Abstract
Segmentation of nuclei in histology images is key in analyzing and quantifying morphology changes of nuclei features and tissue structures. Conventional diagnosis, segmenting, and detection methods have relied heavily on the manual-visual inspection of histology images. These methods are only effective on clearly visible cancerous lesions on histology images thus limited in their performance due to the complexity of tissue structures in histology images. Hence, early detection of breast cancer is key for treatment and profits from Computer-Aided-Diagnostic (CAD) systems introduced to efficiently and automatically segment and detect nuclei cells in pathology. This paper proposes, an automatic watershed segmentation method of cancerous lesions in unsupervised human breast histology images. Firstly, this approach pre-processes data through various augmentation methods to increase the size of dataset images, then a stain normalization technique is applied to these augmented images to isolate nuclei features from tissue structures. Secondly, data enhancement techniques namely; erosion, dilation, and distance transform are used to highlight foreground and background pixels while removing unwanted regions from the highlighted nuclei objects on the image. Consequently, the connected components method groups these highlighted pixel components with similar intensity values and, assigns them to their relevant labeled component binary mask. Once all binary masked groups have been determined, a deep-learning recurrent neural network from the Keras architecture uses this information to automatically segment nuclei objects with cancerous lesions and their edges on the image via watershed filling. This segmentation method is evaluated on an unsupervised, augmented human breast cancer histology dataset of 11,151 images. This proposed method produced a significant evaluation result of F1-accuracy score.
1. Introduction
Early diagnosis of breast cancer (BC) relies heavily on how cancerous lesions spread on nuclei and tissue glands and thus offers a need for a prognosis. Traditionally, the Bloom-Richardson grading system is used by clinicians to determine the grade of how tumor cells are morphing into normal nuclei the degree of morphing, and how the tumors are increasing in number [1]. The grading system is done by examining cells through a microscope, which is limited in observing other important features. With this deficiency, automatic-aided imaging is preferred and assists in analyzing nuclei cells in BC histology images.
Research authors in [2] propose an automatic segmentation method for BC via pre-processing images through median filtering, rotation, and identification of pectorals via highlighting the region of interest (ROI). The watershed algorithm is then used to divide the image into foreground and background via sobel filters, thresholding, and distance maps to remove uninterested regions of the image. Overlapping of objects is observed with this method and authors in [3] reiterate the need for early breast cancer detection through their proposed method. The proposed method uses image processing and watershed algorithms to segment breast mammogram images through density estimation, contour extraction, and region segmentation to assist BC detection.
In [4], image pre-processing is handled via median filtering to remove noise, morphology operations using gray scaling, active contours to highlight and accentuate foreground and background regions, and watershed segmentation to merge different image regions and their boundaries. Image pre-processing based on color, texture, and gradient intensity through canny, Sobel, and blob edge detectors is presented by the method in [5]. Watershed segmentation separates overlapping/touching objects by dilation and erosion morphology operations, and cancerous tumors extracted from the remaining object features. A CAD system proposed by [6] pre-processes ultra-sound breast images to improve image brightness, and image segmentation through the marker-controlled watershed to visualize topographically interested lesion regions and their edge boundaries on the image.
Segmentation of cells can also be used to locate tumor regions of interest (ROI) within the image. However, nuclei segmentation has several challenges namely; improper image pre-processing, the complexity of tissue structures, overlapping cells, uneven color distribution caused by the staining procedure, and scanning equipment irregularities [7]. Conventionally, thresholding and morphology operations have been the most widely used techniques for segmenting medical images. For instance, ref. [8] proposes an adaptive deconvolution algorithm to color normalize images in a dataset, use morphology operations to pre-process the images, threshold them, and further post-process these images to detect edges. Consequently, ref. [9] proposes an automated segmentation technique that uses gray scaling, median filtering, and bottom-top hat filters as pre-processing steps to enhance image contrast. Thresholding is used to identify regions of interest, while post-processing morphology techniques namely; dilation, area opening, and filling in holes are used to improve final segmentation results.
Further, a novel method proposed by [10] uses wavelet transform decomposition to highlight and enhance regions of interest and edge boundaries on mammogram images. Thresholding and binary morphological operations namely; dilation and erosion are used to identify the breast ROI, masking the ROI to isolate it from unwanted pectoral muscle regions. In [11], image pre-processing is handled through normalization, segmentation through color deconvolution for nuclei enhancement, and data augmentation to increase dataset size. A binary threshold was used to aid in detecting nuclei edges in the images.
Watershed segmentation is a common remedy for overlapping objects on medical images resulting from data preparation and pre-processing. In [12], a convolution neural network (CNN) model is used to identify and label nuclei cells into classes namely; nuclei, edges, and image background to form binary masks. The binary mask generated by the CNN model separates overlapping nuclei and non-overlapping objects, the Euclidean distance transform is used to segment edges and nuclei on the image.
Watershed segmentation is used to segment nuclei on glands. This method uses markers to diminish and accentuate images ROI at specific locations as either foreground or background. A nuclei segmentation method proposed by [13] uses stain separation to obtain foreground pixels of an image. After the staining separation procedure, the watershed segmentation method isolates overlapping nuclei based on various morphology changes observed on the histology images. These morphology changes are key to highlighting the size and spread of BC in the nucleus.
In [14], a fully convolution-deep-neural architecture is utilized to segment nuclei cells. This technique uses image color normalization and standardization to highlight and balance the brightness, of nuclei features and tissue structure regions respectively. The Link-Net architecture is used to encode and decode images for nuclei segmentation.
Hematoxylin and Eosin (H&E)image staining procedure presented by [15], normalizes and highlights the nucleus and tissue regions respectively. Image contrast is adjusted to highlight and extract the nucleus from the image background and other uninterested regions. Watershed segmentation is done on the extracted nucleus region to remove, overlapping and unrelated objects not captured by the previous contrast adjustment step. A novel architecture by [16] excludes intersecting pixels of overlapped nuclei objects to form a new foreground image. Morphological operations are applied to the new image to ascertain edges, nuclei are labeled using markers, and the final result is segmented via watershed.
Content-based image retrieval algorithm proposed by [17], pre-processes images by normalization techniques; histogram equalization, and object size kernel to identify nuclei objects. Thresholding and iterative morphology operations remove overlapping objects highlighting the nucleus centers and edges. The final image is a segmented region combining the nuclei, nuclei centers, and their edges/boundaries. While watershed segmentation is efficient computationally, it is devoid of over-segmentation when used on histology images.
A technique proposed by [18] introduces improvements to the watershed segmentation method to deal with the over-segmentation of nuclei cells in histology images. In their pre-processing stage, Gaussian filters were used to remove noise and enhancement of nuclei cells. Morphological operations namely; opening and closing were used to highlight and accentuate both background and foreground pixels while maintaining their edges on the image. The image is further enhanced via OTSU thresholding and dilation morphology to outline the foreground and avoid overlapping nuclei objects on the image. Lastly, the foreground and background markers obtained from the previous step are segmented via the watershed segmentation method to obtain nuclei edges.
Another proposed method to deal with over-segmentation is by [19], which initially separates H&E stains on the histology image via color de-convolution methods. Morphological operations namely; OTSU thresholding, opening, and closing to distinguish the foreground from the background and highlight the nuclei objects on the image. Distance transform is further used to identify touching nuclei objects and watershed segmentation to separate the touching nuclei objects while maintaining their edge boundaries. In [20], a nuclei segmentation method is used that pre-processes images via color unmixing to separate H&E stains on images. Morphology operations namely; opening to remove unconnected foreground, and background pixels, while closing to deal with irregular shapes of nuclei objects. Foreground and background markers are transformed from morphology-operated images which then are used to identify nuclei objects. Lastly, watershed segmentation was used to segment and maintain nuclei and their edges.
In [21], ROIs are manually selected, and Gaussian filters are used to remove noise while enhancing image features. Consequently, the K-means clustering method was used to segment and identify the ROI. After ROI identification, image thresholding was performed to obtain nuclei regions, and morphological operations namely; opening and closing were used to remove unconnected pixel objects. Lastly, the watershed segmentation method was used to segment overlapping nuclei objects and highlight their edges. A global-local thresholding approach proposed by [22], distinguishes the foreground from the background on an image. This approach introduces overlapping nuclei objects from the extracted foreground pixels leading to over-segmentation. Binary masks were created from the overlapping objects for the foreground and background. A seed-controlled watershed segmentation technique was utilized on these masks to segment nuclei objects, highlight their edges, and remove unrelated parts.
Our proposed segmentation method is similar to [13], which assists automatic segmentation of cancerous lesions in unsupervised BC histology images. Image pre-processing is handled through data augmentation, a staining normalization procedure, and morphology operations to isolate nuclei and distinguish foreground from background pixels on the image Region of Interest (ROI). The connected components analysis groups pixels with similar intensity values resulting in labeled binary masks. A recurrent deep-learning neural network uses these binary masks to separate overlapping nuclei and non-overlapping objects. Additionally, watershed filling within the recurrent neural network (RNN) is used to segment and maintain nuclei and their edges automatically thus making the entire process efficient computation-wise. Figure 1, shows a flow diagram of the proposed automatic watershed segmentation method.
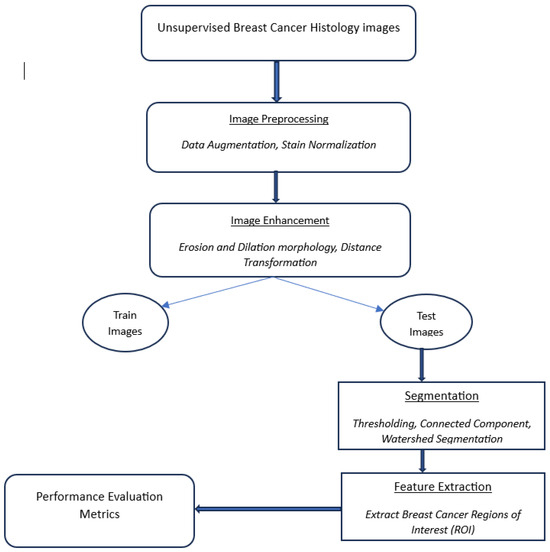
Figure 1.
Breast cancer histology images.
2. Methods and Techniques
This proposed approach achieves pre-processing and segmentation of unsupervised BC histology images through the following steps:
2.1. Dataset
This study uses 24 unsupervised BC histology images from the publicly available Kaggle dataset repository.
2.1.1. Dataset Preparation/Pre-Processing
Data is key for any neural network model to learn and deduce useful information from the data provided [23]. The amount of data required by deep learning model networks is huge specifically, medical imaging data which is limited or difficult to obtain from publicly available datasets. Therefore, the data augmentation method has been mostly preferred to deal with data scarcity.
2.1.2. Data Augmentation
Data augmentation entails, artificially transforming existing images in a dataset by rotation, scaling, cropping, flipping, height, and width shifts to create more mages. Augmentation is more utilized because of its effectiveness in training different deep learning models [24]. It assists in solving data scarcity by increasing the size and variety of images in datasets useful for training models, without collecting new samples [25]. With the increase in dataset size, it is necessary to maintain image quality [26].
In this study, data augmentation methods namely; rotation, scaling, height, and width shifts are used to increase the dataset size from 24 unsupervised BC histology images to 11,151 images.
2.1.3. Data Stain Normalization
During the preparation and pre-processing stages, images undergo various distortions and inconsistencies. These are attributed to the H&E staining procedure used to magnify minute nuclei features, tissue structures, and image transformations resulting from data augmentation. Laboratory slide preparation, examination, analysis, and digitalization of scanning samples are other factors that lead to image variations [27]. These factors, negatively impact the training and testing of neural networks. Hence, a stain normalization technique is needed to remove color irregularities in medical images and improve the model’s efficacy. In this proposed study, the Macenko et al. [28] stain normalization technique is used for BC images to separate nuclei features from tissue structures. Images in the dataset images are first converted from the BGR to RGB color space to enable smooth stain normalization.
Macenko stain normalization: is used to prepare tissue slides. Image colors are converted to their optical density (OD) equivalent via a simple logarithmic transformation shown below,
with I as the RGB color vector and individual components normalized to [0, 1].
A value , is used as a threshold value to remove data with higher OD intensity.
Single value decomposition (SVD) is applied to optical density tuples to create a plane. The plane corresponds to the largest singular values. OD-transformed pixels are then projected onto the plane to determine the angle at each point related to the first SVD direction. The color space transformation is applied to the original BC histology image. An image histogram is stretched such that the range covers the lower of the data.
Minimum and maximum vectors are calculated and projected back to the OD space. The Hematoxylin stain corresponds to the minimum vector while the Eosin stain is the maximum vector. Stain concentrations are determined to form a matrix representing RGB channels and OD intensities respectively. This study has values of and set at 1 and 0.15 respectively. Figure 2 shows original, augmented, normalized H&E, normalized H, and normalized E breast cancer histology images respectively. After this stain normalization process, our proposed approach focuses on the normalized H image (image with only nuclei objects), having isolated from the greater normalized H&E image set.
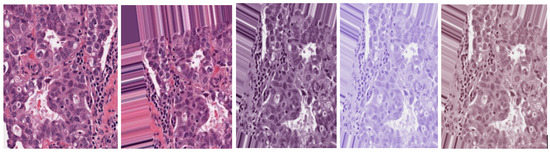
Figure 2.
Original H&E image, Augmented H&E image, Normalized H&E image, Normalized H image, Normalized E image respectively.
2.2. Image Enhancement
Enhancement of dataset images is by improving their brightness, contrast, and scaling to compensate for the non-uniformity resulting from image illumination. This proposed study handles this step through thresholding, morphology operations, and distance transform.
2.2.1. Thresholding
OTSU thresholding is used to capture the outline of the BC ROI on the (normalized H) histology image/images and is shown by Figure 3.
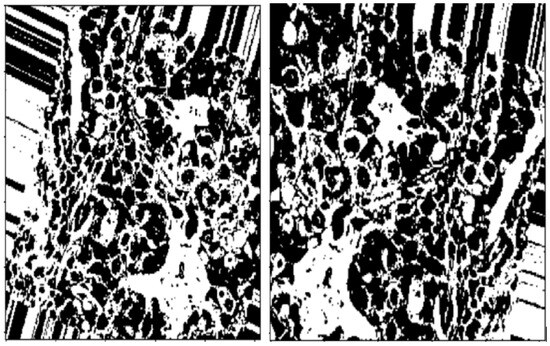
Figure 3.
Images after OTSU thresholding.
2.2.2. Morphology Operations
These include dilation and erosion operations to remove noise, remove overlapping edges, and extract certain regions on BC (normalized H) histology images. Opening and closing operations are used to distinguish between the background and the foreground in an image. These regions are distinguished via diminishing and accentuating image pixels and edges. These operations also highlight the unknown area between the background and the foreground. Figure 4, Figure 5 and Figure 6, show images resulting from morphology operations.
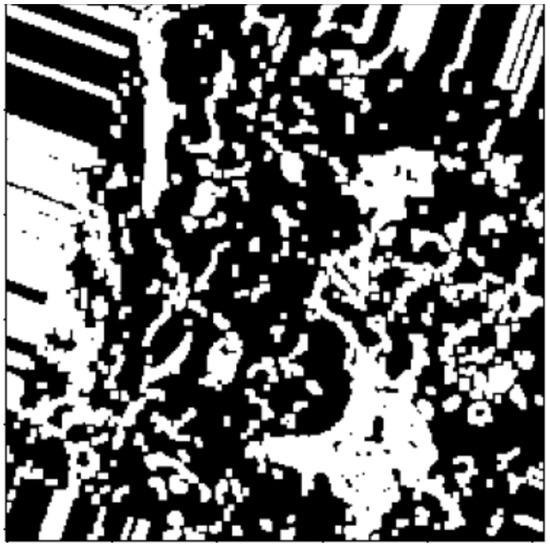
Figure 4.
Image after noise removal via thresholding.
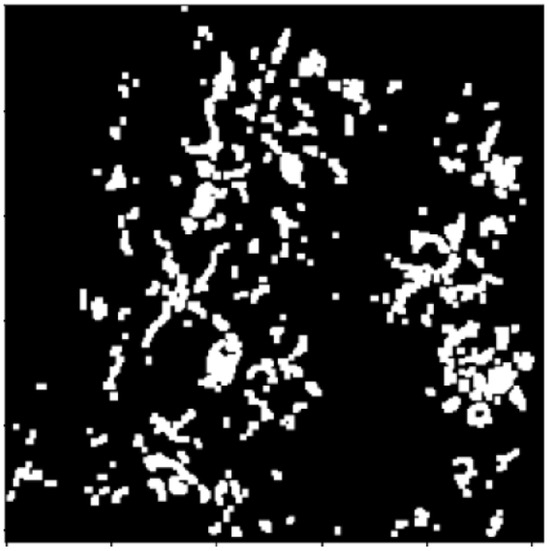
Figure 5.
Enter Image after clearing borders via opening morphology operation.
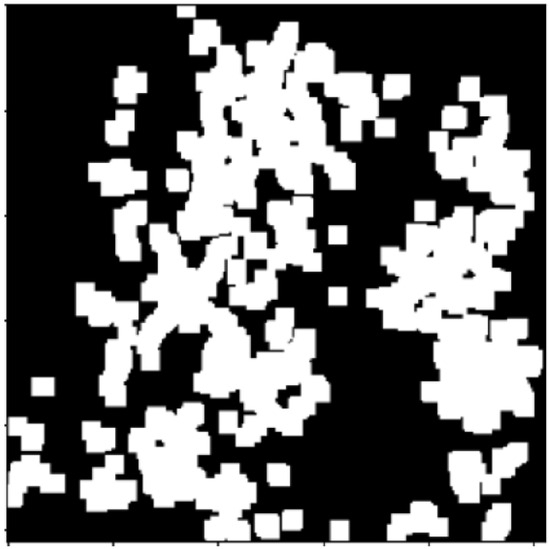
Figure 6.
Sure background image after dilation morphology.
2.2.3. Distance Transform
This method isolates nuclei objects on (normalized H) image and, finds the sure foreground by removing the remaining uninterested ROIs. After the distance transformation step, further thresholding is done to highlight and emphasize the sure foreground objects and sure background on the BC (normalized H) histology image. Figure 7 and Figure 8 show image results from distance transformation and thresholding respectively.
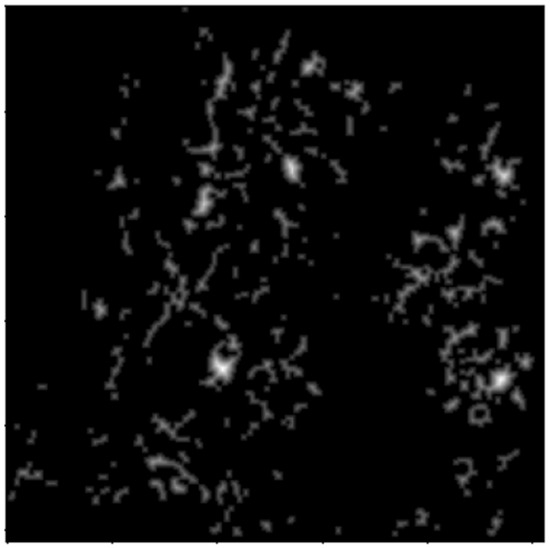
Figure 7.
Distance transform image.
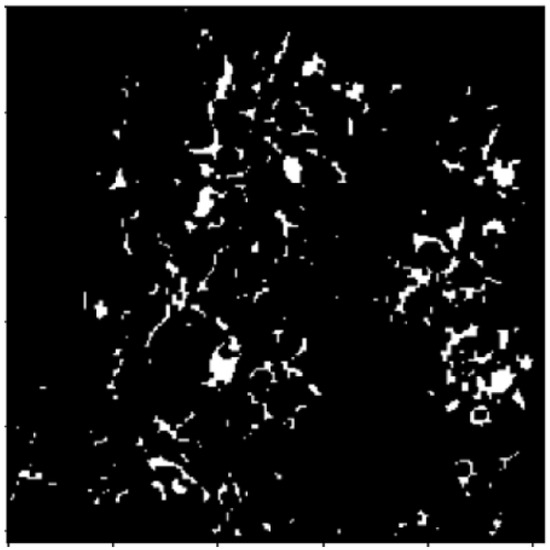
Figure 8.
Thresholding after distance transformation.
2.3. Segmentation
2.3.1. Connected Components
This method groups pixels with their, neighborhoods of similar intensity values into either darker (background) connected components or, brighter (foreground) connected components. It uses binary masks to label sure foreground and background regions with positive numbers, and the unknown region with 0 on an image. The connected component method extracts the ROI of a (normalized H) image as shown in Figure 9.
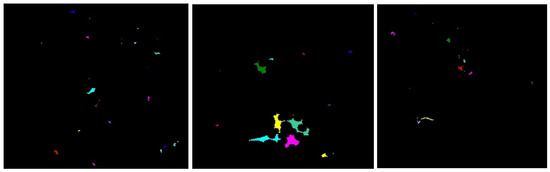
Figure 9.
Connected component images.
2.3.2. Watershed Segmentation
Once all pixel groups have been determined and labeled according to their assigned components, the watershed filling is used to segment and maintain nuclei objects and their edges by color-coding(mask) to distinguish the different regions namely; background, foreground, and edge boundaries. Finally, we have an RGB image with color-coded labels painted over the image. Figure 10 shows image results of the proposed watershed segmentation method.
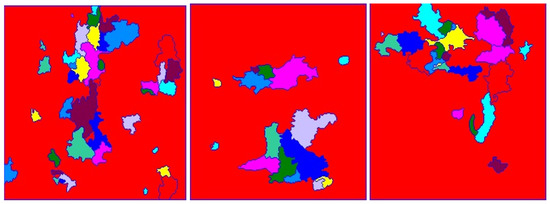
Figure 10.
Unsupervised BC (normalized H) histology images result from this proposed watershed segmentation method.
The impact of automatic watershed segmentation on unsupervised (BC normalized H) histology images was evaluated using a deep-learning recurrent neural network model. The neural network architecture consisted of eight layers: one input dense layer, three hidden dense layers, one output dense layer, and three dropout layers. The cross-entropy was minimized using categorical cross-entropy with the Adam optimizer, learning rate of 0.001, batch size of 32, and 30 epochs. We settled on the above hyper-parameters from iterative model experiments.
3. Results and Discussion
The experimental results of this study are based on an analysis of the entire automatic watershed segmentation process. Experiments were carried out, on 11,151 unsupervised BC (normalized H) histology images split into 8079 training and 2694 testing set images. Processing of the unsupervised BC dataset to aid data analysis has been discussed in previous sections of this work.
Figure 1 shows an overview of the processing stages of the proposed segmentation method namely; pre-processing, image enhancement, Segmentation, and feature extraction. Additionally, the performance evaluation of the proposed approach is based on its automatic ability to segment cancerous lesions on (normalized H) breast histology images shown by Figure 10.
Figure 11 illustrates visual examples of images before and after the application of our proposed segmentation method. These image results demonstrate the effectiveness of the proposed method on other human glands histology images gathered from the publicly available Warwick QU Dataset in the Kaggle dataset repository. Therefore, this proposed method is efficient in segmenting and maintaining nuclei objects and their edges on histology images, with this clearly illustrated from the various image results of other human glands namely; kidney and colon H&E images among others provided in the publicly available Warwick dataset. Breast cancerous lesions are detected via their spread on tissues signifying nuclei objects irregularity on histology images. The H&E stain separation and normalization technique aids the dividing of image pixels into hematoxylin (blue color) denoting nuclei features, whereas eosin (pink color) denotes tissue structures. Therefore, any other breast lesion on either lobules or ducts will be detected by separating nuclei objects from non-nuclei objects in histology images, thus the applicability of the proposed approach.
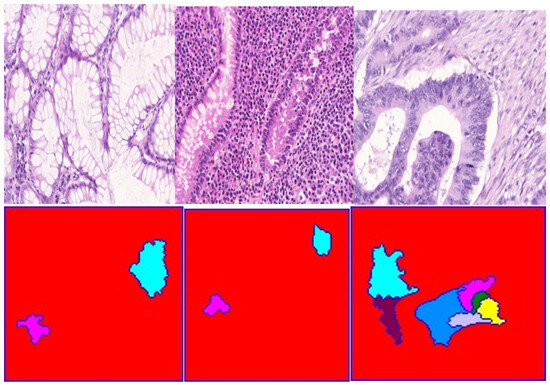
Figure 11.
First Row: Original histology images of different human glands provided by the Warwick QU Dataset in the Kaggle dataset repository. Second Row: Resultant images after segmentation application using the proposed watershed method.
Table 1 shows the performance improvement caused by introducing the proposed watershed segmentation method.

Table 1.
Performance evaluation results before and after applying watershed segmentation method.
Our proposed technique achieved significant results in identifying and highlighting BC lesions throughout the unsupervised (normalized H) histology image dataset. These results are attributed to various reasons namely; dataset augmentation that increases the dataset size, and stain normalization reducing image color inconsistencies. Table 2 shows the comparison between, our proposed approach and other watershed-related techniques that focus on segmenting nuclei objects on human histology images.

Table 2.
Comparison Table of the proposed method to other watershed-related implementations that segment nuclei objects on BC histology images.
Figure 12 and Figure 13 present training loss and accuracy graph curves and validation loss and accuracy graph curves of the proposed method.
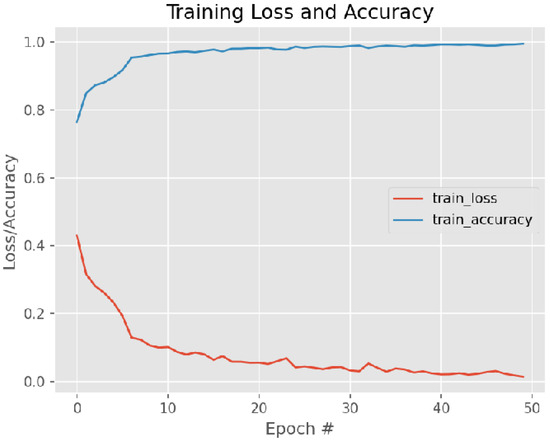
Figure 12.
Training Loss and Accuracy graph curves.
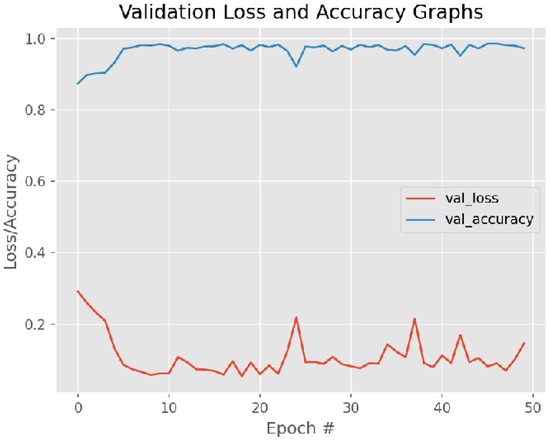
Figure 13.
Validation Loss and Accuracy graph curve.
Limitations
Over-segmentation is the one main drawback of the watershed technique. Most existing watershed segmentation techniques, use markers to label ROIs in human histology images as discussed in the literature review section, instead of connected nuclei objects(masks). Also, most existing nuclei segmentation methods tend to segment nuclei objects on the entire normalized image which contains both nuclei features and tissue structures, thus increasing processing time.
Another limitation is the reluctance of pathologists to invest in CAD systems due to their high number of false positive results. Hence, there is a need to introduce deep-learning neural networks within CAD systems to assist, in the early diagnosis and treatment of breast cancer. Therefore, methods like the automatic watershed segmentation of cancerous lesions in human breast histology images can be used to segment nuclei objects, not on the entire normalized image but rather isolate nuclei features to form a normalized H (nuclei features) dataset different from other tissue structures dataset for easier and faster model processing.
4. Conclusions and Future Work
There is a huge potential use of CAD systems on breast histology images, specifically, work targeting cancerous lesions segmentation and detection. Segmentation is one key aspect of deducing important information from the analysis of medical images, explicitly human histology images. Several computer-aided diagnosis systems have emerged that aid the extraction of ROIs that identify objects of similar features and characteristics on images for exploration purposes. These systems allow medical practitioners to interpret medical human-related images and thus improve the efficacy of diagnosis and treatment tasks. Segmentation is made quite elementary and faster with the introduction of these automatic diagnostics systems.
This study discusses implementing the automatic watershed segmentation method of cancerous lesions in unsupervised BC (normalized H) histology images. It breaks it down into, pre-processing, data augmentation, image enhancement, and segmentation steps. Data augmentation is used to increase the size of the limited image dataset provided herein. Stain normalization removes color inconsistencies and irregularities resulting from the data augmentation process. Thresholding and morphology operations enhance and highlight BC lesions with their edges on histology images, the connected components method groups components with similar characteristics into binary masks. The binary masks are then used within the RNN to separate overlapping and non-overlapping objects. Lastly, the watershed filling is used to segment, maintain, and topographically distinguish nuclei and their edges in different image regions namely; background, and foreground.
Accuracy performance evaluation protocol is used to gauge the efficiency of the proposed model in meeting its segmentation task. From the reviewed literature, our study has made several observations that are key in the segmentation and detection of breast cancer lesions in histology images. Pre-processing images through data augmentation is important in cases of limited or lack of publicly available datasets to increase dataset size. Stain normalization depending on the stain procedure used on images is necessary in removing color irregularities on images caused by data augmentation and assists in nucleus identification. Thresholding and morphological operations are needed to improve the quality of the image by highlighting and accentuating nuclei objects, and opening and closing operations are preferred. Notably, the image pre-processing and morphology operations used in a study will decide the segmentation method to use thus offering a ripple effect on the ultimate performance of the model.
Future Work
Therefore, there is a need for automatic early diagnostics systems in place of manual methods to assist in easier, faster, and act as a second opinion for clinicians to detect BC lesions in histology images. This proposed study provides, great solutions for learning models to correctly identify, separate, and detect nuclei objects and their edges on BC (normalized H) histology images compared to other methods. Despite the significant outcome of this proposed watershed segmentation method with supervised models, a series of encouraging future perspectives pop up namely; publicly available datasets, introducing hybrid approaches, and weight regularization methods fine-tuned for unsupervised models to increase their efficacy and reduce over-fitting. These perspectives are discussed meticulously below:
- Data Availability and Integrity—most deep learning approaches require significantly large dataset sizes to achieve meaningful and effective performance results. Therefore, there is a need for more publicly available BC histology image datasets to aid deep learning.
- Regularization methods—to improve the performance of models. This can be done through model hyper-parameter tuning such as optimizing learning rates, dropout, loss functions, activation functions, and early stopping methods.
- Blended Approaches—combining various/several methods to form a hybrid method that improves overall evaluation performance. This combination can occur at any step of the model namely; pre-processing, combining various attributes of different models to form one that will enhance the training, extraction, detection, and classification of nuclei objects. Additionally, in the future, our work can expand to reach out and diagnose image datasets of other human glands histology images, not limited to just BC histology images.
Author Contributions
V.M. and E.M. contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the Department of Computer Science, University of South Africa, Preller Street, Muckleneuk Ridge, Pretoria 1709 South Africa.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used to support the findings of this study can be obtained from the corresponding author upon request.
Conflicts of Interest
The authors declare they have no conflicts of interest.
References
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Alshanbari, H.; Amain, S.; Shuttelworth, J.; Slman, K.; Muslam, S. Automatic segmentation in breast cancer using watershed algorithm. Int. J. Biomed. Eng. Sci. 2015, 2, 1–6. [Google Scholar]
- Nayak, T.; Bhat, N.; Bhat, V.; Shetty, S.; Javed, M.; Nagabhushan, P. Automatic segmentation and breast density estimation for cancer detection using an efficient watershed algorithm. In Data Analytics and Learning: Proceedings of DAL 2018; Springer: Berlin/Heidelberg, Germany, 2019; pp. 347–358. [Google Scholar]
- Kaur, A.; Rashid, M.; Bashir, A.K.; Parah, S.A. Detection of breast cancer masses in mammogram images with watershed segmentation and machine learning approach. In Artificial Intelligence for Innovative Healthcare Informatics; Springer: Berlin/Heidelberg, Germany, 2022; pp. 35–60. [Google Scholar]
- Shahin, O.R.; Ayadi, R.; Ghorbel, O. Mammogram Breast Cancer Detection using Fast Watershed Segmentation. Int. J. 2020, 9, 1–6. [Google Scholar]
- Sadad, T.; Hussain, A.; Munir, A.; Habib, M.; Ali Khan, S.; Hussain, S.; Yang, S.; Alawairdhi, M. Identification of breast malignancy by marker-controlled watershed transformation and hybrid feature set for healthcare. Appl. Sci. 2020, 10, 1900. [Google Scholar] [CrossRef]
- Veta, M.; Huisman, A.; Viergever, M.A.; van Diest, P.J.; Pluim, J.P. Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; pp. 618–621. [Google Scholar]
- Lal, S.; Desouza, R.; Maneesh, M.; Kanfade, A.; Kumar, A.; Perayil, G.; Alabhya, K.; Chanchal, A.K.; Kini, J. A robust method for nuclei segmentation of H&E stained histopathology images. In Proceedings of the 2020 7th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 27–28 February 2020; pp. 453–458. [Google Scholar]
- Kaushal, C.; Singla, A. Automated segmentation technique with self-driven post-processing for histopathological breast cancer images. CAAI Trans. Intell. Technol. 2020, 5, 294–300. [Google Scholar] [CrossRef]
- Zebari, D.A.; Zeebaree, D.Q.; Abdulazeez, A.M.; Haron, H.; Hamed, H.N.A. Improved threshold based and trainable fully automated segmentation for breast cancer boundary and pectoral muscle in mammogram images. IEEE Access 2020, 8, 203097–203116. [Google Scholar] [CrossRef]
- Kiran, I.; Raza, B.; Ijaz, A.; Khan, M.A. DenseRes-Unet: Segmentation of overlapped/clustered nuclei from multi organ histopathology images. Comput. Biol. Med. 2022, 143, 105267. [Google Scholar] [CrossRef] [PubMed]
- Kowal, M.; Żejmo, M.; Skobel, M.; Korbicz, J.; Monczak, R. Cell nuclei segmentation in cytological images using convolutional neural network and seeded watershed algorithm. J. Digit. Imaging 2020, 33, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Qiao, S.; Hao, Y.; Bai, Y.; Cheng, R.; Zhang, W.; Zhang, G. Breast cancer histopathological images recognition based on two-stage nuclei segmentation strategy. PLoS ONE 2022, 17, e0266973. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, V.A.; Kumar, M.S.; Patan, R.; Kallam, S.; Mohamed, M.Y.N. Segmentation of nuclei in histopathology images using fully convolutional deep neural architecture. In Proceedings of the 2020 IEEE International Conference on Computing and Information Technology (ICCIT-1441), Tabuk, Saudi Arabia, 9–10 September 2020; pp. 1–7. [Google Scholar]
- Guatemala-Sanchez, V.R.; Peregrina-Barreto, H.; Lopez-Armas, G. Nuclei segmentation on histopathology images of breast carcinoma. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 26–30 July 2021; pp. 2622–2628. [Google Scholar]
- Xie, L.; Qi, J.; Pan, L.; Wali, S. Integrating deep convolutional neural networks with marker-controlled watershed for overlapping nuclei segmentation in histopathology images. Neurocomputing 2020, 376, 166–179. [Google Scholar] [CrossRef]
- Kurmi, Y.; Chaurasia, V. Content-based image retrieval algorithm for nuclei segmentation in histopathology images: CBIR algorithm for histopathology image segmentation. Multimed. Tools Appl. 2021, 80, 3017–3037. [Google Scholar] [CrossRef]
- Vahadane, A.; Sethi, A. Towards generalized nuclear segmentation in histological images. In Proceedings of the 13th IEEE International Conference on BioInformatics and BioEngineering, Chania, Greece, 10–13 November 2013; pp. 1–4. [Google Scholar]
- Shen, P.; Qin, W.; Yang, J.; Hu, W.; Chen, S.; Li, L.; Wen, T.; Gu, J. Segmenting multiple overlapping nuclei in H&E stained breast cancer histopathology images based on an improved watershed. In Proceedings of the 2015 IET International Conference on Biomedical Image and Signal Processing (ICBISP 2015), Beijing, China, 19 November 2015; pp. 1–4. [Google Scholar]
- Veta, M.; Van Diest, P.J.; Kornegoor, R.; Huisman, A.; Viergever, M.A.; Pluim, J.P. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS ONE 2013, 8, e70221. [Google Scholar]
- Baker, Q.B.; Banat, S.; Eaydat, E.; Alsmirat, M. Automated detection of benign and malignant in breast histopathology images. In Proceedings of the 2018 IEEE/ACS 15th International Conference on Computer Systems and Applications (AICCSA), Aqaba, Jordan, 28 October–1 November 2018; pp. 1–5. [Google Scholar]
- Shu, J.; Fu, H.; Qiu, G.; Kaye, P.; Ilyas, M. Segmenting overlapping cell nuclei in digital histopathology images. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5445–5448. [Google Scholar]
- Majanga, V.; Viriri, S. Dental images’ segmentation using threshold connected component analysis. Comput. Intell. Neurosci. 2021, 2021, 2921508. [Google Scholar] [CrossRef] [PubMed]
- Hussain, Z.; Gimenez, F.; Yi, D.; Rubin, D. Differential data augmentation techniques for medical imaging classification tasks. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: Bethesda, MD, USA, 2017; Volume 2017, p. 979. [Google Scholar]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152, 106391. [Google Scholar] [CrossRef] [PubMed]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef] [PubMed]
- Veta, M.; Pluim, J.P.; Van Diest, P.J.; Viergever, M.A. Breast cancer histopathology image analysis: A review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).