Abstract
The binding of fluoroquinolones, the most commonly prescribed antibiotics, with melanin is well explored. However, their binding patterns and exact mechanism of interaction with tyrosinase, a key enzyme in melanogenesis, are not explored yet. Thus, in the present study, seven fluoroquinolone drugs were selected to characterize their interactions with the tyrosinase enzyme: ciprofloxacin, enoxacin sesquihydrate, ofloxacin, levofloxacin, sparfloxacin, moxifloxacin and gemifloxacin. The results confirmed that all the drugs execute excellent enzyme activity, with an inhibition range from IC50 = 28 ± 4 to 50 ± 1.9 μM, outperforming the standard hydroquinone (IC50 = 170 μM). Later, kinetic studies revealed that all the drugs showed irreversible, but mixed-type, tyrosinase inhibition, with a preferentially competitive mode of action. Further, 2D and 3D docked complexes and binding analyses confirmed their significant interactions in the active region of the target enzyme, sufficient for the downstream signaling responsible for the observed tyrosinase inhibition. Thus, this is the first report demonstrating their mechanism of tyrosinase inhibition, critical for melanin-dependent responses, including toxicity.
1. Introduction
Fluoroquinolones are a group of synthetic, broad-spectrum antibiotics patented in the late nineties, including norfloxacin (1978), pefloxacin (1979), enoxacin (1980), ciprofloxacin (1981), fleroxacin (1981) and ofloxacin (1982) [1], and are still used today. In Figure 1, the general structure of fluoroquinolones, with an accepted numbering scheme for positions on the molecule, is shown. In the 1980s, there was a big revolution when enoxacin, an analog of nalidixic acid, was derived with a significantly increased spectrum of antibacterial activity [2]. Ciprofloxacin, the most successful and widely accepted fluoroquinolone, was marketed in 1986; since then, fluoroquinolones have become recognized as promising agents for controlling a wide range of infections, including gastrointestinal, urinary, respiratory, skin and ocular infections [2,3,4,5]. This class of compounds has enhanced pharmacokinetic properties, with extensive and potent activities against parasites, bacteria and mycobacteria, including resistant strains that outperform the bactericidal drugs existing previously [5,6]. However, in an attempt to retain their effectiveness, the present guidelines mostly recommend this class as second-line agents to prescribe when narrow-spectrum antibiotics have failed [7].
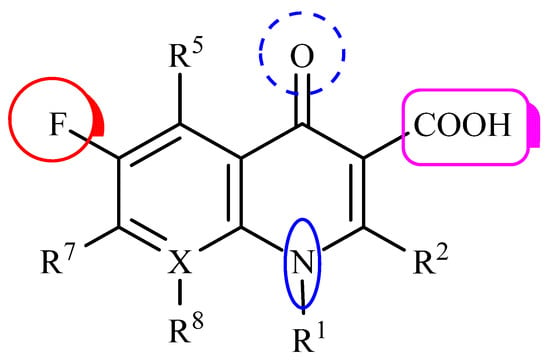
Figure 1.
General structure of fluoroquinolones, using the accepted numbering scheme for positions on the molecule. The radicals R1, R2, R5, R7 and R8 indicate possible positions for structural modification; X usually corresponds to a C or N atom. Adapted with permission from [8].
Despite their excellent antibacterial properties, phototoxicity is a rare, but characteristic, side effect of fluoroquinolone antibiotic use [9], which is believed to be associated with UVA-induced drug degradation, resulting in the formation of reactive oxygen species (ROS), causing oxidative damage to cell components, including lipids, proteins and nucleic acids [10,11,12,13]. Fluoroquinolone-induced skin phototoxicity may appear in vivo, with different degrees of severity, as exaggerated sunburn-like lesions, blisters, scaling, edema, erythema, eczematosus and bullous eruptions in areas of the skin exposed to sunlight or artificial UVA radiation [9,14,15,16,17].
Melanocytes, a heterogeneous group of cells, have the ability to produce a pigmented protein called melanin, primarily responsible for skin, hair and eye color. Tyrosinase, being a limiting enzyme for melanogenesis, affects melanin biosynthesis significantly. It catalyzes the hydroxylation of L-tyrosine to 3,4-dihydroxyphenylalanine (L-DOPA) and L-DOPA to dopaquinones [18], resulting in the accumulation of melanin pigments. There are two types of epidermal melanin, eumelanin and pheomelanin, known for their photo-protective and photo-toxic properties [19,20]. Melanin bears antioxidant properties, and a strong affinity for drugs and other chemical substances. Therefore, it efficiently filters toxic substances and protects body tissues from oxidative and chemical stress. The affinity of melanin for drugs restricts their access to cell receptors, and then releases them from the complexes in non-toxic concentrations, protecting the tissues from their undesirable side effects. However, the affinity of melanin for drugs may result in them becoming over accumulated in cells. In particular, their long-term intake and the slow release of their metabolites may elevate the level of noxious substances stored on melanin, causing degeneration in the melanin-containing cells (present in the eye and skin) and surrounding tissues [21,22,23]. It was suggested that the toxic reactions of fluoroquinolones [24,25,26], tetracyclines [27,28] and phenothiazines [29], directed to pigmented tissues, may be related to the accumulation of drugs in melanin biopolymers. The effect of fluoroquinolone antibiotics, such as ciprofloxacin [26], lomefloxacin [30], sparfloxacin [26], norfloxacin and moxifloxacin [31,32], on the antioxidant status and melanin synthesis in normal human melanocytes (HEMa-LP) is extensively studied, with all suggesting melanin-associated drug-induced toxicity. It has been demonstrated that fluoroquinolones bind well to melanin-rich tissues [33,34]; the relation between the affinity of fluoroquinolones to melanin and skin or eye toxicity is well documented, but the mechanism of their interaction with tyrosinase, a key player in melanin synthesis, is not reported.
Thus, the following fluoroquinolone drugs with a single active compound were purchased and screened for comparative tyrosinase activity: ciprofloxacin, enoxacin sesquihydrate, ofloxacin, levofloxacin, sparfloxacin, moxifloxacin and gemifloxacin. Later, to explore the mechanism, type of enzyme inhibition and possible interactions of the test drugs with tyrosinase, enzyme kinetic in vitro studies and molecular docking in silico studies were performed, adding in the existing understanding of drug-induced, tyrosinase rooted, melanin-associated toxicity.
2. Materials and Methods
2.1. Chemicals
Mushroom tyrosinase, 3,4-dihydroxyphenylalanine (L-DOPA), mono-sodium phosphate monohydrate, di-sodium phosphate dihydrate, hydroquinone, sodium hydroxide and dimethyl sulfoxide (Sigma Aldrich, St. Louis, MI, USA) were used. Seven fluoroquinolone drugs with a single active compound were purchased from the local pharmacy.
2.2. In Vitro Anti-Tyrosinase Activity
The mushroom tyrosinase was used to screen the anti-tyrosinase activity of the tested antibiotics, following the method described previously [34]. Briefly, 20 μL of tyrosinase (30 U/mL), 140 μL of phosphate buffer (20 mM, pH 6.8) and 20 μL of the test drug’s solution were mixed in a 96-well plate (SPL, Gyeonggi-do, Korea) and incubated at 25 °C for 10 min. Later, 20 μL of 0.85 mM 3,4-dihydroxyphenylalanine (L-DOPA) was added and the plate was incubated again at 25 °C for 20 min. Finally, the formation of dopachrome was tracked at 450 nm using a plate reader (BioTek, ELX800, Winooski, VT, USA) and the experiment was performed in triplicate. Hydroquinone was used as a standard. The following formula was used to find the percentage of tyrosinase inhibition necessary to calculate 50% inhibition (IC50). We used the following formula to calculate the % inhibition:
ODsample means the optical density of the test drugs, while ODcontrol means the optical density of the blank (without a test drug and without a standard drug).
2.3. Kinetic Analysis of Tyrosinase Inhibition Activity
The kinetic mechanism of enzyme inhibition was determined by following the method reported previously [35]. Different drug concentrations, as indicated in the graphs, were used for the Lineweaver–Burk plot and Dixon plot; however, 0.0625 to 2 mM substrate L-DOPA concentration was used in all the kinetic studies. The pre-incubation period was the same as in the mushroom tyrosinase inhibition assay protocol. After enzyme addition, the formation of dopachrome was tracked for 5 min with 30 s intervals at a wavelength of 450 nm.
The type of enzyme inhibition was assessed by Lineweaver–Burk graph plotting the inverse of velocities 1/V versus the inverse of substrate concentration 1/[S] mM−1, and the inhibition constant Ki was determined by Dixon graph plotting 1/V versus the drug concentrations. Later, series of experiments were performed to find the mode of reversible or irreversible inhibitory enzyme behavior, and complexes formed between the drugs and enzyme.
2.4. Molecular Docking Studies
2.4.1. Repossession of Mushroom Tyrosinase from PDB
The crystal structure of mushroom tyrosinase was retrieved from the Protein Data Bank (PDB), with the code PDBID 2Y9X [36]. Energy minimization of the target structure was carried out by using the conjugate gradient algorithm and AMBER force field in UCSF Chimera 1.10.1 [37]. The stereo-chemical properties, and Ramachandran graph and values of the mushroom tyrosinase structure were assessed by the MolProbity server [38], while the hydrophobicity graph was generated by Discovery Studio 4.1 Client [39] (https://discover.3ds.com/discovery-studio-visualizer-download, accessed on 21 April 2021). Moreover, the online tool VADAR 1.8 [40] (http://vadar.wishartlab.com/, accessed on 21 April 2021) was used to find the protein architecture and statistical percentage values.
2.4.2. Preparation of Ligands and Molecular Docking Simulation
The chemical structures of ciprofloxacin, gemifloxacin, enoxacin, levofloxacin, moxifloxacin, ofloxacin and sparfloxacin were sketched using the ACD/ChemSketch tool and energy minimized using the visualizing software UCSF Chimera 1.10.1 [38]. The PyRx docking tool was used to perform the molecular docking experiment for all the ligands against mushroom tyrosinase. The grid box center values of X = −0.4045, Y = 21.8847 and Z = −42.2150, and the size values, were adjusted to X = 30.0478, Y = 28.5792, and Z = 21.2800 for a better conformational position in the active region of mushroom tyrosinase. All the synthesized ligands were docked separately against mushroom tyrosinase, with a default exhaustiveness value of 8. The predicted docked complexes were evaluated on the basis of the lowest binding energy (Kcal/mol) values and structural activity relationship (SAR) analyses. The three-dimensional (3D) graphical depictions of all the docked complexes were created using Discovery Studio (2.1.0) and UCSF Chimera 1.10.1.
3. Results and Discussion
The present study was designed to investigate the unexplored mechanism of selected fluoroquinolone drug–enzyme interactions and their binding patterns, critical for melanin-dependent responses and important for the cosmetic, medicine and agriculture industries.
Our result confirmed that all the selected antibiotics (Figure 2) i.e., ciprofloxacin, enoxacin sesquihydrate, gemifloxacin, levofloxacin, moxifloxacin, ofloxacin and sparfloxacin showed excellent tyrosinase inhibitory activity with IC50 ± SEM values 37 ± 2, 28 ± 4, 34 ± 2, 39 ± 2, 50 ± 1.9, 40 ± 2 and 30 ± 2.8 µM, respectively. Interestingly, all the drugs outperformed the standard hydroquinone (IC50 = 170 μM). Additionally, norfloxacin was tested for comparison, and it was found to be a tyrosinase inhibitor with IC50 = 820 μM. The selected fluoroquinolones are known for their broad-spectrum antibacterial activity. These drugs were selected due to their excellent bioavailability, easy synthesis, low toxicity, ability to kill different clinical pathogens and, most importantly, they have been used as antibiotics for a long time. The fluoroquinolones are synthetic fluorinated analogues of naldixic acid, which are important for cosmetics, medicine and agriculture [41,42,43]. A cyclopropyl moiety at position 1 (ciprofloxacine, gemifloxacin, moxifloxacine, and sparfloxacine) is considered to be the most potent modification against anaerobic bacteria [44], and any bulky substituent at position 7 (gemifloxiacine and enoxacine) is very close to the topoisomerase IV binding site, resulting in a lower level of microbiological activity [45,46]. Moreover, 3-carboxylate (ciprofloxacine, levofloxacine, moxifloxacine, ofloflaxacine and sparfloxacine), fluoro groups (enoxacine and gemifloxacine) and 4-carbonyl groups are important for antimicrobial activity [47]. Substituents at position 5 (sparflaxacine) change the overall steric configuration of the molecule. The addition of an amino group, as in the case of sparfloxacine at position 5, can remarkably increase the in vitro activity against Gram-positive bacteria and Toxoplasma gondii [47,48]. The addition of fluorine at position 6 (ciprofloxacin, levofloxacin, moxifloxacin, ofloxfloxacine and sparfloxacine) is used widely nowadays, due to their clinical importance, which improves antimicrobial activity as compared to the original quinolone compound [49]. Any kind of substitution at positions 1, 7 and 8 plays a key role in the overall biological activities of the compounds under development [50]. It was observed that enoxacin exhibited the most potent tyrosinase inhibitory activity, with the lowest IC50 of 28 μM. Position 8 is important, as it affects the steric configuration, similarly to position 5, by substitution, accessing the binding sites of enzymes [51]. Thus, the presence of an ethyl group at position 1 plays a very important role in tyrosinase inhibitory activity, as it was shown to have higher activity than the reference hydroquinone. The role of the 1-cyclo propyl group in ciprofloxacin and 1,8 cyclo compounds in ofloxacin and levofloxacin was also studied, which exhibited a higher tyrosinase inhibition potential than moxifloxacin, but lower than gemifloxacin. Hence, the IC50 values of all the tested drugs were found to be 3–6 times lower than the standard hydroquinone. An important observation is that sparfloxacine exhibits comparable inhibition activity to the most potent enoxacine drug, which is because of the bulkiness at position 7, and the bulk here also increases the anti-anaerobic activity [52]. It is worth noting that antibiotics already on the market show excellent anti-tyrosinase activity, which may give them an advantage over any new synthetic drug that requires a medical trial before considerations. Their interactions, binding patterns and dose-dependent effect on melanin have been studied widely [26,27,28,29,30,31,32]. However, to study their unexplored binding mechanism with tyrosinase, a key enzyme for melanogenesis, a kinetic study is performed.
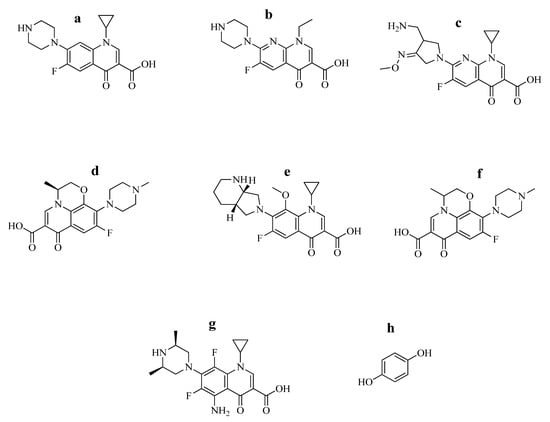
Figure 2.
Study drugs (a) ciprofloxacin, (b) enoxacin sesquihydrate, (c) gemifloxacin, (d) levofloxacin, (e) moxifloxacin, (f) ofloxacin, (g) sparfloxacin and (h) hydroquinone. Their structures are adopted with permission [8,53].
3.1. Mechanism of Enzyme Kinetics
To understand the mechanism of enzyme inhibition, oxidation of the L-DOPA indicator of diphenolase activity was determined from Lineweaver–Burk and Dixon plots. The Lineweaver–Burk plots, in the presence of most potent drugs, i.e., enoxacin sesquihydrate, ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin, moxifloxacin and gemifloxacin, are shown in Figure 3 and Figures S1–S6, respectively. Additionally, a kinetic study for norfloxacin was also performed (Figure S7). The plots (1/V versus 1/[S]) showed straight lines with different slopes. Their analysis revealed that Vmax reduces with Km shift and increasing drug concentration, which indicated that they inhibit tyrosinase by dual distinct channels and show a mixed type of inhibition. This revealed that antibiotics can bind with a free enzyme (E), as well as with the enzyme–substrate (ES) complex [54]. The insightful analysis of EI and ESI binding affinities showed their mixed-type mode of inhibition. The secondary replots of slope versus concentration of the test drugs showed the EI dissociation constant (Ki) (Figure 3b1 and Figures S1–S7(b2–b8)), while the ESI dissociation constant (Ki’) was shown by the secondary replots of intercept versus concentration of the test drugs (Figure 3c1 and Figures S1–S7(c2–c8)). Both the Dixon plot (Figure 3d1 and Figures S1–S7(d2–b8)) and secondary replot of the Lineweaver–Burk plot were used to calculate the Ki. The Ki values ranged from 0.1 to 0.4 mM (Figure 3b1 and Figures S1–S7(b2–b8)) and the Ki’ values ranged from 0.8 to 2 mM (Figure 3c1 and Figures S1–S7(c2–c8)). The analysis showed lower dissociation constant values for Ki compared to Ki’, demonstrating more prosperous binding between the enzyme and antibiotics [55], justifying their preferred competitive mode of inhibition.
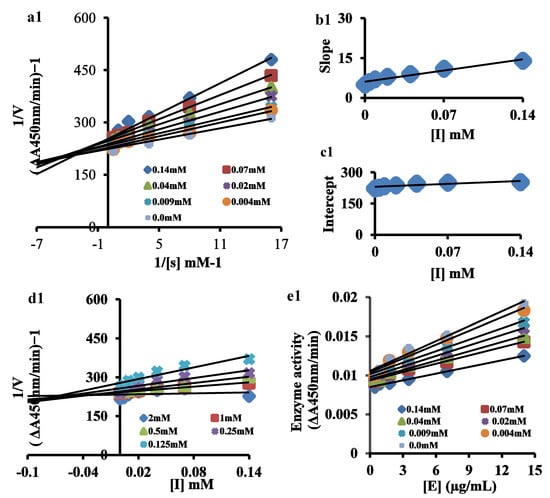
Figure 3.
(a1) Lineweaver–Burk plot for inhibition of tyrosinase enzyme in the presence of various concentrations of enoxacin sesquihydrate (as indicated) and L-DOPA concentrations (0.0625, 0.125, 0.25, 0.5, 1 mM and 2 mM). (b1) The insets represent the plot of the slope from Lineweaver–Burk plot versus inhibitor. (c1) The secondary replot of the Lineweaver–Burk plot, 1/V (y-intercept) versus various concentrations of inhibitor. (d1) The Dixon plot of the reciprocal of the initial velocities versus various concentrations of the test drug. (e1) Relationship between the catalytic activity of tyrosinase and various concentrations of the test drug.
3.2. The Inhibitory Effect of Drugs on Diphenolase Activity of Tyrosinase
The tyrosinase inhibition behavior by the most potent drugs, i.e., enoxacin sesquihydrate, ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin, moxifloxacin, gemifloxacin and norfloxcin, was assessed by reversibility plots (Figure 3e1 and Figure S8(a2–a8)). For the catalysis of L-DOPA, the plots of enzyme activity versus the concentration of the enzyme (0.44, 0.88, 1.75, 3.5, 7 and 14 μg/mL) in the presence of antibiotics produced a family of straight lines. The series of parallel straight lines, which had the same slopes for all the antibiotics, indicated their irreversible binding behavior [56,57] with the enzyme. Thus, our results confirmed the irreversible binding behavior of the tested drugs with the binuclear active site of tyrosinase.
3.3. Molecular Docking and Structural Assessment of Mushroom Tyrosinase
Mushroom tyrosinase (Agaricus bisporus) belongs to an oxidoreductase copper-containing protein that comprises 391 residues. The mushroom tyrosinase structure has 39% α-helices (154 residues), 14% β-sheets (57 residues) and 46% coils (180 residues). In the Ramachandran plots, the residues in favored regions and the residues in allowed regions were 95.90% and 100.0%, respectively (Figure S9). The values of the Ramachandran graph indicate the good precision for phi (φ) and psi (ψ) angles among the coordinates of the receptor molecules, and most of the residues plummeted into an acceptable region (Figure S9).
The docked complexes of all the drugs against mushroom tyrosinase were assessed individually, and were compared on the basis pattern of the drug interactions and minimum energy values. The results confirmed that all the drugs showed good binding energy values in the active region of the target protein (Figure 4). The autodock standard error is testified as 2.5 kcal/mol; however, the predicted energy value difference is observed to be less for all the docking complexes.

Figure 4.
Molecular docking energy graph of test drugs.
3.4. Binding Analyses of Test Drugs and Tyrosinase Enzyme
The in vitro and in silico results showed that enoxacin sesquihydrate has more therapeutic potential and is confined in the active binding pocket of the target protein (Figure 5A,B). The docking result of the enoxacin sesquihydrate–mushroom tyrosinase docked complex showed two hydrogen bonds at the His85 and His263 residues. The oxygen atom of enoxacin formed a hydrogen bond against His85, with a bond length of 4.65 Å, and His263, with a bond length of 4.17 Å. Our docking results showed good correlation with the published research, which strengthens our work and its efficacy [34,58,59].
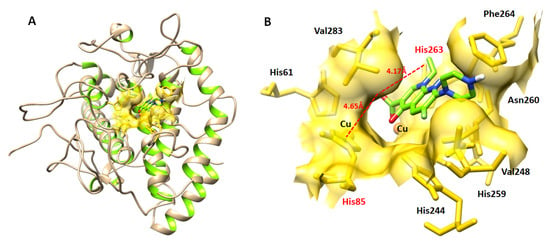
Figure 5.
(A,B) Docking complex (3D) of enoxacin sesquihydrate and tyrosinase enzyme.
In ciprofloxacin docking, hydrogen and hydrophobic interactions have been observed at different residue positions. The oxygen atoms in the piperazine ring form a couple of hydrogen bonds with His85 and Asn81; whereas, val283 and Pro284 hydrophobically interacted with appropriate bond distances. In enoxacin sesquihydrate docking, a single hydrogen bond was observed at position Asn260, through an oxygen atom. Moreover, a couple of hydrophobic interactions were reported at His85, His263, His269, Val283 and His244, respectively. Most importantly, enoxacin sesquihydrate also formed a hydrophobic interaction with Cu401, through π-π stacking. In the gemifloxacin docking results, three hydrogen bonds were observed at positions Asn81, His244 and Asn260, through oxygen atoms of the drug. Moreover, Ala323 and Val283 also interacted through hydrophobic interactions. In levofloxacin tyrosinase docking, a couple of hydrogen bonds and hydrophobic interactions have been observed at different residue positions. The oxygen atoms of levofloxacin formed two hydrogen bonds at positions His85 and Asn81, respectively. Moreover, Val283 and Pro284 interacted by forming hydrophobic interactions. In the moxifloxacin and ofloxacin docking results, a couple of hydrogen bonds were reported at positions His85 and Asn81, respectively. Moreover, His244 and Val283 interacted through hydrophobic interactions.
In the sparfloxacin docking complex, a single hydrogen bond was observed at position Met280, and different hydrophobic interactions were reported at different residue positions, such as Gly281, Asn260, Phe264, His85, His259, His263, Val283 and ser282, respectively. Furthermore, both copper ions interacted hydrophobically with the drug. The comparative results showed that all the drugs bind at binding pocket residues, having slightly different conformational positions inside the binding pocket, and they showed good correlation with the published data, which ensures the credibility of the docking analysis. The 2D conformations, binding poses, and interactions with binding residues of all the drugs are mentioned in Supplementary Figure S10.
4. Conclusions
In the present study, seven fluoroquinolone drugs were used and their mechanism of tyrosinase inhibition was explored for the first time. All the test drugs showed excellent tyrosinase inhibition that outperformed the standard. Among all, enoxacin sesquihydrate showed the highest tyrosinase inhibitory activity in vitro. The kinetic analysis revealed that all the drugs showed irreversible, but mixed-type, tyrosinase inhibition, with a preferentially competitive mode of action. Further, the 2D and 3D docked complexes and their binding analysis confirmed the significant interactions of all the tested drugs in the active region of the target enzyme, sufficient for the downstream signaling responsible for tyrosinase inhibition. Thus, this is the first report demonstrating the mechanism, type of enzyme inhibition and possible interactions of the test drugs with tyrosinase. This is critical for melanin-dependent biological responses, and is important to consider during cosmetic, medicine and agricultural applications in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app12104849/s1.
Author Contributions
Conceptualization, M.H.M., B.A.A., A.O.A. and A.A.; methodology, Q.J. and M.H; software, M.H.; validation, T.T. and M.M.; formal analysis, Q.J. and T.T.; investigation, Q.J.; writing—original draft preparation, A.A., M.H. and Q.J.; writing—review and editing, B.A.A. and K.K.-M.; visualization, T.T.; supervision, A.O.A., M.H.M., B.A.A., Y.S.A., K.K.-M. and A.A.; project administration, M.H.M. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
Authors would like to acknowledge the support of the Deputy for Research and Innovation—Ministry of Education, Kingdom of Saudi Arabia for this research through a grant (NU/IFC/ENT/01/005), under the institutional Funding Committee at Najran University, Kingdom of Saudi Arabia. M.H. acknowledges the Ohio State University for providing the “President’s Postdoctoral Scholars Program (PPSP)” award for financial support to complete this computational research.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Appelbaum, P.C.; Hunter, P.A. The fluoroquinolone antibacterials: Past, present and future perspectives. Int. J. Antimicrobial. Agents 2000, 16, 5–15. [Google Scholar] [CrossRef]
- Patrick, G.L. An Introduction to Medicinal Chemistry; Oxford University Press: Oxford, UK, 2013; pp. 379–435. [Google Scholar]
- Dalhoff, A. Global fluoroquinolone resistance epidemiology and implications for clinical use. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 976273. [Google Scholar] [CrossRef] [PubMed]
- Linder, J.A.; Huang, E.S.; Steinman, M.A.; Gonzales, R.; Stafford, R. Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 2005, 118, 259–268. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, M.V.; Saraiva, M.F.; de Souza, M.V.; da Costa, C.F.; Vicente, F.R.; Lourenço, M.C. Synthesis and antitubercular activity of lipophilic moxifloxacin and gatifloxacin derivatives. Bioorganic Med. Chem. Lett. 2007, 17, 5661–5664. [Google Scholar] [CrossRef]
- Anquetin, G.; Greiner, J.; Mahmoudi, N.; Santillana-Hayat, M.; Gozalbes, R.; Farhati, K.; Derouin, F.; Aubry, A.; Cambau, E.; Vierling, P. Design, synthesis and activity against Toxoplasma gondii, Plasmodium spp., and Mycobacterium tuberculosis of new 6-fluoroquinolones. Eur. J. Med. Chem. 2006, 41, 1478–1493. [Google Scholar] [CrossRef]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Pham, T.D.; Ziora, Z.M.; Blaskovich, M.A. Quinolone antibiotics. Medchemcomm 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Oliveira, H.S.; Gonçalo, M.; Figueiredo, A.C. Photosensitivity to lomefloxacin. A clinical and photobiological study. Photodermatol. Photoimmunol. Photomed. Figueiredo. Photodermatol. Photoimmunol. Photomed. 2000, 16, 116–120. [Google Scholar] [CrossRef]
- De Guidi, G.; Bracchitta, G.; Catalfo, A. Photosensitization reactions of fluoroquinolones and their biological consequences. Photochem. Photobio. 2011, 87, 1214–1229. [Google Scholar] [CrossRef]
- Viola, G.; Facciolo, L.; Canton, M.; Vedaldi, D.; Dall’Acqua, F.; Aloisi, G.G.; Amelia, M.; Barbafina, A.; Elisei, F.; Latterini, L. Photophysical and Phototoxic Properties of the Antibacterial Fluoroquinolones Levofloxacin and Moxifloxacin. Chem. Biodivers. 2004, 1, 782–801. [Google Scholar] [CrossRef]
- Dwivedi, A.; Mujtaba, S.F.; Yadav, N.; Kushwaha, H.N.; Amar, S.K.; Singh, S.K.; Pant, M.C.; Ray, R.S. Cellular and molecular mechanism of ofloxacin induced apoptotic cell death under ambient UV-A and sunlight exposure. Free Radic. Res. 2014, 48, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Ray, R.S.; Farooq, M.; Pant, A.B.; Hans, R.K. Photosensitizing Potential of Ciprofloxacin at Ambient Level of UV Radiation. Photochem. Photobiol. 2007, 83, 1226–1236. [Google Scholar] [CrossRef] [PubMed]
- Owen, K. Comparative grepafloxacin phototoxicity in mouse skin. J. Antimicrob. Chemother. 1998, 42, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Takahashi, Y.; Kawaguchi, H.; Utsunomiya, S.; Miura, N.; Izumi, H.; Tanimoto, A. A dermal phototoxicity study following intravenous infusion administration of ciprofloxacin hydrochloride in the novel microminipigs. Toxicol. Pathol. 2013, 41, 109–113. [Google Scholar] [CrossRef]
- Scholar, E.M. Fluoroquinolines: Past, present and future of a novel group of antibacterial agents. Am. J. Pharm. Educ. 2002, 66, 164–171. [Google Scholar]
- Ferguson, J.; Dawe, R. Phototoxicity in quinolones: Comparison of ciprofloxacin and grepafloxacin. J. Antimicrob. Chemother. 1997, 4, 93–98. [Google Scholar] [CrossRef][Green Version]
- Cooksey, C.J.; Garratt, P.J.; Land, E.J.; Pavel, S.; Ramsden, C.A.; Riley, P.A.; Smit, N.P. Evidence of the indirect formation of the catecholic intermediate substrate responsible for the autoactivation kinetics of tyrosinase. J. Biol. Chem. 1997, 272, 26226–26235. [Google Scholar] [CrossRef]
- Rozanowska, M.; Sarna, T.; Land, E.J.; Truscott, T. Free radical scavenging properties of melanin: Interaction of eu- and pheo-melanin models with reducing and oxidising radicals. Free Radic. Biol. Med. 1999, 26, 518–525. [Google Scholar] [CrossRef]
- Simon, J.D.; Peles, D.; Wakamatsu, K.; Ito, S. Current challenges in understanding melanogenesis: Bridging chemistry, biological control, morphology, and function. Pigment Cell Melanoma Res. 2009, 22, 563–579. [Google Scholar] [CrossRef]
- Hu, D.N.; Savage, H.E.; Roberts, J.E. Uveal melanocytes, ocular pigment epithelium, and Müller cells in culture: In vitro toxicology. Int. J. Toxicol. 2002, 21, 465–472. [Google Scholar] [CrossRef]
- Knorle, R.; Schniz, E.; Feuerstein, T.J. Drug accumulation in melanin: An affinity chromatographic study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1998, 714, 171–179. [Google Scholar] [CrossRef]
- Larsson, B.S. Interaction between Chemicals and Melanin. Pigment Cell Res. 1993, 6, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Beberok, A.; Buszman, E.; Wrześniok, D. Interaction of norfloxacin and sparfloxacin with melanin in relation to phototoxic reactions. Ann. Univ. Mariae. Curie. Skłodowska Sectio. DDD Pharm. 2009, 4, 87–92. [Google Scholar]
- Beberok, A.; Buszman, E.; Zdybel, M.; Pilawa, B.; Wrześniok, D. EPR examination of free radical properties of DOPA–melanin complexes with ciprofloxacin, lomefloxacin, norfloxacin and sparfloxacin. Chem. Phy. Lett. 2010, 497, 115–122. [Google Scholar] [CrossRef]
- Beberok, A.; Buszman, E.; Wrześniok, D.; Otręba, M.; Trzcionka, J. Interaction between ciprofloxacin and melanin: The effect on proliferation and melanization in melanocytes. Eur. J. Pharmacol. 2011, 669, 32–37. [Google Scholar] [CrossRef]
- Rok, J.; Buszman, E.; Beberok, A.; Delijewski, M.; Otręba, M.; Wrześniok, D. Modulation of Melanogenesis and Antioxidant Status of Melanocytes in Response to Phototoxic Action of Doxycycline. Photochem. Photobiol. 2015, 91, 1429–1434. [Google Scholar] [CrossRef]
- Rok, J.; Buszman, E.; Delijewski, M.; Otręba, M.; Beberok, A.; Wrześniok, D. Effect of tetracycline and UV radiation on melanization and antioxidant status of melanocytes. J. Photochem. Photobiol. B Biol. 2015, 148, 168–173. [Google Scholar] [CrossRef]
- Buszman, E.; Beberok, A.; Rózańska, R.; Orzechowska, A. Interaction of chlorpromazine, fluphenazine and trifluoperazine with ocular and synthetic melanin in vitro. Die Pharm. J. Pharma. Sci. 2008, 63, 372–376. [Google Scholar]
- Beberok, A.; Otręba, M.; Wrześniok, D.; Buszman, E. Cytotoxic effect of lomefloxacin in culture of human epidermal melanocytes. Pharmacol. Rep. 2013, 65, 689–699. [Google Scholar] [CrossRef]
- Beberok, A.; Wrześniok, D.; Otręba, M.; Miliński, M.; Buszman, E. Effect of norfloxacin and moxifloxacin on melanin synthesis and antioxidant enzymes activity in normal human melanocytes. Mol. Cell Biochem. 2015, 401, 107–114. [Google Scholar] [CrossRef]
- Beberok, A.; Wrześniok, D.; Otręba, M.; Buszman, E. Impact of sparfloxacin on melanogenesis and antioxidant defense system in normal human melanocytes HEMa-LP—An in vitro study. Pharmacol. Rep. 2015, 67, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Hamanaka, H.; Mizutani, H.; Asahig, K.; Shimizu, M. Melanocyte melanin augments sparfloxacin-induced phototoxicity. J. Dermatol. Sci. 1999, 21, 27–33. [Google Scholar] [CrossRef]
- Ono, C.; Tanaka, M. Binding characteristics of fluoroquinolones to synthetic levodopa melanin. J. Pharm. Pharmacol. 2003, 55, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, Z.; Rafiq, M.; Nadeem, H.; Hassan, M.; Afzal, S.; Waseem, M.; Afzal, K.; Latip, J. Carvacrol derivatives as mushroom tyrosinase inhibitors; synthesis, kinetics mechanism and molecular docking studies. PLoS ONE 2017, 12, e0178069. [Google Scholar] [CrossRef]
- A Structural View of Biology. Available online: www.rcsb.org (accessed on 21 April 2021).
- Pettersen, E.F. TD Goddard: CC Huang: GS Couch: DM Greenblatt: EC Meng: TE Ferrin. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III; De Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, D.C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct. Funct. Bioinform. 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Free Download: BIOVIA Discovery Studio Visualizer. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 21 April 2021).
- Single (or Multiple) Model Protein Structure Analysis. Available online: http://vadar.wishartlab.com/ (accessed on 21 April 2021).
- O’Donnell, J.A.; Gelone, S.P. Fluoroquinolones. Infect. Dis. Clin. N. Am. 2000, 14, 489–513. [Google Scholar] [CrossRef]
- Guneysel, O.; Onur, O.; Erdede, M.; Denizbasi, A. Trimethoprim/sulfamethoxazole resistance in urinary tract infections. J. Emerg. Med. 2009, 36, 338–341. [Google Scholar] [CrossRef]
- Blandeau, J.M. Expanded activity and utility of the new fluoroquinolones: A review. Clin. Ther. 1999, 21, 3–40. [Google Scholar] [CrossRef]
- Quintero, B.; Miranda, M.A. Mechanisms of photosensitization induced by drugs: A general survey. Ars Pharm. 2000, 41, 27–46. [Google Scholar]
- Domagala, J.M. Structure-activity and structure-side-effect relationships for the quinolone antibacterials. J. Antimicrob. Chemother. 1994, 33, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, E. History of Quinolones and Their Side Effects. Chemotherapy 2001, 47, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Tillotson, G.S. Quinolones: Structure-activity relationships and future predictions. J. Med. Microbiol. 1996, 44, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Fluit, A.C.; Schmitz, F.J. Fluoroquinolones: Structure and target sites. Curr. Drug Targets 2003, 4, 181–190. [Google Scholar] [CrossRef]
- Cecchetti, V.; Filipponi, E.; Fravolini, A.; Tabarrini, O.; Bonelli, D.; Clementi, M.; Cruciani, G.; Clementi, S. Chemometric methodologies in a quantitative structure− activity relationship study: The antibacterial activity of 6-aminoquinolones. J. Med. Chem. 1997, 40, 1698–1706. [Google Scholar] [CrossRef]
- Nawaz, M.S.; Bodla, R.; Kant, R.; Singh, S.P.; Bhutani, R.; Kapoor, G. Fluoroquinolone as antimicrobial agent: A Review. Int. J. Pharm. Sci. Res. 2017, 2, 57. [Google Scholar]
- Daneshtalab, M.; Ahmed, A. Nonclassical Biological Activities of Quinolone Derivatives. J. Pharm. Pharm. Sci. 2012, 15, 52–72. [Google Scholar] [CrossRef]
- Shandil, R.K.; Jayaram, R.; Kaur, P.; Gaonkar, S.; Suresh, B.L.; Mahesh, B.N.; Jayashree, R.; Nandi, V.; Bharath, S.; Balasubramanian, V. Moxifloxacin, ofloxacin, sparfloxacin, and ciprofloxacin against Mycobacterium tuberculosis: Evaluation of in vitro and pharmacodynamic indices that best predict in vivo efficacy. Antimicrob. Agents Chemother. 2007, 51, 576–582. [Google Scholar] [CrossRef]
- Wang, P.; Martin, B.D.; Parida, S.; Rethwisch, D.G.; Dordick, J.S. Multienzymic synthesis of poly (hydroquinone) for use as a redox polymer. J. Am. Chem. Soc. 1995, 117, 12885–12886. [Google Scholar] [CrossRef]
- Yi, W.; Wu, X.; Cao, R.; Song, H.; Ma, L. Biological evaluations of novel vitamin C esters as mushroom tyrosinase inhibitors and antioxidants. Food Chem. 2009, 117, 381–386. [Google Scholar] [CrossRef]
- Rattanangkool, E.; Kittikhunnatham, P.; Damsud, T.; Wacharasindhu, S.; Phuwapraisirisan, P. Quercitylcinnamates, a new series of antidiabetic bioconjugates possessing α-glucosidase inhibition and antioxidant. Eur. J. Med. Chem. 2013, 66, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cai, D.; Mou, D.; Yan, Q.; Sun, Y.; Pan, W.; Wan, Y.; Song, H.; Yi, W. Design, synthesis and biological evaluation of hydroxy- or methoxy-substituted 5-benzylidene(thio) barbiturates as novel tyrosinase inhibitors. Bioorganic Med. Chem. 2014, 22, 3279–3284. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Chen, Q.-X.; Wang, Q.; Huang, H.; Song, K.-K. Irreversibly inhibitory kinetics of 3,5-dihydroxyphenyl decanoate on mushroom (Agaricus bisporus) tyrosinase. Bioorganic Med. Chem. 2005, 13, 6206–6211. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Ashraf, Z.; Abbas, Q.; Raza, H.; Seo, S.-Y. Exploration of Novel Human Tyrosinase Inhibitors by Molecular Modeling, Docking and Simulation Studies. Interdiscip. Sci. Comput. Life Sci. 2018, 10, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Vanjare, B.D.; Mahajan, P.G.; Dige, N.C.; Raza, H.; Hassan, M.; Han, Y.; Kim, S.J.; Seo, S.-Y.; Lee, K.H. Novel 1,2,4-triazole analogues as mushroom tyrosinase inhibitors: Synthesis, kinetic mechanism, cytotoxicity and computational studies. Mol. Divers. 2020, 25, 2089–2106. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).