Abstract
Clinical guidelines, which represent the normative process models for healthcare organizations, are typically available in a textual, unstructured form. This issue hampers the application of classical conformance-checking algorithms to the medical domain, which take in input of a formalized and computer-interpretable description of the process. In this paper, (i) we propose overcoming this problem by taking advantage of a Large Language Model (LLM), in order to extract normative rules from textual guidelines; (ii) we then check and quantify the conformance of the patient event log with respect to such rules. Additionally, (iii) we adopt the approach as a means for evaluating the quality of the models mined by different process discovery algorithms from the event log, by comparing their conformance to the rules. We have tested our work in the domain of stroke. As regards conformance checking, we have proved the compliance of four Northern Italy hospitals to a general rule for diagnosis timing and to two rules that refer to thrombolysis treatment, and have identified some issues related to other rules, which involve the availability of magnetic resonance instruments. As regards process model discovery evaluation, we have assessed the superiority of Heuristic Miner with respect to other mining algorithms on our dataset. It is worth noting that the easy extraction of rules in our LLM-assisted approach would make it quickly applicable to other fields as well.
1. Introduction
Conformance checking is one of the main tasks of process mining [] and aims to verify whether the actual behavior of an organization conforms to its normative behavior. Specifically, actual behavior is recorded within the event log, a collection of process traces [] that store the activities executed during the organization’s everyday work, each one with its time stamp. In particular, in the case of a healthcare organization, a trace maintains the ordered sequence of activities executed on a specific patient during their hospital stay. Normative behavior, in the medical field, is usually represented by a clinical guideline.
Conformance-checking algorithms take in input of an event log and a technical description of the normative process, to verify their alignment. The technical description is typically a graphical representation showing the activity control flow allowed by the normative model. Several modeling languages have been developed to this end (and a conversion into a Petri Net [] is usually possible). Declarative modeling languages—defining behavior that is (not) allowed—exist as well. In any case, a formalization of the normative model is required.
In medical practice, however, clinical guidelines are typically expressed in natural language and describe the normative behavior in the form of very long and complex texts. Several approaches to the representation of Computer Interpretable Guidelines (CIGs) exist (see, for example, []), but they require a significant knowledge acquisition effort.
In this work, we aim at supporting conformance checking in medicine, by verifying the conformance of patient traces directly with respect to textual guidelines, without requiring the acquisition of a CIG.
- In particular, our work is articulated into three main contributions:
- We adopt a Large Language Model (LLM)-based approach to
- Extract a set of normative behaviors from a textual guideline;
- Formalize such an output into executable rules.
- We check the conformance of patient traces to the extracted rules and quantify it. To this end, we define the Trace Conformance Indicator (TCI), a metric measuring the percentage of log traces that satisfy the rules.
- We also exploit our conformance-checking approach as a means for assessing the quality of process model discovery algorithms []. To this end,
- We mine a process model from the event log by means of the algorithm we wish to evaluate, obtaining a graphical representation (a Petri Net).
- We check the conformance of the paths in the Petri Net to the extracted rules and quantify it. To this end, we define the Path Conformance Indicator (PCI), a metric measuring the percentage of model paths that satisfy the rules.
As regards contribution (2), the TCI can easily highlight the number of compliant traces in an event log, with respect to a given rule, helping physicians or hospital administrators to quickly identify clinical or organizational issues, such as the presence of bottlenecks. As regards contribution (3), we propose using the PCI as a novel dimension for comparing different process model discovery algorithms, which can complement existing ones []; this new dimension focuses on the conformance of the mined model to the guideline. As a matter of fact, different miners extract different process models from the same log, according to the way they operate, and their adherence to the normative behavior may vary. The information contained in the log may be captured correctly by some miners, but not by others, depending on the log at hand and on the algorithmic strategy. By calculating the PCI, we obtain a score for the mined model. The miner which produces the model with the PCI closest to the TCI calculated on the same event log can be considered the best one: indeed, the concordance of the two indicators represents the concordance of the information mined in the process model, with respect to the information contained in the log, as regards the adherence to normative knowledge, summarized by the rules.
In this work, we report our experiments in the domain of stroke. It is, however, worth noting that the easy LLM-assisted extraction of the rules would make it quickly applicable to other fields as well. We made some first tests in this direction and foresee a more extensive study in the future.
The paper is organized as follows: in Section 2, we present related work; in Section 3, we present the technical details of the approach; in Section 4, we provide experimental results, and in Section 5, we discuss issues and challenges. Finally, Section 6 is devoted to the conclusions and future work.
2. Related Work
In this section, we present related work. In particular, we first discuss the classical approaches to conformance checking (which require the presence of a formalized and computer-interpretable normative model). We then survey the use of LLMs in process mining. Finally, we make some concluding remarks.
2.1. Conformance Checking
In process-mining literature (see, for example, []), two main families of conformance-checking algorithms exist, known as log replay algorithms and trace alignment algorithms.
Log replay algorithms try to rerun each available event log trace on the input normative process model. When the model is represented as a Petri Net, a token-based approach can be applied [], which generates a new token whenever the model reaches a dead end, to advance towards the next state, in order to complete the trace replay. The final number of tokens left in the model is calculated, and conformance is evaluated on the basis of the superfluous generated tokens. More generally, when the model is not necessarily represented as a Petri Net, the ratio between the replayable traces and the total number of traces provides a measure of conformance, known as fitness [].
Race alignment algorithms, on the other hand, evaluate deviations at the level of activities and identify asynchronous moves, i.e., situations where the trace can not make the same step of the model, or vice versa. Every asynchronous move provides a cost (somehow similarly to the notion of edit distance), and an optimal alignment, which minimizes the cost, can be calculated []. Computing fitness at the level of activities (instead of the level of traces, as in log replay algorithms) is more precise but computationally intensive, since, in the case of a big event log, the number of potential alignment paths can be huge.
In addition to the already-mentioned ones, a third family of algorithms exists, which is the one of constraint-based conformance checking (see, for example, []). It translates process logic into rules and subsequently verifies whether each trace satisfies the rules. Our approach can be considered as belonging to this family. Notably, we are not aware of other literature contributions in constraint-based conformance checking that adopt an LLM for rule extraction from a free text model.
2.2. LLMs in Process Mining
LLMs such as BERT [] and ChatGPT-4o [] have significantly advanced natural language processing (NLP) [] capabilities, making them powerful tools for a variety of applications in different research areas, including process mining.
The work in [] reviews the most recent applications of LLMs to process mining.
As shown in such a survey, on the one hand, LLMs can generate natural language text: indeed, they have been used to produce textual descriptions from event logs or from process models [,], or from visual data such as dotted charts []. In particular, in [], proper abstraction techniques are proposed, to reduce the size of the event log or abstract from the artifacts, still keeping the meaningful information, while in [], advanced prompt engineering techniques are adopted to guide LLMs to comprehend the various process structures, with the final goal of enabling users to better understand the process. Querying strategies starting from natural language questions are also adopted to enhance the quality of the process, e.g., by identifying root causes of performance issues [], or unfairness [].
On the other hand, LLMs have been adopted to extract a process model from a textual document, incorporating advanced prompt engineering, error handling, and code generation techniques []. In [], the authors show how an LLM can be exploited for mining imperative declarative process models from text, or how it can assess the suitability of process tasks for robotic process automation. In this way, LLMs are able to overcome previous approaches based on Natural Language Processing techniques that were specific to single tasks and were unable to act as general-purpose instruments in the field of process mining. In [], users are also allowed to interact with the tool, for optimizing and refining the generated process model.
LLMs have also been used to generate code in [], for performing SQL queries (which can be useful, e.g., to filter the content of the event log, given that it is stored in a database), or to obtain other insights from the event log itself.
None of the approaches mentioned above address the task of conformance checking, with the only exception of [], where the LLM was asked to identify anomalies in some event logs, i.e., paths that were illogical, had wrong activity ordering, or missed key activities, through the query “Can you pinpoint the central anomalies of the process from this data?”. However, while satisfactory in other domains, the responses for medical event logs were below expectations. Moreover, no elicitation of rules from domain knowledge was proposed in that paper.
To the best of our knowledge, our approach is thus the first one which resorts to LLMs for extracting rules for conformance checking, by analyzing a normative model which is not computer-interpretable but is expressed as a natural language (unstructured) text, focusing on the medical domain.
2.3. Final Considerations
As a final consideration, it is worth noting that our approach is (loosely) coupled with the Computer Interpretable Guidelines (CIGs) [] area of research. CIGs normally require a significantly time-consuming knowledge acquisition effort, where the domain expert, possibly supported by a knowledge engineer, converts the guideline textual information into a computer-interpretable language, usually supported by a user-friendly Graphical User Interface. The acquired guideline can then be executed on real patients, or adopted for simulations or for education purposes. Once the CIG is available, since it is typically a graph, it can also be used as a normative model, where classical process conformance-checking approaches can be applied. The manual creation of a CIG will normally require several days of work by the domain expert. Our approach can be hardly compared to the acquisition of a CIG: indeed, we do not aim at realizing all the objectives mentioned above (e.g., the execution of the guideline on real patients and simulations). Instead, we wish to verify whether a process trace/model is compliant with the guideline. The use of an LLM helps us obtain the key rules to be verified in a few seconds; our tool then allows us to verify compliance by exploiting the executable Python version of the rules, as detailed in the following sections. In conclusion, the knowledge formalization and the implementation efforts of our approach are significantly lower than the ones required to acquire a CIG, but the objective is also narrower/more specific, so that a fair comparison between the two areas would not be possible.
3. LLM-Assisted Conformance Checking
In this section, we first illustrate our tool architecture; we then provide further details of its main steps.
3.1. Architecture
Figure 1 describes the architecture of our LLM-assisted conformance checking tool.
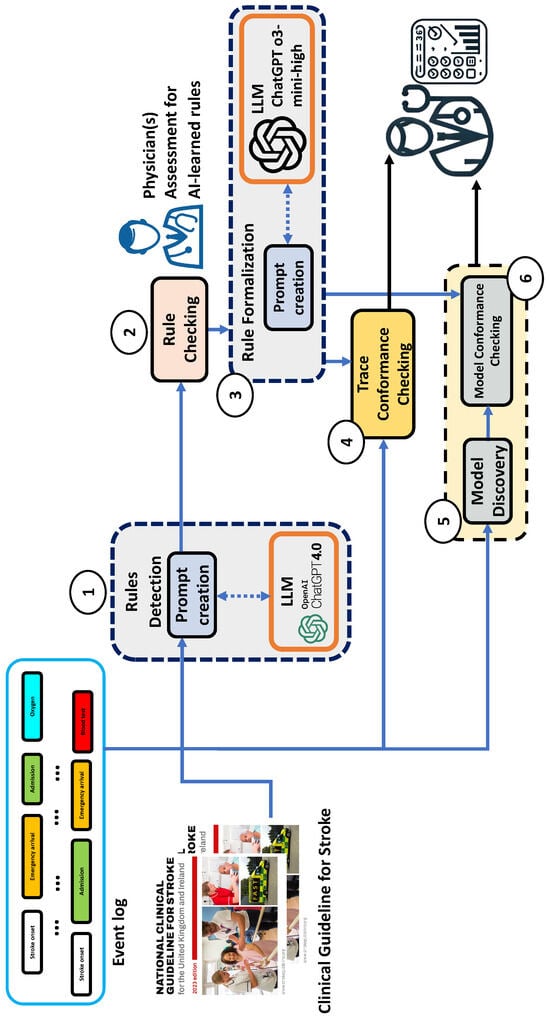
Figure 1.
The tool architecture (the circled numbers represent the workflow steps).
The entire architecture was developed using Python v.3.12.3 and the PM4Py library [], integrated with OpenAI’s ChatGPT-4o APIs (https://platform.openai.com/docs/models/gpt-4o (accessed on 20 July 2025)) [] and ChatGPT o3-mini-high model (https://openai.com/index/openai-o3-mini/ (accessed on 20 July 2025). We plan to integrate the recently published ChatGPT-5 in the future, waiting for it to stabilize after the initial criticism).
- The tool operates according to the following steps:
- The Rule Detection module takes in input of the clinical guideline in a textual format in natural language. It implements an algorithm that makes an HTTP-REST call to the ChatGPT-4o API using Python to query the ChatGPT-4o LLM, passing the guideline as an input. This enables the LLM to extract rules in natural language.
- The Rule Checking module shows the extracted rules to a medical expert, who is in charge of checking the validity of the rules on the basis of domain knowledge.
- The Rule Formalization module takes in input of the validated rules along with an example trace from the available event log, and automatically converts the rules from natural language into Python script code by using the ChatGPT o3-mini-high model (optimized for coding).
- The Trace Conformance Checking module takes in input of the event log and the formalized rules, and automatically checks the log conformance with respect to the rules, trace by trace, outputting the TCI, defined as the percentage of compliant traces for each rule.
- The Model Discovery module allows us to call different process discovery algorithms (e.g., Heuristic Miner [] and SIM []), taking into account the event log, and outputting a Petri Net, representing the mined process model.
- The Model Conformance Checking module takes in input of a process model and the formalized rules, and outputs the PCI, i.e., the percentage of paths compliant with each rule. In particular, the best model is the one where the PCI is the closest to the TCI calculated on traces in step 4.
In the next subsection, we will illustrate the functionality of the Trace Conformance Checking and Model Conformance Checking modules in further detail.
3.2. Trace Conformance Checking
In this section, we detail how our tool operates when we need to check the conformance of the logged traces with respect to a rule (already extracted and formalized by the LLM).
First, we identify an anchor activity (referred to as ), mentioned in the rule. As an example, consider the text of Rule 1 (which has been extracted in the Rule Detection step during our experiments, and which will be further discussed in Section 4): “If patients have acute ischemic stroke and treatment can be started within 4.5 h of known onset, then they should be considered for thrombolysis with alteplase or tenecteplase”. According to such a rule, we need to check whether thrombolysis, a life-saving procedure for stroke patients, is executed before 4.5 h from stroke onset. In this situation, the anchor activity is thrombolysis, and is set accordingly. A rule can involve other activities, temporal constraints (as in this case with respect to the stroke onset), or even a branching logic condition (such as the prescription to execute an activity only if a certain condition—e.g., a parameter value—holds), but the anchor activity provides the context to be considered: in the example, only traces involving the thrombolysis procedure will be examined for conformance checking, as the others are out of scope. Obviously, more complex rules, as the branching logic ones, can be checked only if the log maintains the required information (e.g., it stores the parameter value to be tested in the branching condition, as an attribute of a proper data collection activity).
Once all the traces involving the anchor activity have been identified, they are stored in the set .
Referring to Algorithm 1, the function then returns the TCI, calculated as the percentage of traces among those provided in that satisfy a given rule. At this stage, all the activities mentioned in the rule and all the temporal constraints have to be taken into account and checked. The function in the algorithm is in charge of this.
| Algorithm 1 Function returns the percentage of traces that satisfy a rule |
|
Figure 2 provides the Python code of function exploited in Algorithm 1, automatically generated by ChatGPT o3-mini-high, when the rule at hand is Rule 1.
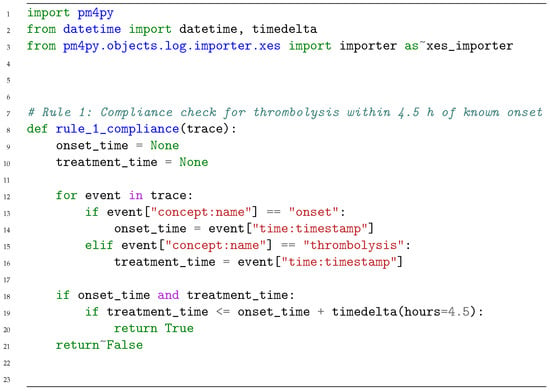
Figure 2.
Python code generated with ChatGPT o3-mini-high model for Rule 1.
Figure 3 shows an example trace, where Rule 1 is satisfied. The example trace belongs to , since it contains the anchor activity (thrombolysis), and also belongs to , as thrombolysis takes place 3 h after onset (no matter how many activities are completed in between).
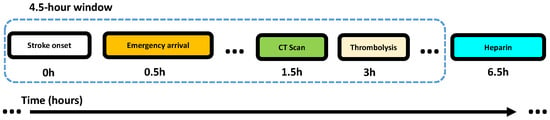
Figure 3.
Example trace compliant with Rule 1.
3.3. Model Conformance Checking
In this section, we describe how the Model Conformance-Checking module operates.
We require models to be represented as Petri Nets. Petri Nets are the oldest and best-investigated process modeling language allowing for the modeling of concurrency. Although the graphical notation is intuitive and simple, Petri Nets are executable and many analysis techniques can be used to analyze them [,,]. A Petri Net is formally defined as a tuple where
- is a finite set of places.
- is a finite set of transitions, with .
- is a set of arcs (flow relation).
- is a weight function that assigns a positive integer weight to each arc.
- is the initial marking, a function that assigns a number of tokens to each place.
Also in this case, we first need to identify all the paths in the process model (i.e., in the Petri Net) which contain the anchor activity, mentioned in the rule, which provides the context of interest. In order to identify such paths, by taking into account Petri Net semantics, we first complete an unfolding step, generating an occurrence net, which is a net without cycles, self conflicts, or backward conflicts. In particular, we refer to [] for the construction of a finite initial part of the unfolding which contains full information about all the reachable states. We then visit the resulting graph structure and build the set .
The function in Algorithm A1, detailed in Appendix A for the interested reader, works similarly to the function , which was described in Algorithm 1 above, but operates on paths instead of traces. Such a function returns the PCI, i.e., the percentage of all paths in that satisfy a particular rule . Note that, to complete such a task, we rely on a method (https://pm4py-source.readthedocs.io/en/latest/pm4py.objects.petri.html (accessed on 20 July 2025)) (available from the open-source Python library PM4Py []) which decorates the model according to temporal performance information (aggregated by mean) obtained by applying trace replay. The resulting model thus specifies the average traversal times for each path that a set of traces took to travel between particular pairs of nodes. This information is necessary to verify whether the rules containing temporal constraints are satisfied by the path at hand.
It is worth noting that the model conformance-checking step, when exploring the process model, may find a path never appearing in the real traces. This is expected and, to some extent, desirable. Indeed, in real-world processes, it is unlikely that the instances in the event log cover all possible executions, such that a certain degree of generalization is needed in the process model, to allow for more behavior than the one recorded in the log itself. However, the extension of the generalization degree only depends on the mining algorithm being adopted [] (not on our conformance-checking approach).
4. Experimental Results
We conducted our experiments in the field of stroke management. Stroke is a very critical medical condition, characterized by an insufficient blood flow to the brain, which can lead to cell death. This can be due to ischemia (a lack of glucose and oxygen supply) caused by a thrombosis or embolism, or to a hemorrhage. As a consequence, in the acute phase, the patient’s life is threatened; moreover, stroke survivors can experience serious adverse events which can lead to permanent disability. Stroke is the leading cause of adult disability in the United States and Europe and the number-two cause of death worldwide.
In the following, we present the results of our experiments, referring to the architecture presented in Figure 1, step by step.
According to step 1 of our architecture, we prompted ChatGPT-4o by asking it in natural language to extract rules using the following prompt: “Extract and formalize rules from <text> by using if-then semantics”, where <text> is the emergency phase management section of one of the most recently published stroke clinical guidelines []. We therefore followed the paradigm defined as “direct provision of insights” in []. The dimension of the emergency phase section of the guideline is of 109,905 characters, including spaces. ChatGPT-4o outputted 28 rules, organized into 23 groups by the type of diagnostic or therapeutic procedure they address (e.g., brain imaging, thrombolysis). Each group is thus composed of one or more items, and every item expresses an IF–THEN rule, in natural language.
In this paper, according to the advice of our medical collaborator, we will mainly focus on three rules that refer to thrombolysis. Thrombolysis is a life-saving procedure that has become the most important instrument for effectively preserving physical and cognitive functions after stroke. For this reason, verification of the accurate execution of thrombolysis is essential when considering the acute treatment of patients. However, this life-saving activity can only be executed in certain conditions and with very precise timing. The rules we are exploiting refer specifically to timing in the administration of thrombolysis, to the imaging procedures needed to enable the thrombolysis treatment, and to the antiplatelet treatment that should follow thrombolysis administration. ChatGPT-4o expressed such rules as reported below:
- If patients have acute ischemic stroke and treatment can be started within 4.5 h of known onset, then they should be considered for thrombolysis with alteplase or tenecteplase;
- If patients have acute ischemic stroke and were last known to be well more than 4.5 h earlier, then they should be considered for thrombolysis with alteplase if treatment can be started between 4.5 and 9 h of known onset or within 9 h of the midpoint of sleep when they have woken with symptoms, and they have evidence from CT/MR perfusion (core-perfusion mismatch) or MRI (DWI-FLAIR mismatch) of the potential to salvage brain tissue;
- If patients with acute ischemic stroke are treated with thrombolysis, then they should be started on an antiplatelet agent after 24 h unless contraindicated, once significant hemorrhage has been excluded.
Note that Rule 1 and Rule 2 are mutually exclusive, since Rule 1 refers to patients whose stroke onset took place less than 4.5 h ago, while Rule 2 refers to patients whose onset took place between 4.5 and 9 h ago. There is no interplay between the two rules, as they are intended for separate groups of patients. In particular, the second group of patients is still eligible for thrombolysis, provided that imaging showed the potential for preserving brain tissues. This category of patients thus requires more specific diagnostic evidence, which can be obtained only by means of timely availability of the imaging instruments. Rule 3, on the other hand, refers to all kinds of patients which underwent thrombolysis treatment.
In step 2 of the architecture described in Figure 1, our medical collaborator checked these rules.
The rules were judged as semantically correct (since they summarize medical knowledge available in the guideline) and relevant (since they must be applied in practice to provide a correct and timely treatment to the patients).
According to the expert’s opinion, the extracted rules were not affected by hallucinations or inaccuracies.
During the rule formalization step (step 3 of our architecture), the rules were converted into a computer-interpretable format. The interested reader can find in the Appendix A an example of the prompt used to generate the Python code for Rule 1 (while the Python code itself is reported in Figure 2). For the construction of the prompt, an input tuple was provided containing the rule text and an example trace from our dataset, for the purpose of providing the LLM with information about process trace data structures in the XES (https://www.xes-standard.org/ (accessed on 15 July 2025)) format. An example XES process trace can also be found in the Appendix A.
We then considered real patient traces from four different hospitals in Northern Italy, for a total of 639 traces. All the data come from a research activity conducted with a set of hospitals in Northern Italy that was approved by ethical committees. However, due to privacy reasons, patients’ data cannot be made publicly available. On the other hand, our code can be accessed at https://osf.io/zbnd2/?view_only=ff4cd41b8cd348e9822ade6ade5049e. This repository includes a Python implementation of Rule 1, along with a synthetic event log in the XES format, useful for testing.
The traces exhibit 17 activities on average (see Table 1 for some more detailed statistics, hospital-by-hospital).

Table 1.
Summary statistics by hospital.
According to step 4 of the architecture in Figure 1, we calculated the TCI. Table 2 reports on the results for Rule 1, Rule 2, and Rule 3 described above.

Table 2.
Trace conformance indicator () on Rule 1, Rule 2, and Rule 3 for four different hospitals.
As can be observed, the TCI is always low for Rule 2: this finding shows that it is not easy to complete thrombolysis on time (i.e., within 9 h) in such patients, probably because their identification also requires the completion of different imaging exams, and access to the imaging instruments is often a bottleneck in hospitals, especially in small/medium-size ones, due to organizational issues. The values of this indicator can be useful for suggesting whether hospital administrators should take the opportunity to increment the availability of imaging instruments (in particular, magnetic resonance ones) and/or reschedule patient priorities. On the other hand, conformance for Rule 3 is always very good, demonstrating a correct implementation of the antiplatelet therapy.
For the sake of generalizability, in the following, we will also present the results for two additional, very general rules that we extracted by means of our approach: Rule 4 states that all patients affected by stroke should be investigated through a brain imaging exam within one hour from stroke onset; Rule 5 states that an MR scan should be preferred to a CT scan (when possible). Such rules apply to all patients and should be respected at every healthcare center.
Table 3 shows the TCI for the four hospitals under examination for these two additional rules. As can be observed, the TCI is always very high for Rule 4, testifying that all hospitals typically respect the critical temporal constraint on the first diagnostic brain imaging exam. On the other hand, Rule 5 is seldom respected, reinforcing the hypothesis, already emerging with Rule 2, that the availability of magnetic resonance instruments may often be suboptimal.

Table 3.
Trace conformance indicator () on Rule 4 and Rule 5.
The next steps of the architecture in Figure 1 involve the generation of process models using different model discovery algorithms (step 5) and the calculation of the PCI for each of them (step 6).
In the rest of this section, we show how we have exploited step 5 and step 6 as a means of comparing different process-mining algorithms, in the dimension of conformance.
To this end, we adopted the PCI as a new measure (to be possibly added to the well-known Replay Fitness, Generalization, Simplicity and Precision []), useful for comparing the performance of different mining algorithms. If the PCI was close to the TCI in the same event log for a particular mined model, this would testify that the miner did not lose too much information with respect to the event log traces, and was reliable as regards the conformance to the normative model.
Note that the mined process model is not the normative model (it is not the guideline); therefore, we did not wish to verify whether a trace was compliant with it (e.g., by using a log replay algorithm on the mined model): instead, the goal was the one of assessing the quality of the mined model itself, as a means of correctly describing the actual behavior reported in the log, from the point of view of conformance.
The following tables show the values of the PCI, referring to the thrombolysis rules, obtained on three different model sets: the first set (see Table 4) groups the four hospital models mined by resorting to the Heuristic Miner algorithm []; the second set (see Table 5) was obtained by the Alpha Miner algorithm [], while the third set (see Table 6) was obtained by the SIM algorithm []. SIM is a process-mining tool recently developed by our group which can discover the process model incrementally, supporting the interaction with domain experts, who can selectively merge parts of the model to achieve compactness and reduced redundancy. All the algorithms were applied with their default parameter settings in these experiments. In the tables, the TCI is also reported, in brackets, for comparison. The best results (where the PCI is very close to the TCI and better/equal with respect to other miners) are highlighted in bold.

Table 4.
Path conformance indicator () for Rule 1, Rule 2, and Rule 3 for four different hospitals models, mined by the Heuristic Miner algorithm. The best results are highlighted in bold.

Table 5.
Path conformance indicator () for Rule 1, Rule 2, and Rule 3 for four different hospitals models, mined by the Alpha Miner algorithm. The best results are highlighted in bold.

Table 6.
Path conformance indicator () for Rule 1, Rule 2, and Rule 3 for four different hospitals models, mined by the SIM algorithm. The best results are highlighted in bold.
According to the PCI and its closeness to the TCI, as described above, and considering the results in Table 4, Table 5 and Table 6, in our validation study, we obtained that Heuristic Miner was the most reliable algorithm: indeed, our tables have 12 entries (corresponding to 3 rules multiplied by 4 hospitals), and in 9 of 12 cases, Heuristic Miner obtained a PCI value which was equal to the TCI value or closer to the TCI value with respect to the PCI value obtained by a different miner. Alpha Miner was better than Heuristic Miner only on Rule 3 applied to hospital H2 (with the PCI = 100%, identical to the TCI, while Heuristic Miner only scored 99.23%). SIM was also better than Heuristic Miner on Rule 1 applied to hospital H3 (with PCI = 92.12%, closer to TCI = 83.33% with respect to Heuristic Miner, which obtained TCI = 100.00%, which is too high).
We applied a test of a hypothesis concerning a system of two proportions [], and we found that Heuristic Miner is significantly more accurate than Alpha Miner with a confidence of 95%, while it is more accurate than SIM with a confidence of 80%.
To complete the comparison, we report in Table 7 the results for the Replay Fitness, Generalization, Simplicity, and Precision [] for the four process models learned by Alpha Miner, Heuristic Miner, and SIM, respectively. All four quality dimensions are important when conducting process discovery. However, the Replay Fitness, indicating to what extent the model can reproduce the traces in the log, is considered more important than the other measures []. Heuristic Miner also outperforms the Alpha Miner and SIM from the point of view of the Replay Fitness (see the average result highlighted in yellow in Table 7).

Table 7.
Results for Replay Fitness, Generalization, Simplicity, and Precision [] for the four process models learned by Alpha Miner, Heuristic Miner, and SIM.
Interestingly, Heuristic Miner is known to be well suited for noisy logs (such as medical ones) and has been successfully adopted in medical process mining []. The results in Table 4, Table 5, Table 6 and Table 7, therefore, strengthen existing findings and provide a positive validation outcome for our study. More experiments (on more miners) will help us to further assess the utility of our approach.
In fact, in the future, we plan to extend the tests performed so far, working on more rules and on more mining algorithms. We also plan to adopt the approach in different application domains.
5. Discussion
Despite the encouraging results we obtained in our experiments, we acknowledge that our approach is not free from issues and challenges to be considered.
One issue is the need to test the robustness of rule extraction across different LLMs. As a first step in this direction, we have established an experiment relying on Google’s Gemini 2.5 flash, by using the same prompt and the same guideline excerpt we described in Section 4. Gemini outputted only 16 rules, organized into 3 groups. Interestingly, Rule 1, Rule 2, Rule 4, and Rule 5 presented in Section 4 were extracted by Gemini as well. Rule 3, on the other hand, was missing: the LLM identified a couple of generic rules about antiplatelet treatment but not correlated to thrombolysis. Our current choice of relying on GPT is therefore more appropriate. Additional investigations, however, are needed, to assess the performance of the various LLMs. In particular, we would like to test tools that have been specifically trained on healthcare data, such as Google’s Med-Gemini (https://research.google/blog/advancing-medical-ai-with-med-gemini/ (accessed on 20 July 2025)) [], which is part of a growing suite of AI models specifically trained and developed for medical applications.
We also foresee adopting a Retrieval Augmented Generation (RAG) [] approach to increase the number and completeness of the extracted rules, and to address specific issues, such as the use of less standardized terminology within the guideline text. In this last case, in particular, the LLM could take advantage of a RAG technique to incorporate the information from an ontology of standardized medical terms, such as SNOMED-CT (https://www.snomed.org/ (accessed on 20 July 2025)), that could augment the quality of text comprehension and of rule extraction.
Another strategy for improving rule quality could be the one of adopting more specific prompts. Prompt engineering is in fact a very active area of research [], able to enhance model efficacy without modifying the model parameter values. Prompts enable the smooth adaptation of pre-trained models to downstream tasks by shaping their behavior through the prompt alone. These prompts may take the form of natural language instructions that give context for guidance, or learned vector representations that trigger relevant knowledge. The already-mentioned RAG, for instance, can be seen as a prompting approach: it interprets user input, formulates a specific query, and searches a pre-constructed knowledge base for relevant material. The retrieved content is then added to the original prompt, enhancing it with additional context. We may also consider the very interesting Automatic Prompt Engineer (APE) approach []: APE overcomes the constraints of fixed, manually designed prompts by dynamically creating and selecting the most effective ones. It processes user input, generates candidate instructions, and applies reinforcement learning to identify the best prompt, adapting in real time to different contexts.
On a different note, for the sake of generalizability, it would be important to test our tool in different application domains and on guidelines of different complexities. We performed some initial tests on rule extraction from a particularly articulated guideline text about Multiple Sclerosis (MS) (https://www.neuro.it/web/eventi/NEURO/lineeguida.cfm (accessed on 2 September 2025)), which involves branching logic choices and complex flowcharts. We selected this pathology since we have some previous experience on this domain []. We obtained encouraging results: as an example, in the following, we show an articulated rule, which correctly captures some steps of the workflow for Multiple Sclerosis patients under natalizumab treatment.
IF treating patients with MS with natalizumab, THEN periodically evaluate the risk of Progressive Multifocal Leukoencephalopathy (PML) by measuring anti-JCV antibody titer, AND discuss the risk/benefit ratio of continuing therapy with the patient; after initiating therapy, consider switching to a 6-week interval dosing regimen to minimize PML risk.
Additional experiments are of course needed and are one of our future plans. As soon as we collect an event log related to a different disease, in particular, we will be able to complete the experiments on trace conformance checking and process model conformance checking as well.
6. Conclusions and Future Research Directions
In this paper, we have introduced an LLM-assisted approach to support conformance checking in medical process mining. Our contribution is innovative according to different dimensions:
- It adopts an LLM to extract normative behaviors from a textual guideline and to formalize it into computer-interpretable rules; while this task is relatively straightforward (and, in fact, could be easily adopted to different domains as well), to the best of our knowledge, our approach is the first of its kind in medicine.
- It evaluates the conformance of patient traces to the extracted rules and quantifies it through the TCI. The TCI allows us to assess the conformance of the actual behavior of a given hospital with respect to the guideline, in a quality assessment perspective, without requiring a time-consuming formalization of the guideline itself.
- It evaluates the conformance of a process model to the rules: it can therefore be adopted as a new dimension to compare different process models (discovered by different algorithms), focusing on their adherence to the prescribed normative behavior.
Our experiments in the domain of stroke have provided insight about the quality of the actual processes implemented by four Northern Italy hospitals, referring to thrombolytic treatment. In particular, while the timing prescribed by Rule 1 (referring to the start of thrombolysis within 4.5 h from stroke onset) and Rule 3 (referring to the administration of antiplatelet agents after thrombolysis treatment) is typically respected, the TCI is always low for Rule 2: this finding suggests that such small/medium-sized hospitals may suffer from the presence of a bottleneck in the access to advanced imaging instruments, which are needed for patients whose onset took place more than 4.5 h ago, but who still have the potential to have their brain tissue preserved. Such a bottleneck has also been verified by the analysis of two additional, more general rules. Moreover, our experiments have proved the high quality of Heuristic Miner as a process discovery algorithm in this medical field, confirming a strength already recognized elsewhere [].
The collaboration with a medical expert, who has checked the alignment of the rules to medical knowledge, has guaranteed the semantic correctness of our approach in its current version. In any case, as already commented on in Section 5, we believe that it would be worth testing rule extraction by resorting to different LLMs and, in particular, to tools that have been trained on healthcare data, such as Google’s Med-Gemini []. We will verify whether Med-Gemini is able to learn more rules from the guideline and/or to extract rules that are semantically more correct (possibly making the expert rule-checking phase faster). On the same note, we would like to move towards the (partial) automation of rule checking itself. To this end, we will consider the research area known as “LLM-as-a-Judge” [], where LLMs are employed as evaluators for complex tasks, executed by other LLMs. While human evaluation can be slow and costly, LLMs can guarantee faster turnaround and consistent evaluation standards, provided that a robust evaluation criterion is defined. While the LLM-as-a-judge technology must be considered carefully in the medical field, it could at least speed up the rule-checking process, assigning a first score to the rules, (some of) which may be further verified by experts if needed.
Author Contributions
Conceptualization, G.L., S.M. and M.S.; Methodology, G.L., S.M. and M.S.; Software, G.L., S.M. and M.S.; Validation, G.L., S.M. and M.S.; Data curation, G.L., S.M. and M.S.; Writing—original draft, G.L., S.M. and M.S.; Writing—review & editing, S.M. and M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was approved by the Ethics Committee of Comitato Etico Territoriale (CET) (Northern Italy) (protocol code 216/CE and date of approval 19 March 2025).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Due to privacy reasons, patients’ data cannot be made publicly available. On the other hand, our code can be accessed at https://osf.io/zbnd2/?view_only=ff4cd41b8cd348e9822ade6ade5049e. This repository includes a Python implementation of Rule 1, along with a synthetic event log in the XES format, useful for testing.
Acknowledgments
We acknowledge the contribution of our medical collaborators for their work in semantically checking the LLM-extracted rules, and for her advice on focusing the experiments on the thrombolysis treatment phase.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
In this Appendix, we provide some additional technical material, for the interested reader.
The function in Algorithm A1 works similarly to the function , which was described in Algorithm 1, but operates on paths instead of traces. Such a function returns the PCI, i.e., the percentage of all paths in the occurrence net that satisfy a particular rule .
The listing in Figure A1 shows an example of the prompt used to generate the Python code for Rule 1, discussed in Section 4. The listing in Figure A2 shows an extract of a process trace in XES format, to be given as input along with the prompt.
| Algorithm A1 Function returns the percentage of paths in the model that satisfy a particular rule . |
|


Figure A2.
Part of an example process trace in XES format.
References
- van der Aalst, W.M.P. Process Mining—Data Science in Action, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Reichert, M.; Weber, B. Enabling Flexibility in Process-Aware Information Systems—Challenges, Methods, Technologies; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar] [CrossRef]
- Desel, J.; Reisig, W.; Rozenberg, G. (Eds.) Lectures on Concurrency and Petri Nets, Advances in Petri Nets; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2004; Volume 3098. [Google Scholar] [CrossRef]
- de Clercq, P.A.; Kaiser, K.; Hasman, A. Computer-interpretable Guideline Formalisms. In Computer-Based Medical Guidelines and Protocols: A Primer and Current Trends; ten Teije, A., Miksch, S., Lucas, P.J.F., Eds.; Studies in Health Technology and Informatics; IOS Press: Amsterdam, The Netherlands, 2008; Volume 139, pp. 22–43. [Google Scholar] [CrossRef]
- Buijs, J.C.A.M.; van Dongen, B.F.; van der Aalst, W.M.P. On the Role of Fitness, Precision, Generalization and Simplicity in Process Discovery. In On the Move to Meaningful Internet Systems: OTM 2012, Confederated International Conferences: CoopIS, DOA-SVI, and ODBASE 2012, Rome, Italy, 10–14 September 2012; Proceedings, Part I; Meersman, R., Panetto, H., Dillon, T.S., Rinderle-Ma, S., Dadam, P., Zhou, X., Pearson, S., Ferscha, A., Bergamaschi, S., Cruz, I.F., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7565, pp. 305–322. [Google Scholar] [CrossRef]
- Dunzer, S.; Stierle, M.; Matzner, M.; Baier, S. Conformance checking: A state-of-the-art literature review. In Proceedings of the 11th International Conference on Subject-Oriented Business Process Management, S-BPM ONE 2019, Seville, Spain, 26–28 June 2019; Betz, S., Ed.; ACM: New York, NY, USA, 2019; pp. 1–10. [Google Scholar] [CrossRef]
- Rozinat, A.; van der Aalst, W.M.P. Conformance checking of processes based on monitoring real behavior. Inf. Syst. 2008, 33, 64–95. [Google Scholar] [CrossRef]
- Leemans, S.J.J.; Fahland, D.; van der Aalst, W.M.P. Scalable process discovery and conformance checking. Softw. Syst. Model. 2018, 17, 599–631. [Google Scholar] [CrossRef] [PubMed]
- Adriansyah, A.; Munoz-Gama, J.; Carmona, J.; van Dongen, B.F.; van der Aalst, W.M.P. Alignment Based Precision Checking. In Proceedings of the Business Process Management Workshops—BPM 2012 International Workshops, Tallinn, Estonia, 3 September 2012; Rosa, M.L., Soffer, P., Eds.; Revised Papers; Lecture Notes in Business Information Processing. Springer: Berlin/Heidelberg, Germany, 2012; Volume 132, pp. 137–149. [Google Scholar] [CrossRef]
- Borrego, D.; Barba, I. Conformance checking and diagnosis for declarative business process models in data-aware scenarios. Expert Syst. Appl. 2014, 41, 5340–5352. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 3–5 June 2019; Burstein, J., Doran, C., Solorio, T., Eds.; Long and Short Papers. Association for Computational Linguistics: Minneapolis, MN, USA; 2019; Volume 1, pp. 4171–4186. [Google Scholar] [CrossRef]
- OpenAI; Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; et al. OpenAI: GPT-4 technical report CoRR. arXiv 2024, arXiv:2303.08774. [Google Scholar] [CrossRef]
- Khurana, D.; Koli, A.; Khatter, K.; Singh, S. Natural language processing: State of the art, current trends and challenges. Multim. Tools Appl. 2023, 82, 3713–3744. [Google Scholar] [CrossRef] [PubMed]
- Berti, A.; Kourani, H.; Hafke, H.; Li, C.Y.; Schuster, D. Evaluating Large Language Models in Process Mining: Capabilities, Benchmarks, and Evaluation Strategies. arXiv 2024, arXiv:2403.06749. [Google Scholar] [CrossRef]
- Berti, A.; Schuster, D.; van der Aalst, W.M.P. Abstractions, Scenarios, and Prompt Definitions for Process Mining with LLMs: A Case Study. In Proceedings of the Business Process Management Workshops—BPM 2023 International Workshops, Utrecht, The Netherlands, 11–15 September 2023; Weerdt, J.D., Pufahl, L., Eds.; Revised Selected Papers; Lecture Notes in Business Information Processing. Springer: Berlin/Heidelberg, Germany, 2023; Volume 492, pp. 427–439. [Google Scholar] [CrossRef]
- Kourani, H.; Berti, A.; Hennrich, J.; Kratsch, W.; Weidlich, R.; Li, C.Y.; Arslan, A.; Schuster, D.; van der Aalst, W.M.P. Leveraging Large Language Models for Enhanced Process Model Comprehension. arXiv 2024, arXiv:2408.08892. [Google Scholar]
- Qafari, M.S.; van der Aalst, W. Fairness-Aware Process Mining. In Proceedings of the on the Move to Meaningful Internet Systems: OTM 2019 Conferences, Rhodes, Greece, 21–25 October 2019; Panetto, H., Debruyne, C., Hepp, M., Lewis, D., Ardagna, C.A., Meersman, R., Eds.; Springer: Berlin/Heidelberg, Germany; Cham, Switzerland, 2019; pp. 182–192. [Google Scholar]
- Kourani, H.; Berti, A.; Schuster, D.; van der Aalst, W.M. ProMoAI: Process Modeling with Generative AI. In Proceedings of the Thirty-Third International Joint Conference on Artificial Intelligence. International Joint Conferences on Artificial Intelligence Organization, Jeju, Republic of Korea, 3–9 August 2024. [Google Scholar]
- Grohs, M.; Abb, L.; Elsayed, N.; Rehse, J.R. Large Language Models can accomplish Business Process Management Tasks. arXiv 2023, arXiv:2307.09923. [Google Scholar] [CrossRef]
- Jessen, U.; Sroka, M.; Fahland, D. Chit-Chat or Deep Talk: Prompt Engineering for Process Mining. arXiv 2023, arXiv:2307.09909. [Google Scholar] [CrossRef]
- Berti, A.; van Zelst, S.; Schuster, D. PM4Py: A process mining library for Python. Softw. Impacts 2023, 17, 100556. [Google Scholar] [CrossRef]
- Weijters, A.J.; van Der Aalst, W.M.; De Medeiros, A.A. Process Mining with the HeuristicsMiner Algorithm; Technische Universiteit Eindhoven: Eindhoven, The Netherlands, 2006. [Google Scholar]
- Bottrighi, A.; Guazzone, M.; Leonardi, G.; Montani, S.; Striani, M.; Terenziani, P. Integrating ISA and Part-of Domain Knowledge into Process Model Discovery. Future Internet 2022, 14, 357. [Google Scholar] [CrossRef]
- Jensen, K.; Kristensen, L.M. Coloured Petri Nets—Modelling and Validation of Concurrent Systems; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar] [CrossRef]
- Reisig, W.; Rozenberg, G. (Eds.) Lectures on Petri Nets I: Basic Models, Advances in Petri Nets; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1998; Volume 1491. [Google Scholar] [CrossRef]
- van der Aalst, W.M.P.; Stahl, C. Modeling Business Processes—A Petri Net-Oriented Approach; Cooperative Information Systems Series; MIT Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Esparza, J.; Römer, S.; Vogler, W. An Improvement of McMillan’s Unfolding Algorithm. Form. Methods Syst. Des. 2002, 20, 285–310. [Google Scholar] [CrossRef]
- van der Aalst, W.M.P.; Rubin, V.; Verbeek, H.M.W.; van Dongen, B.F.; Kindler, E.; Günther, C.W. Process mining: A two-step approach to balance between underfitting and overfitting. Softw. Syst. Model. 2010, 9, 87–111. [Google Scholar] [CrossRef]
- National Clinical Guideline for Stroke for the UK and Ireland. London: Intercollegiate Stroke Working Party. Available online: https://www.strokeguideline.org (accessed on 8 April 2025).
- van der Aalst, W.M.P.; van Dongen, B.F. Discovering Workflow Performance Models from Timed Logs. In Proceedings of the Engineering and Deployment of Cooperative Information Systems, First International Conference, EDCIS 2002, Beijing, China, 17–20 September 2002; Proceedings. Han, Y., Tai, S., Wikarski, D., Eds.; Lecture Notes in Computer Science. Springer: Berlin/Heidelberg, Germany, 2002; Volume 2480, pp. 45–63. [Google Scholar] [CrossRef]
- Johnson, R.A. Miller and Freund’s Probability and Statistics for Engineers, 8th ed.; Prentice Hall International: Hoboken, NJ, USA, 2011. [Google Scholar]
- Rojas, E.; Munoz-Gama, J.; Sepúlveda, M.; Capurro, D. Process mining in healthcare: A literature review. J. Biomed. Inform. 2016, 61, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, S.; Sellergren, A.; Kohlberger, T.; Zhou, Y.; Ktena, I.; Kiraly, A.; Ahmed, F.; Hormozdiari, F.; Jaroensri, T.; et al. Advancing Multimodal Medical Capabilities of Gemini. arXiv 2024, arXiv:2405.03162. [Google Scholar] [CrossRef]
- Lewis, P.; Perez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Küttler, H.; Lewis, M.; tau Yih, W.; Rocktäschel, T.; et al. Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: New York, NY, USA, 2020; Volume 33, pp. 9459–9474. [Google Scholar]
- Sahoo, P.; Singh, A.K.; Saha, S.; Jain, V.; Mondal, S.; Chadha, A. A Systematic Survey of Prompt Engineering in Large Language Models: Techniques and Applications. arXiv 2025, arXiv:2402.07927. [Google Scholar]
- Zhou, Y.; Muresanu, A.I.; Han, Z.; Paster, K.; Pitis, S.; Chan, H.; Ba, J. Large Language Models Are Human-Level Prompt Engineers. arXiv 2023, arXiv:2211.01910. [Google Scholar] [CrossRef]
- Francia, R.; Leone, M.; Leonardi, G.; Montani, S.; Pennisi, M.; Striani, M.; D’Alfonso, S. AutoML-Med: A Framework for Automated Machine Learning in Medical Tabular Data. arXiv 2025, arXiv:2508.02625. [Google Scholar]
- Gu, J.; Jiang, X.; Shi, Z.; Tan, H.; Zhai, X.; Xu, C.; Li, W.; Shen, Y.; Ma, S.; Liu, H.; et al. A Survey on LLM-as-a-Judge. arXiv 2025, arXiv:2411.15594. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).