini, M.; Marche, P.N.; Cottier, M.; Forest, V. Impact of silica nanoparticle surface chemistry on protein corona formation and consequential interactions with biological cells. Mater. Sci. Eng. C 2017, 75, 16–24. [Google Scholar] [CrossRef]
- Salvati, A.; Pitek, A.S.; Monopoli, M.P.; Prapainop, K.; Bombelli, F.B.; Hristov, D.R.; Kelly, P.M.; Aberg, C.; Mahon, E.; Dawson, K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nano 2013, 8, 137–143. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Pitek, A.S.; Lynch, I.; Dawson, K.A. Formation and characterization of the nanoparticle-protein corona. Methods Mol. Biol. 2013, 1025, 137–155. [Google Scholar] [PubMed]
- Carrillo-Carrion, C.; Carril, M.; Parak, W.J. Techniques for the experimental investigation of the protein corona. Curr. Opin. Biotechnol. 2017, 46, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Konduru, N.V.; Molina, R.M.; Swami, A.; Damiani, F.; Pyrgiotakis, G.; Lin, P.; Andreozzi, P.; Donaghey, T.C.; Demokritou, P.; Krol, S.; et al. Protein corona: Implications for nanoparticle interactions with pulmonary cells. Part. Fibre Toxicol. 2017, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Liu, R.; Deng, Z.; Ge, G.; Liu, Y.; Xie, L. Quantitative study of protein coronas on gold nanoparticles with different surface modifications. Nano Res. 2014, 7, 345–352. [Google Scholar] [CrossRef]
- Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C.W. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef]
- Bastus, N.G.; Comenge, J.; Puntes, V. Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: Size focusing versus ostwald ripening. Langmuir 2011, 27, 11098–11105. [Google Scholar] [CrossRef]
- Hühn, J.; Carrillo-Carrion, C.; Soliman, M.G.; Pfeiffer, C.; Valdeperez, D.; Masood, A.; Chakraborty, I.; Zhu, L.; Gallego, M.; Yue, Z.; et al. Selected Standard Protocols for the Synthesis, Phase Transfer, and Characterization of Inorganic Colloidal Nanoparticles. Chem. Mater. 2017, 29, 399–461. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.K.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV-Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles. the effect of their size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef]
- Gossmann, R.; Fahrländer, E.; Hummel, M.; Mulac, D.; Brockmeyer, J.; Langer, K. Comparative examination of adsorption of serum proteins on HSA- and PLGA-based nanoparticles using SDS-PAGE and LC-MS. Eur. J. Pharm. Biopharm. 2015, 93, 80–87. [Google Scholar] [CrossRef]
- Polo, E.; Araban, V.; Pelaz, B.; Alvarez, A.; Taboada, P.; Mahmoudi, M.; del Pino, P. Photothermal effects on protein adsorption dynamics of PEGylated gold nanorods. Appl. Mater. Today 2019, 15, 599–604. [Google Scholar] [CrossRef]
- Rahme, K.; Chen, L.; Hobbs, R.G.; Morris, M.A.; O’Driscoll, C.; Holmes, J.D. PEGylated gold nanoparticles: Polymer quantification as a function of PEG lengths and nanoparticle dimensions. RSC Adv. 2013, 3, 6085–6094. [Google Scholar] [CrossRef]
- Cucci, L.M.; Naletova, I.; Consiglio, G.; Satriano, C. A hybrid nanoplatform of graphene oxide/nanogold for plasmonic sensing and cellular applications at the nanobiointerface. Appl. Sci. 2019, 9, 676. [Google Scholar] [CrossRef]
- Guerrini, L.; Alvarez-Puebla, R.; Pazos-Perez, N. Surface Modifications of Nanoparticles for Stability in Biological Fluids. Materials 2018, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Santos-Martinez, M.J.; Rahme, K.; Corbalan, J.J.; Faulkner, C.; Holmes, J.D.; Tajber, L.; Medina, C.; Radomski, M.W. Pegylation increases platelet biocompatibility of gold nanoparticles. J. Biomed. Nanotechnol. 2014, 10, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Piella, J.; Bastús, N.G.; Puntes, V. Size-dependent protein-nanoparticle interactions in citrate-stabilized gold nanoparticles: The emergence of the protein corona. Bioconjug. Chem. 2017, 28, 88–97. [Google Scholar] [CrossRef]
- Johnston, B.D.; Kreyling, W.G.; Pfeiffer, C.; Schäffler, M.; Sarioglu, H.; Ristig, S.; Hirn, S.; Haberl, N.; Thalhammer, S.; Hauck, S.M.; et al. Colloidal Stability and Surface Chemistry Are Key Factors for the Composition of the Protein Corona of Inorganic Gold Nanoparticles. Adv. Funct. Mater. 2017, 27. [Google Scholar] [CrossRef]
- Sasidharan, A.; Riviere, J.E.; Monteiro-Riviere, N.A. Gold and silver nanoparticle interactions with human proteins: Impact and implications in biocorona formation. J. Mater. Chem. B 2015, 3, 2075–2082. [Google Scholar] [CrossRef]
- Huang, G.; Liu, J.; Jin, W.; Wei, Z.; Ho, C.-T.; Zhao, S.; Zhang, K.; Huang, Q. Formation of Nanocomplexes between Carboxymethyl Inulin and Bovine Serum Albumin via pH-Induced Electrostatic Interaction. Molecules 2019, 24, 3056. [Google Scholar] [CrossRef]
- Gref, R.; Lück, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Müller, R.H. “Stealth” corona-core nanoparticles surface modified by polyethylene glycol (PEG): Influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B Biointerfaces 2000, 18, 301–313. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).