Abstract
Obstructive Sleep Apnea (OSA) is a prevalent condition that disrupts sleep quality and contributes to significant health risks, necessitating accurate and efficient diagnostic methods. This study introduces a machine learning-based framework aimed at detecting apnea events through analysis of polysomnographic (PSG) and oximetry data. The core component is a Long Short-Term Memory (LSTM) network, which is particularly suited to processing sequential time-series data, capturing complex temporal relationships within physiological signals such as oxygen saturation, heart rate, and airflow. Through extensive feature engineering and preprocessing, the framework optimizes data representation by normalizing, scaling, and encoding input features to enhance computational efficiency and model performance. Key results demonstrate the model’s effectiveness, achieving a balanced accuracy of 79%, precision of 68%, and recall of 76% on the test dataset, with validation set metrics similarly high, supporting the model’s ability to generalize effectively. Comprehensive hyperparameter tuning further contributed to a stable, robust architecture capable of accurately identifying apnea events, providing clinicians with a valuable tool for the early detection and tailored management of OSA. This data-driven framework offers an efficient, reliable solution for OSA diagnostics with the potential to improve clinical decision making and patient outcomes.
1. Introduction
Obstructive Sleep Apnea (OSA) affects approximately 2% to 26% of adults with a substantial number of cases going undetected. This condition leads to frequent disruptions in sleep and lower oxygen levels, which are caused by repeated upper airway collapse during rest. Such occurrences are associated with serious health risks, including cardiovascular issues and heightened daytime drowsiness, which in turn raises accident susceptibility [1,2].
Consequently, managing and continually monitoring OSA is essential. Polysomnography (PSG), the established standard for diagnosing sleep-related breathing disorders, plays a vital role. This diagnostic technique tracks various physiological indicators, such as brain activity, heart rate, and respiratory patterns through multiple sensors, enabling a comprehensive assessment of sleep cycles and disturbances [3]. Additionally, standardized screening questionnaires are valuable for initial assessment, helping to identify individuals who may require further diagnostic procedures.
The primary treatment for OSA is Continuous Positive Airway Pressure (CPAP), which acts as a pneumatic support to maintain an open airway during sleep [4]. While CPAP generally enhances airflow and breathing for many individuals, its effectiveness can differ among patients and it may not fully address all anatomical factors; in some cases, it may even cause collapses at specific locations, like the epiglottis. Consequently, alternatives such as oral devices or positional therapies are sometimes needed for those who experience discomfort or struggle with adherence to CPAP.
Given the complexity and variability of OSA, machine learning (ML) has become an increasingly important tool in its detection. ML algorithms are particularly skilled at managing and analyzing large, intricate datasets typical in sleep studies, including those produced by polysomnography [5]. These algorithms can detect complex patterns and subtle data features that traditional methods may miss, thereby enhancing the accuracy and efficiency of OSA diagnoses [6]. This ability is especially critical due to OSA’s high prevalence and its associated health risks, such as cardiovascular disease and diabetes. Platforms integrating artificial intelligence not only improve diagnostics but also support the detection of critical future events, such as apnea episodes. Detection modeling of future apnea events enables a proactive approach, providing clinicians the opportunity to intervene preemptively if potential issues are detected. By forecasting these events, healthcare providers could respond in a timely manner, potentially mitigating the onset of acute episodes and optimizing patient management. Furthermore, the integration of AI in healthcare not only expedites treatment decisions but also optimizes therapeutic strategies and reduces healthcare costs [7].
Section 2 provides an overview of machine learning models applied in diagnosing and treating OSA as well as detecting apnea events and current treatment methods. Section 3 describes in detail the architecture of the data-based framework for processing medical data, including the proposed integration of an LSTM model into the framework. Section 4 then presents the developed deep learning-based framework for detecting apnea events, which can aid clinicians in improving patient diagnosis. Results and validation will be presented in Section 5, while future directions and a discussion of the findings will be provided in Section 6. The paper will conclude in Section 7.
2. Related Work
Recent advancements in sleep medicine and machine learning (ML) have significantly transformed the diagnosis and management of Obstructive Sleep Apnea (OSA). While polysomnography (PSG) remains the gold standard for diagnosing OSA, it is resource-intensive and inconvenient for patients. As a result, alternative approaches leveraging ML techniques have been developed to enhance diagnostic accuracy, improve patient experience, and reduce costs.
Machine learning frameworks have demonstrated substantial promise in automating OSA diagnosis. For instance, Gutiérrez-Tobal et al. [8] proposed an ensemble-learning regression model to estimate OSA severity using at-home oximetry data. Their results showed that ML could provide accurate diagnostic outcomes while improving patient comfort. Similarly, Dutta et al. [9] employed decision tree models to evaluate key physiological traits derived from PSG data, enabling personalized therapy strategies for OSA. This personalized approach aligns with the need for tailored treatments, particularly in complex cases.
Deep learning models have been at the forefront of this research. Vaquerizo-Villar et al. [10] employed convolutional neural networks (CNNs) to diagnose OSA in pediatric populations, reducing reliance on expensive PSG procedures. El-Moaqet et al. [11] demonstrated the effectiveness of recurrent neural networks (RNNs) for processing single-channel respiration signals, highlighting their ability to extract temporal dependencies critical for apnea detection. Similarly, Chang et al. [12] utilized a one-dimensional CNN to process single-lead ECG data, achieving high accuracy in distinguishing apnea from non-apnea events.
Single-channel oximetry has emerged as a valuable diagnostic tool. Behar et al. [13] demonstrated the feasibility of using single-channel oximetry for the mass screening of OSA, offering a low-cost, accessible solution. Random forest algorithms, as shown by Deviaene et al. [14], enhance computational efficiency by selecting critical features, enabling real-time clinical applications.
The integration of multiple physiological signals has further improved diagnostic accuracy. Sharma et al. [15] combined blood oxygen saturation (SpO2) and pulse rate data, utilizing deep learning models to detect apnea events with enhanced precision. Nasir et al. [16] explored advanced architectures such as Xception and residual networks, demonstrating significant improvements in classification performance.
Non-invasive and wearable technologies have also gained traction. Kim et al. [17] reviewed smartphone-based applications, highlighting their potential in correlating the apnea–hypopnea index (AHI) with oxygen desaturation levels, enabling early detection in home settings. Similarly, Lee et al. [18] emphasized the role of wearable devices in monitoring sleep patterns and cardiovascular health, showcasing their real-time diagnostic capabilities.
Probabilistic models have proven effective for respiratory event detection. Sadoughi et al. [19] utilized layered Hidden Markov Models (HMMs) to analyze PSG data, while Romero and Jané [20] employed dynamic Bayesian networks to identify obstructive apnea episodes from ECG-based time-series data.
Upper Airway Stimulation (UAS) has emerged as a promising treatment for patients intolerant to Continuous Positive Airway Pressure (CPAP) therapy. Veugen et al. [21] demonstrated its long-term effectiveness in reducing respiratory events and improving patient adherence. Positional therapies, oral devices, hypoglossal nerve stimulation, and myofunctional therapy, by enhancing the function and coordination of oral and facial muscles, further diversify treatment options for conditions such as sleep apnea, orthodontic concerns, and speech disorders, as highlighted by Aboussouan et al. [22].
Research into home-based solutions has expanded diagnostic accessibility. Espinosa et al. [23] reviewed advancements in home monitoring devices, emphasizing the role of ML algorithms in enhancing their accuracy and reliability. Waseem et al. [24] highlighted the detection value of the Oxygen Desaturation Index (ODI) derived from oximetry data for detecting moderate to severe OSA in patients using opioids.
The framework presented in this study aligns with these advancements, introducing a Long Short-Term Memory (LSTM)-based model optimized for time-series PSG data. By capturing complex temporal dependencies, this model achieved a balanced accuracy of 79%, precision of 68%, and recall of 76%, demonstrating its robustness in apnea event detection. Feature engineering and advanced preprocessing techniques, including SpO2 variability and patient-specific data, further enhanced model performance. These contributions underscore the potential of ML frameworks to revolutionize OSA diagnosis and management, offering scalable, personalized solutions for diverse patient populations.
3. Data-Based Framework for Analyzing Sleep Data
A data-based framework has been developed to assist clinicians in making more accurate diagnoses of patients’ health status. With a growing number of individuals diagnosed with OSA, PSG remains a key diagnostic method. This overnight test gathers a range of physiological signals, such as brain activity, SpO2 levels, heart rate, respiratory metrics, and eye and limb movement, depending on the equipment used. In this study, the Philips Alice 6 LDx PSG device was utilized [25]. Such comprehensive data are essential in determining the severity of OSA in each patient.
Alongside PSG, patients complete the standardized STOP questionnaire, which provides additional context for assessing their condition. This combined approach captures a detailed view of each patient’s sleep patterns and potential abnormalities, making it invaluable for identifying various sleep disorders. For each patient during sleep, two additional files are recorded. The csv_event file logs various types of events that occur throughout the night. Each event entry includes the associated sleep stage, timestamp, epoch number, body position, and event validity. The csv_hypnogram file records epoch numbers, the start time of each epoch, and the corresponding sleep stage.
Applying machine learning and sophisticated statistical analysis to these data enables a more nuanced understanding of each patient’s health profile by uncovering patterns that may not be easily observed otherwise. This allows for tailored treatments, better resource allocation, improved diagnostic accuracy, and the ability to track patient-specific trends.
The platform developed here consolidates this data-driven approach by incorporating PSG data, stored in .edf files with additional patient-specific files containing sleep data and STOP questionnaire responses. Central to this platform is a suite of Python-based analyses that generate individualized health insights, integrating both statistical evaluation and machine learning for more dynamic data processing. The data and code are stored in Google Sheets and accessible via Node.js requests, while the web application’s React.js front-end retrieves and visualizes patient data by referencing their unique patient ID.
Figure 1 illustrates the overall system structure, and Figure 2 displays the application’s user interface, which allows for visualization upon input of a patient ID. This comprehensive data framework offers a transformative approach in sleep medicine, making diagnostics more efficient, precise, and tailored to individual needs.
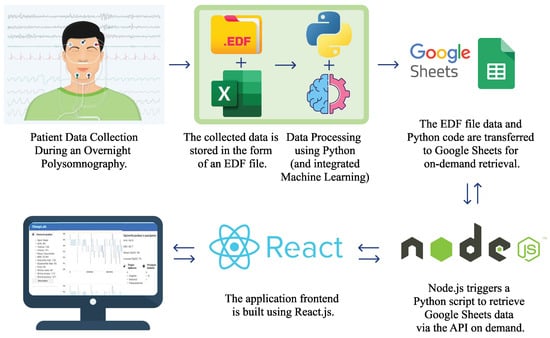
Figure 1.
System architecture: from data collection by recording patients to visualization through a web application, which provides doctors with a more interactive approach to analyzing the collected data.
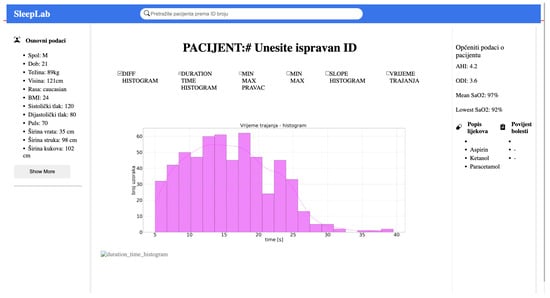
Figure 2.
The web application interface is developed using React.js. In the menu, the user inputs the patient ID and sends a request, which is handled by Node.js. The request triggers the execution of Python code to generate graphs as shown in the image.
Integration of the LSTM Model into the Framework for Analyzing Sleep Data
In this expanded version of the framework, an LSTM (Long Short-Term Memory) model is integrated to enhance the detection of apnea events and better understand the temporal dynamics of sleep disorders. The LSTM model is particularly well suited for time-series data like polysomnographic signals, which involve sequences of data points over time. Appendix A gives a detailed explanation of LSTM model and architecture.
To integrate the LSTM model, the existing framework is first adjusted to ensure compatibility with sequential data inputs. This involves preprocessing the polysomnographic signals by extracting relevant features such as heart rate, oxygen saturation, and respiratory rate, which are then formatted into sequences that the LSTM model can process. The data are normalized and split into training and test sets with temporal dependencies taken into account.
The LSTM model is then trained to recognize patterns indicative of apnea events, which can be difficult to detect with traditional methods. This new addition enables the framework to detect not only anomalies in isolated data points but also to capture complex temporal relationships between various sleep parameters. The result is a more accurate and dynamic system for monitoring sleep disorders, providing personalized insights and improving the overall diagnostic process.
4. Model and Methods
Before feeding data into the LSTM network, comprehensive preprocessing steps were undertaken. The data used in the research were PSG collected from 58 patients sampled in the sleep medicine laboratory, SleepLab, University Hospital of Split, and saved in .EDF format. Initially, we analyzed data from 16 patients, each with 8–9 h of recordings and an average of 300 events, including hypopnea, obstructive apnea, mixed apnea, and central apnea. In the final phase, data from 58 patients were used, showing similar recording durations but greater variability in the number of events with many patients having fewer than 100 events and some fewer than 10. The data were first downsampled from 500 Hz to 1 sample per second and reduced from float64 to float32 to optimize computational efficiency. Before implementation, an analysis of variance was conducted to ensure that there was no significant loss of information when transitioning to 1 Hz and converting to float32. The input data of the network includes key physiological signals such as SpO2, heart rate, airflow channel 0, and airflow channel 1. During data processing and preparation for further use, some artifacts were detected primarily at the beginning and end of the patient recordings. Additionally, a few artifacts were present during the recordings. The approach applied was to remove sequences with missing data to ensure data quality.
In addition to PSG data file, different types of events that occurred during the night were also collected. Each event is associated with a sleep stage, the time it occurred, the epoch number, the body position, and event validity. Please note that a standard epoch in PSG is typically 30 s long and is chosen because it balances detailed signal resolution with manageable data chunks for manual or automated scoring. For this reason, the recorded data were cut into fixed length chunks, each 30 s long, and assigned with a label indicating whether an apnea event occurred. Figure 3 shows the distribution of apneas and hypapneas by sleep stage for a single patient. Due to the simplicity of our model, apnea (central, mixed, obstructive) or hypopnea events were marked as 1, while other events were marked as 0, based on the SpO2 signal processing, as shown in Figure 4.
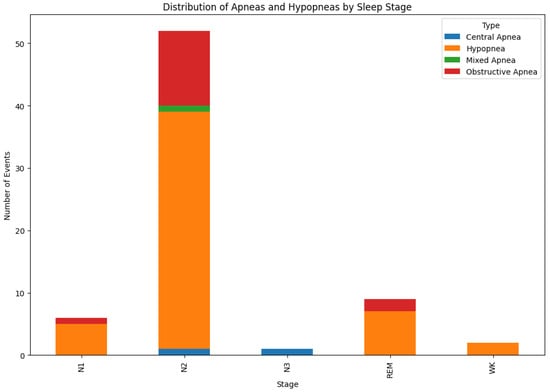
Figure 3.
Distribution of apnoeas event by sleep stage for a single patient.
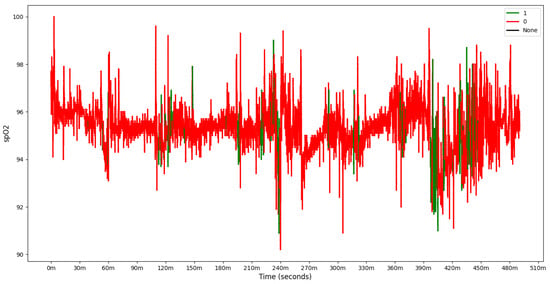
Figure 4.
SpO2 with label of one patient, 1 denoting detected apnea, and 0 no apneic event.
The dataset was then divided into training, validation and testing sets. Specifically, 15% of the data was allocated for validation, 15% for testing, and the remaining 70% for training. This split was repeated for each patient using random sampling with segments from different parts of the night assigned as 15% validation, 15% test, and the rest for training. To ensure balanced sampling during training, a Weighted Random Sampler was used due to class imbalances in event labels. A patient-based data split was tested during the data division process; however, due to the limited number of samples and the variability in patients’ clinical conditions—some exhibiting better health and others more severe cases—it was necessary to adopt a random sampling approach. Data designated for testing, training, and validation were scaled to a range of 0–1 or −1 to 1. Various scaling methods including MinMaxScaler, StandardScaler, and RobustScaler were tested, with implementation options explored on a per-sequence or per-patient basis, or as specified in a configuration file. As an optional feature, a unique, encoded ID was assigned to each patient, providing the model with an additional feature for patient-specific insights.
To enhance model performance, several features were engineered to provide additional context for the model. Specifically, we calculated statistical features such as standard deviation, gradient, minimum, maximum, and amplitude for each SpO2 and heart rate sequence. These features were chosen based on their potential to capture signal variability, which is known to correlate with apnea events. Among these, standard deviation and amplitude features contributed significantly to recall improvement by providing the model with a sense of recent signal fluctuation. Feature selection experiments indicated that the addition of these derived features increased recall by approximately 5% in the validation set, indicating their effectiveness in aiding in the detection of apnea events.
Further preprocessing steps included the removal of SpO2 values close to zero to eliminate potential measurement errors. For each signal parameter, statistical features such as standard deviation, gradient, minimum, maximum, and amplitude were calculated. A sliding window approach was employed to generate sequences with the window size defining the length of each sequence. Additionally, a time feature was computed by dividing each row index (in seconds) by 43,200 (the total number of seconds in 12 h) to incorporate temporal context into the model.
Each sequence was labeled based on the occurrence of an apnea event in the subsequent epoch, which was recorded in the csv_events file. This approach, along with preprocessing adjustments, ensured that the data were well prepared and appropriately structured to optimize the detection performance and robustness of the model.
Model Preparation and Hyperparameters Testing
The model architecture in this study was designed to be flexible, accommodating a range of hyperparameter adjustments specified via a configuration file. The base structure consists of a single LSTM layer followed by a dense layer, with provisions for expanding the architecture by adding layers or units as necessary, as illustrated in Figure 5. While bidirectional layers were considered for enhancing feature extraction, their inclusion significantly increases model complexity and size, underscoring the importance of balancing model depth with available data and task-specific requirements.
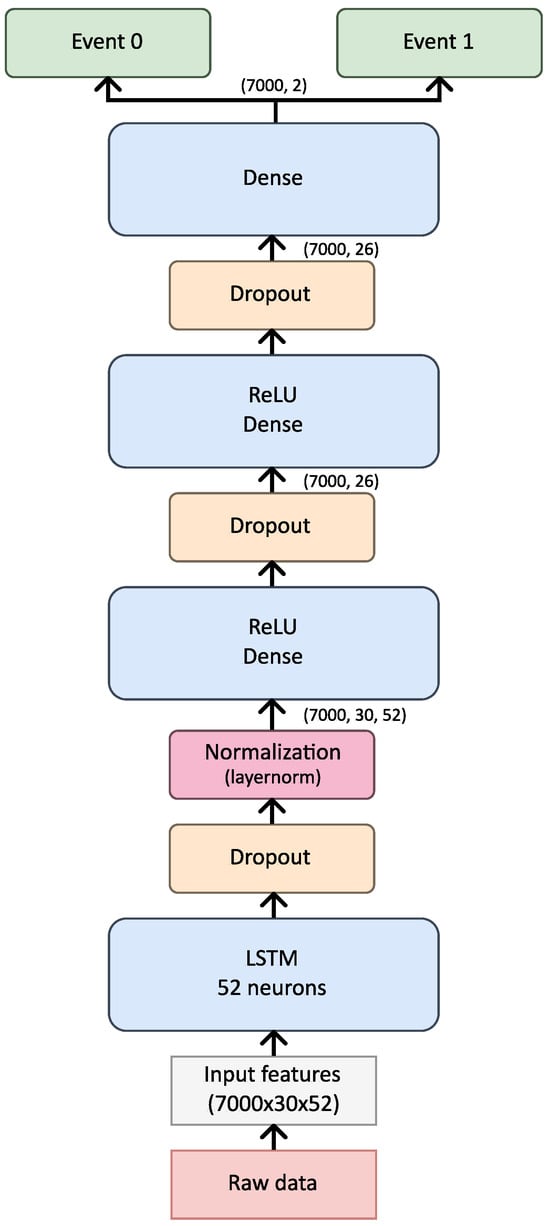
Figure 5.
Architecture of the used model.
Several critical training parameters were fine tuned for optimal performance. The learning rate controls the step size for weight updates, while the number of epochs determines the total passes through the training data. Batch size was selected to optimize training efficiency and memory utilization, particularly on GPU-based systems, achieving a balance between processing speed and memory allocation. The AdamW optimizer was chosen based on its proven performance and fast convergence. Additionally, a learning rate scheduler, ReduceLROnPlateau, was employed to dynamically adjust the learning rate during training, enhancing the model’s capacity for fine tuning.
The model optimization was conducted in three distinct sweeps, each progressively narrowing the search space for hyperparameters and refining the architecture. In the first sweep, over one million combinations of hyperparameters were explored, testing configurations with multiple LSTM and dense layers as well as Bi-LSTM layers. This comprehensive search provided insights into optimal ranges for parameters such as hidden size, layer count, dropout rate, and normalization techniques. Due to computational demands, training was performed on a high-performance RTX 4090 GPU with the most complex models containing over 666,000 parameters.
Insights from the initial sweep led to refinements in the model’s structure. While Bi-LSTM layers were retained for their positive effect on recall, other parameters were adjusted to achieve a balance between model complexity and performance. Dropout rates were increased to mitigate overfitting, and the hidden size was reduced to prevent unnecessary parameter inflation. In the second sweep, the search space was reduced to 500,000 combinations, further refining the architecture. Here, the Bi-LSTM layer configuration proved beneficial for recall, while LayerNorm was retained for its favorable impact on both recall and loss. Modifications included reducing the number of dense layers to avoid overfitting and employing only a single LSTM layer to balance recall and loss.
In the third and final sweep, the model configuration was refined by testing a targeted range of hyperparameters based on previous findings, narrowing the search space to approximately 50,000 combinations. This sweep included testing a variety of parameters to achieve the best performance in detecting apneic events. Specifically, the configurations tested included sequence sizes of 30, 60, and 90, with both balanced and unbalanced datasets, as well as with and without pulse rate data. Feature engineering was applied consistently, while time inclusion varied (True/False). Preprocessing was tested with both StandardScaler and RobustScaler, and hidden sizes were set to range from 1 to 3 units.
During data preprocessing, we removed SpO2 values close to zero to avoid potential measurement errors. This decision was based on observations of irregular, near-zero SpO2 values that are not typical in clinical settings and likely indicate sensor errors rather than genuine physiological states. Additionally, to reduce potential bias, we used a balanced dataset where apnea and non-apnea events were represented proportionally. These preprocessing steps ensure that the model is trained on data that accurately reflects real patient conditions, minimizing the risk of misleading patterns due to artifact-laden signals.
The architectural configurations spanned 1–2 LSTM layers and 2–5 dense layers with dropout rates between 0.2 and 0.5. For normalization, three options were tested: batch normalization, layer normalization, and none. Bidirectional layers were included to assess their impact on feature extraction. The learning rate was varied between 0.0001 and 0.001 to find the optimal convergence speed, which is visualized in Table 1.

Table 1.
Hyperparameters for LSTM.
This refined exploration led to the final model configuration, which included a hidden size multiplier of 2, one LSTM layer, three dense layers, a dropout rate of 0.3, and LayerNorm for normalization. The final model, consisting of 10,766 parameters, demonstrated stability and efficiency over 200 epochs of training, showing optimal recall and loss balance. Table 2 summarizes these hyperparameters, presenting a model specifically optimized for the accurate and efficient detection of apneic events with minimized risk of overfitting.

Table 2.
Hyperparameters selected for model performance testing.
To mitigate overfitting and enhance model generalization, we applied multiple strategies. Dropout layers were included after each dense layer, with dropout rates tuned to 0.3, as this value achieved the optimal trade-off between performance and regularization. Additionally, we monitored the validation set performance across each hyperparameter sweep, ensuring consistency between training and validation results. The final model’s validation balanced accuracy was closely aligned with the test set results (validation recall of 81% vs. test recall of 76%), indicating effective generalization and minimal overfitting despite the model’s complexity.
5. Results and Validation
The final model’s performance was assessed on both the test and validation datasets, promising results that confirm its capacity to generalize well in detecting apnea events.
The model demonstrated strong performance on the test set, achieving an balanced accuracy of 79%, precision of 68%, recall of 76%, and F1 score 72%, are summarized in Table 3 and Figure 6. The confusion matrix indicates that while a small number of apnea events (true positives) were missed, the model correctly identified the majority, achieving a solid recall for apnea detection, as shown in Figure 6. For non-apnea events (label 0), performance was slightly lower in percentage terms, though the majority were classified correctly. This outcome highlights the model’s effectiveness in identifying apnea events while presenting an opportunity to enhance specificity to reduce false positives. Validation metrics, evaluated at the end of the last training epoch, were consistent with test set results. The model achieved an balanced accuracy of 80%, precision of 70%, recall of 81%, and F1 score 75% on the validation set, as summarized in Table 3 and Figure 7. These metrics are slightly elevated relative to the test set, which is a result that aligns with the use of the validation set during training for hyperparameter tuning. The close alignment between test and validation metrics indicates that the model generalizes effectively across different data splits and retains stable performance under varied conditions.

Table 3.
The results for LSTM model.
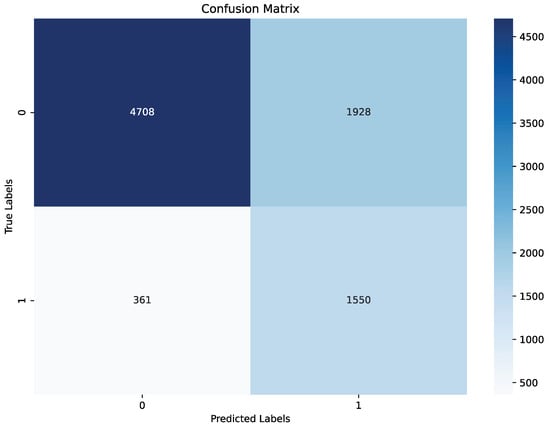
Figure 6.
Confusion matrix of the final model on test set.
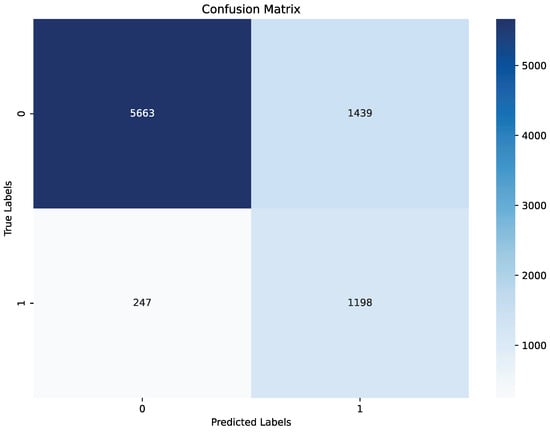
Figure 7.
Confusion matrix of the final model on validation set.
To further analyze model behavior, a feature was enabled to overlay SpO2 data for each patient in the test set with true and predicted labels shown in Figure 8, Figure 9 and Figure 10, providing insight into model detections at a sequence level. For instances showing substantial SpO2 variability, the model tended to predict more apnea events (class 1). It performed accurately when the signal’s variability correlated with apnea phases but occasionally overpredicted apnea during non-apnea phases that exhibited significant SpO2 fluctuations. For instance, in one patient’s SpO2 sequence, the model accurately identified apnea events amidst high-variability periods but showed a tendency to overpredict apnea during other variable sections.
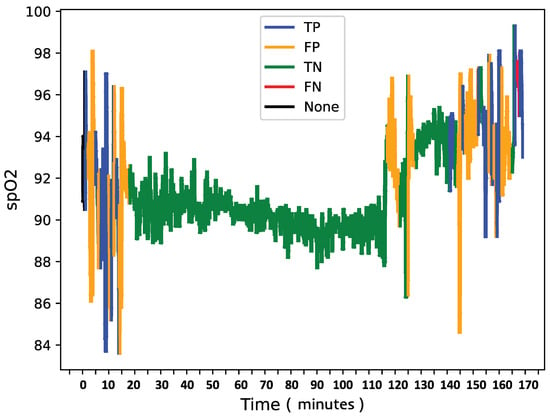
Figure 8.
SpO2 of patient X with confusion matrix labels.
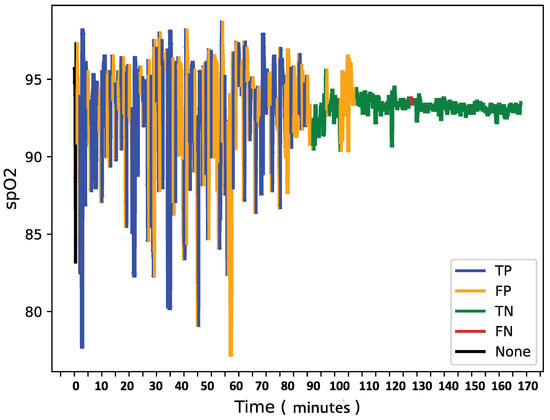
Figure 9.
SpO2 of patient Y with confusion matrix labels.
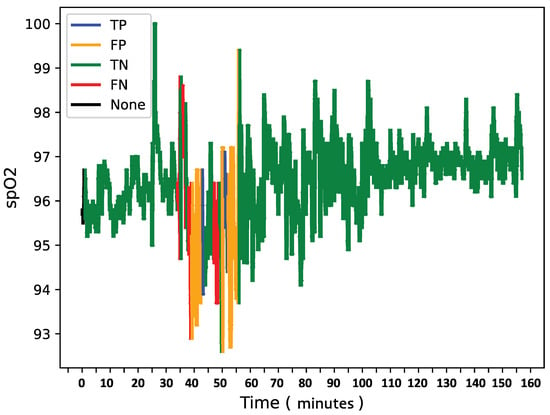
Figure 10.
SpO2 of patient Z with confusion matrix labels.
This pattern suggests that the model might associate high signal variability with apnea phases, which is a tendency that could lead to overdetections.
Overall, the final model demonstrates a robust balance between recall and loss, effectively detecting apnea events with generalizable performance across datasets. Addressing the model’s sensitivity to SpO2 signal variability may further refine its accuracy and reduce overdetection of apnea events during high-variability periods.
6. Discussion
This paper explores the integration of machine learning (ML) techniques in the field of sleep medicine, specifically focusing on the diagnosis and management of Obstructive Sleep Apnea (OSA). OSA’s complexity, variability, and the limitations of traditional diagnostic methods like PSG underscore the need for more efficient, precise, and accessible tools. The proposed digital platform leverages ML models to assist healthcare professionals by streamlining the diagnostic process and supporting the development of personalized treatment plans tailored to each patient’s unique needs. By analyzing data that include genetic, lifestyle, and environmental factors, these models can recommend targeted therapies, improving treatment outcomes and reducing adverse effects.
A significant challenge in developing such systems is the quality and labeling of data. The dataset provided included many epochs without labeled events, necessitating the assignment of default labels such as "no apnea event." However, this approach introduces uncertainty, as undetected or mislabeled apnea events may affect the model’s training. For instance, the labeling of adjacent epochs where apnea events were not confirmed by specialists may have led to misclassifications that impact the model’s learning ability. To improve the model’s accuracy, more robust validation processes for event labels, possibly involving domain experts, are essential. This would provide more reliable training data and improve model detections, contributing to the effectiveness of digital platforms in real-world clinical settings.
Feature engineering also played a critical role in enhancing the model’s performance. By incorporating features such as variance based on past data, the model gained better context for interpreting SpO2 and pulse rate fluctuations, which proved pivotal in detecting apnea events. Features like standard deviation from previous sequences helped improve recall by giving the model more context about the fluctuations in the signal, ultimately leading to better detections. The importance of feature selection and engineering will continue to evolve as more sophisticated data become available, including polysomnography results, genetic data, and real-time inputs from wearable devices.
Despite promising results, the current model still faces challenges, such as reducing false positives and minimizing missed apnea events (false negatives). The model’s tendency to predict apnea events during periods of high variability in the SpO2 signal indicates the need for refined feature engineering and the exploration of more advanced model architectures. Moreover, optimizing hyperparameters and incorporating additional features such as body position could improve the model’s detective accuracy.
In summary, the integration of machine learning into sleep medicine presents a promising advancement in the diagnosis and management of OSA. While the current model shows potential, further efforts will be focused on improving data quality, refining feature engineering, and enhancing model accuracy through collaboration with medical professionals. The future of sleep medicine relies on the seamless integration of these technologies to provide personalized, efficient, and accurate treatments, ultimately leading to better health outcomes for patients with OSA. The continuous refinement and validation of machine learning models, combined with the development of more sophisticated algorithms, will further enhance diagnostic accuracy and support personalized treatment plans in clinical settings.
To address the model’s tendency to overpredict apnea events during periods of high SpO2 variability, future work will explore additional feature engineering techniques, such as incorporating body position and sleep stage as input features. Additionally, integrating an attention mechanism within the LSTM layer could help the model selectively focus on critical parts of the SpO2 signal, potentially reducing false positives by improving the model’s focus on apnea-relevant patterns. These modifications could further refine the model’s accuracy, making it even more valuable in clinical settings.
7. Conclusions
The machine learning model developed in this study demonstrates promising performance in detecting apnea events, with a recall rate of 76% on the test set, indicating high sensitivity in identifying potential apnea episodes. This makes the model a valuable diagnostic tool that helps clinicians identify events requiring further analysis. Although the model tends to detect more apnea events than actually occur (resulting in false positives), this is considered beneficial in a clinical context, since false negatives (missed apnea episodes) pose significant health risks. By reducing the number of false negatives, the model ensures that patients with potential apnea are flagged for additional monitoring or intervention.
The integration of the developed model is envisioned through a web application designed as a platform for healthcare professionals. This platform would provide doctors with an enhanced diagnostic tool, enabling more accurate and reliable disease detection by analyzing input signals using the generated model.
The framework, based on LSTM analysis of polysomnography (PSG) and oximetry data, enhances personalized apnea detection. The LSTM network design, which includes a bidirectional layer with dropout functionality, effectively captures complex temporal patterns, achieving an overall balanced accuracy of 79%, precision of 68%, and recall of 76% on the test set. These results surpass traditional diagnostic methods, offering a balanced approach between recall and specificity.
The model utilizes Long Short-Term Memory (LSTM) networks in combination with dense layers, optimized through hyperparameter tuning using the Weights and Biases (WandB) tool to address the imbalance between apneic and non-apneic events. While the model achieved promising results, it faced challenges related to false positives with confusion matrices showing a trade-off between recall and loss. Priority was given to recall, as the accurate detection of apneic events is critical in the clinical context.
Regarding interpretation, feature engineering played a crucial role in optimizing model performance. By extracting key statistical features such as standard deviation, amplitude, and gradient and applying a sliding window approach for sequence generation, the model’s sensitivity to apnea events was significantly improved. Visualizations such as parallel coordinates plots helped assess the impact of hyperparameters, while configuration management using the Hydra tool enabled systematic experimentation and the efficient handling of complex models.
Hyperparameter optimization, which explored over 500,000 combinations, further refined the model to prevent overfitting and ensure robust performance on diverse patient data.
This framework has the potential to transform clinical practice by enabling early detection, personalized treatment planning, and improved management of patients with OSA. Future improvements will focus on further fine-tuning model parameters, reducing false positives, improving feature extraction, and incorporating real-time data from wearable devices and patient demographics to further enhance accuracy. With continued development, this machine learning approach promises significant advancements in the diagnosis and management of OSA, supporting more effective and data-driven solutions in sleep medicine.
Author Contributions
Conceptualization, K.Z. and Y.S.; methodology, Y.S. and K.Z.; software, Y.S. and K.Z.; validation, P.Š., T.P., I.P.D., R.P. and Z.Đ.; formal analysis, Y.S.; investigation, K.Z. and K.Z.; resources, I.P.D., R.P., P.Š. and Z.Đ.; data curation, K.Z. and Y.S.; writing—original draft preparation, K.Z., Y.S., T.P. and P.Š.; writing—review and editing, K.Z., Y.S., T.P. and P.Š.; visualization, K.Z. and Y.S.; supervision, P.Š., T.P., I.P.D., R.P. and Z.Đ.; project administration, P.Š., T.P., I.P.D., R.P. and Z.Đ. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study and approved by the Institutional Review Board (Ethical Committee of the School of Medicine, University of Split) under protocol number 003-08/23-03/0015, dated 17 October 2023.
Informed Consent Statement
Patient consent was waived due to minimal risk to participants as stated in the ethical committee’s review. The data collection and privacy safeguards are in compliance with GDPR (General Data Protection Regulation 2016/679), ensuring anonymity and confidentiality of all participant data.
Data Availability Statement
The data supporting the reported results are not publicly available due to privacy and ethical restrictions. Access to the data is restricted to authorized personnel only.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A. Machine Learning Model—Long Short-Term Memory (LSTM)
The choice of architecture, specifically one LSTM layer followed by three dense layers, was made after evaluating multiple configurations during preliminary testing. Initial experiments with additional LSTM layers, during which data from the initial 16 patients were exclusively used in the experimentation phase, resulted in overfitting, as indicated by the deterioration of performance on the validation dataset. Using a single LSTM layer preserved temporal dependencies without adding excessive model complexity. The dense layers allowed for progressive feature refinement, enabling the model to capture both high-level and granular features of the SpO2 signal. This configuration demonstrated the best balance between recall and generalizability, which are critical for detecting apnea events in diverse patient data.
Long Short-Term Memory (LSTM) networks are a specialized type of recurrent neural network (RNN) designed to overcome issues such as vanishing and exploding gradients that typically affect traditional RNNs. LSTM networks are distinguished by their ability to store information over extended periods thanks to three main gates: the input gate, forget gate, and output gate. These gates, along with the cell state, allow the LSTM to retain and manage memory over time, preventing the degradation of data that happens in traditional RNNs [26], as visualized in Figure A1. This feature makes LSTM particularly well suited for tasks that involve learning long-term dependencies in sequential data [27].
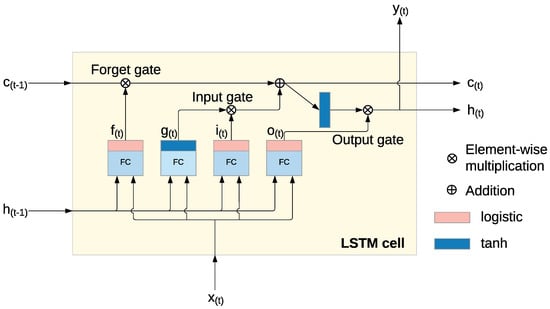
Figure A1.
Long Short-Term Memory (LSTM) cell [28].
Within each LSTM cell, a series of operations control the flow of information through the network, enabling the model to selectively keep or discard information. The following key operations occur [28]:
- The input gate (A1) controls how the current input and the previous hidden state contribute to the memory.
- The forget gate (A2) decides which parts of the previous cell state should be discarded.
- The output gate (A3) determines which elements of the cell state will be used for the output and the next hidden state.
- The cell state (A5) is updated by combining the previous state with the newly processed information, enabling the model to retain the relevant data over time.
The operations within each LSTM cell are governed by the following equations:
where the following apply:
- represents the sigmoid activation function, which outputs values between 0 and 1.
- W represents the weight matrices for both the input and the previous hidden state .
- b is the bias term for each gate.
- (A4) is used to update the cell state using the tanh function, which outputs values between −1 and 1.
This architecture enables the LSTM network to maintain and adjust memory across time steps, making it especially effective for sequential data analysis where the relationships between data points span long periods. Unlike traditional RNNs, which can forget crucial information over time, the LSTM can store and manipulate relevant data, allowing it to make accurate detections for tasks that depend on long-term memory [28].
The choice of architecture, specifically one LSTM layer followed by three dense layers, was made after evaluating multiple configurations during preliminary testing. Initial experiments with additional LSTM layers resulted in overfitting, as indicated by deteriorating validation performance. Using one LSTM layer preserved temporal dependencies without adding excessive model complexity. The dense layers allowed for progressive feature refinement, enabling the model to capture both high-level and granular features of the SpO2 signal. This configuration demonstrated the best balance between recall and generalizability, which are critical for detecting apnea events in diverse patient data. After testing various architectural configurations, we used data from 16 patients to explore the model architecture and feature engineering. We found that a single LSTM layer followed by three dense layers provided the best balance of accuracy, recall, and generalizability. Adding more LSTM layers initially showed improvements in learning but led to overfitting, as the model’s validation performance began to degrade with additional complexity. The use of three dense layers allowed the model to progressively refine features learned by the LSTM layer, enhancing its ability to distinguish apnea events from non-apnea events. This configuration was selected as it minimized overfitting while preserving the model’s capability to capture sequential dependencies in the input data, which is essential for apnea detection.
References
- Sutherland, K.; Vanderveken, O.M.; Tsuda, H.; Marklund, M.; Gagnadoux, F.; Kushida, C.A.; Cistulli, P.A. Oral appliance treatment for obstructive sleep apnea: An update. J. Clin. Sleep Med. 2014, 10, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Abrishami, A.; Khajehdehi, A.; Chung, F. A systematic review of screening questionnaires for obstructive sleep apnea. Can. J. Anesth. 2010, 57, 423. [Google Scholar] [CrossRef] [PubMed]
- Rundo, J.V.; Downey, R. Chapter 25—Polysomnography. In Clinical Neurophysiology: Basis and Technical Aspects; Levin, K.H., Chauvel, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 160, pp. 381–392. [Google Scholar] [CrossRef]
- Van de Perck, E.; Kazemeini, E.; Van den Bossche, K.; Willemen, M.; Verbraecken, J.; Vanderveken, O.; Op de Beeck, S. The effect of CPAP on the upper airway and ventilatory flow in patients with obstructive sleep apnea. Respir. Res. 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Yunpeng, L.; Di, H.; Junpeng, B.; Yong, Q. Multi-step ahead time series forecasting for different data patterns based on LSTM recurrent neural network. In Proceedings of the 2017 14th Web Information Systems and Applications Conference (WISA), Liuzhou, China, 11–12 November 2017; pp. 305–310. [Google Scholar]
- Yang, H.; Lu, S.; Yang, L. Clinical prediction models for the early diagnosis of obstructive sleep apnea in stroke patients: A systematic review. Syst. Rev. 2024, 13. [Google Scholar] [CrossRef]
- Zovko, K.; Šerić, L.; Perković, T.; Belani, H.; Šolić, P. IoT and health monitoring wearable devices as enabling technologies for sustainable enhancement of life quality in smart environments. J. Clean. Prod. 2023, 413, 137506. [Google Scholar] [CrossRef]
- Gutiérrez-Tobal, G.C.; Álvarez, D.; Vaquerizo-Villar, F.; Crespo, A.; Kheirandish-Gozal, L.; Gozal, D.; del Campo, F.; Hornero, R. Ensemble-learning regression to estimate sleep apnea severity using at-home oximetry in adults. Appl. Soft Comput. 2021, 111, 107827. [Google Scholar] [CrossRef]
- Dutta, R.; Delaney, G.; Toson, B.; Jordan, A.; White, D.; Wellman, A.; Eckert, D. A Novel Model to Estimate Key OSA Endotypes from Standard Polysomnography and Clinical Data and Their Contribution to OSA Severity. Ann. Am. Thorac. Soc. 2020, 18. [Google Scholar] [CrossRef]
- Vaquerizo-Villar, F.; Álvarez, D.; Kheirandish-Gozal, L.; Gutiérrez-Tobal, G.C.; Barroso-García, V.; Santamaría-Vázquez, E.; Del Campo, F.; Gozal, D.; Hornero, R. A convolutional neural network architecture to enhance oximetry ability to diagnose pediatric obstructive sleep apnea. IEEE J. Biomed. Health Inform. 2021, 25, 2906–2916. [Google Scholar] [CrossRef] [PubMed]
- ElMoaqet, H.; Eid, M.; Glos, M.; Ryalat, M.; Penzel, T. Deep Recurrent Neural Networks for Automatic Detection of Sleep Apnea From Single Channel Respiration Signals. Sensors 2020, 20, 5037. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Yeh, C.Y.; Lee, C.T.; Lin, C.C. A Sleep Apnea Detection System Based on a One-Dimensional Deep Convolution Neural Network Model Using Single-Lead Electrocardiogram. Sensors 2020, 20, 4157. [Google Scholar] [CrossRef]
- Behar, J.A.; Palmius, N.; Li, Q.; Garbuio, S.; Rizzatti, F.P.; Bittencourt, L.; Tufik, S.; Clifford, G.D. Feasibility of Single Channel Oximetry for Mass Screening of Obstructive Sleep Apnea. EClinicalMedicine 2019, 11, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Deviaene, M.; Testelmans, D.; Borzée, P.; Buyse, B.; Huffel, S.V.; Varon, C. Feature Selection Algorithm based on Random Forest applied to Sleep Apnea Detection. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2580–2583. [Google Scholar] [CrossRef]
- Sharma, P.; Jalali, A.; Majmudar, M.D.; Rajput, K.S.; Selvaraj, N. Deep-Learning Based Sleep Apnea Detection Using SpO2 and Pulse Rate. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Glasgow, UK, 11–15 July 2022. [Google Scholar] [CrossRef]
- Nasir, N.; Barneih, F.; Alshaltone, O.; Al-Shabi, M.; Bonny, T.; Shammaa, A.A. Sleep Apnea Detection Using Xception and Residual Network. In Proceedings of the In Smart Biomedical and Physiological Sensor Technology XX, Orlando, FL, USA, 1 May 2023. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.W.; Hwang, S.H. Diagnostic Value of Smartphone in Obstructive Sleep Apnea Syndrome: A Systematic Review and Meta-Analysis. PLoS ONE 2022, 17, e0268585. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chu, Y.; Ryu, J.; Park, Y.J.; Yang, S.; Koh, S.O. Artificial Intelligence for Detection of Cardiovascular-Related Diseases From Wearable Devices: A Systematic Review and Meta-Analysis. Yonsei Med. J. 2022, 63, S93–S107. [Google Scholar] [CrossRef]
- Sadoughi, A.; Shamsollahi, M.B.; Fatemizadeh, E. Automatic Detection of Respiratory Events During Sleep from Polysomnography Data Using Layered Hidden Markov Model. Physiol. Meas. 2022, 43. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Jané, R. Detecting Obstructive Apnea Episodes Using Dynamic Bayesian Networks and ECG-based Time-Series. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022. [Google Scholar] [CrossRef]
- Veugen, C.C.; Dieleman, E.; Hardeman, J.A.; Stokroos, R.J.; Copper, M.P. Upper airway stimulation in patients with obstructive sleep apnea: Long-term surgical success, respiratory outcomes, and patient experience. Int. Arch. Otorhinolaryngol. 2023, 27, e43–e49. [Google Scholar] [CrossRef] [PubMed]
- Aboussouan, L.S.; Bhat, A.; Coy, T.; Kominsky, A. Treatments for obstructive sleep apnea: CPAP and beyond. Clevel. Clin. J. Med. 2023, 90, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, M.A. Advancements in Home-Based Devices for Detecting Obstructive Sleep Apnea: A Comprehensive Study. Sensors 2023, 23, 9512. [Google Scholar] [CrossRef] [PubMed]
- Waseem, R.; Wong, J.; Ryan, C.M.; Chung, F. Predictive Performance of Oximetry to Detect Sleep Apnea in Patients Taking Opioids. Anesth. Analg. 2021, 133, 500–506. [Google Scholar] [CrossRef]
- Koninklijke Philips, N.V. Alice 6 LDx Diagnostic Sleep System. Available online: https://www.philips.ie/healthcare/product/HC1063315/alice-6-ldx-diagnostic-sleep-system (accessed on 17 November 2024).
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) network. Phys. D Nonlinear Phenom. 2020, 404, 132306. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, M.; Zhu, X.; Zhao, L. Attention-based LSTM for aspect-level sentiment classification. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 606–615. [Google Scholar]
- Rodić, L.D.; Županović, T.; Perković, T.; Šolić, P.; Rodrigues, J.J. Machine learning and soil humidity sensing: Signal strength approach. ACM Trans. Internet Technol. (TOIT) 2021, 22, 1–21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).