Abstract
Lung cancer is one of the common causes of cancer deaths. Early detection and treatment of lung cancer is essential. However, the detection of lung cancer in patients produces many false positives. Therefore, increasing the accuracy of the classification of diagnosis or true detection by computed tomography (CT) is a difficult task. Solving this problem using intelligent and automated methods has become a hot research topic in recent years. Hence, we propose a 2D convolutional neural network (2D CNN) with Taguchi parametric optimization for automatically recognizing lung cancer from CT images. In the Taguchi method, 36 experiments and 8 control factors of mixed levels were selected to determine the optimum parameters of the 2D CNN architecture and improve the classification accuracy of lung cancer. The experimental results show that the average classification accuracy of the 2D CNN with Taguchi parameter optimization and the original 2D CNN in lung cancer recognition are 91.97% and 98.83% on the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) dataset, and 94.68% and 99.97% on the International Society for Optics and Photonics with the support of the American Association of Physicists in Medicine (SPIE-AAPM) dataset, respectively. The proposed method is 6.86% and 5.29% more accurate than the original 2D CNN on the two datasets, respectively, proving the superiority of proposed model.
1. Introduction
Lung cancer is one of the critical diseases with rapidly rising morbidity and mortality rates, being the primary reason for cancer-related mortality around the world, with 1.8 million deaths annually []. Although the early detection rate has increased significantly, distinguishing malignant from benign tumors is one of the most challenging tasks [,]. Therefore, early and accurate diagnosis of lung cancer is essential. In recent years, many researchers applied machine learning models to detect and diagnose lung computed tomography (CT) images with the help of various computer assisted detection (CAD) systems, such as convolutional neural networks (CNNs), which demonstrated classification performance on medical images [,]. Liu et al. [] proposed a multi-view multi-scale CNN for lung nodule type classification from CT images. In another study, a CNN was integrated to assess pulmonary nodule malignancy in an automated existing pipeline of lung cancer detection []. However, this method is restricted to a limited number of visual parameters and the fixed value range. Thus, increasing the accuracy of lung cancer classification through CNN with parametric optimization will play a vital role in lung cancer recognition from CT images.
CT is widely used in screening procedures to reduce high mortality and to obtain many details about lung nodules and the surrounding structures. To effectively detect lung cancer from CT images, CNNs have demonstrated outstanding performance in applications including vision and images in the medical field in CAD systems []. A CNN model was used for classification, including three convolutional layers, three pooling layers, and two fully-connected layers []. Two-dimensional (2D) CNN has been applied in various fields with significant results such as image classification, face recognition, and natural language processing [,]. The design of the existing CAD requires the training of a large number of parameters, but parameters setting is complicated, so the parameters must be optimized to increase the accuracy of the classification []. Yunus and Alsoufi [] applied the Taguchi method to evaluate the output quality characteristics to predict the optimum parameters. In this study, we used the Taguchi method to find optimal parameter combinations based on the design of experiments and to solve the complicated parameters setting problem in CAD systems.
The Taguchi method is widely used for statistical analysis, and has been effectively used to help design experiments and obtain highly reliable results in studies. To optimize factors that affect the performance characteristics of a process or system, the Taguchi method reduces time and material costs []. The 2D CNN contains three convolutional, three max-pooling, and two fully-connected layers, and has the same deep learning architecture, but using a 2D CNN single view to train a large number of parameters might produce unstable predictions []. To alleviate this problem, we used a 2D CNN with the Taguchi method to find optimal parameter combinations.
For the purpose of overcoming the challenges associated with improving the classification performance of benign and malignant tumors from CT images, we propose a 2D CNN with Taguchi parametric optimization to address the complicated parameters setting problem and to increase the accuracy of classification. The objective of this study was to use the 2D CNN with Taguchi parametric optimization to determine the optimum parameters of the 2D CNN architecture and improve the lung cancer classification accuracy. The contribution is finding the optimal parameter combinations in the 2D CNN architecture on proven datasets to improve accuracy for lung cancer recognition with the help of various CAD systems.
2. Methods
The proposed 2D CNN with Taguchi parametric optimization to improve the classification performance and the proposed CAD system framework are shown in Figure 1. For the proposed CAD lung cancer detection system, we used three main steps. First, we use the Taguchi method to evaluate optimal parameter combinations for the 2D CNN model (Section 2.1). Next, we extract the input CT images to the 2D CNN model (Section 2.2). Third, we set the optimization parameters of the 2D CNN model to train and achieve malignant and benign tumor classification (Section 2.3).
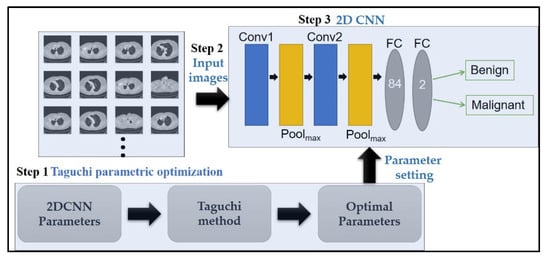
Figure 1.
The framework of the proposed computer assisted detection (CAD) system.
2.1. Taguchi Method
The Taguchi method is a statistical approach using an orthogonal array (OA), consisting of factors and levels, to classify the results to optimize process parameters []. In the Taguchi method, the experimental design using OA tables enables the optimization process. The results are converted to a signal-to-noise (S/N) ratio to determine the combination of control factor levels to improve system stability and reduce quality loss. In this study, considering the factors that were found to be effective on the 2D CNN parameter design, the parameters with eight factors and three levels are listed in Table 1. To reduce the number of experiments and improve the reliability of experiments, the L36(211,312) OA was selected for the experimental design, and 36 experimental runs generated by Minitab® 19 (Scientific Formosa Inc, Taipei, Taiwan) are given in Table 2.

Table 1.
Levels of control factors.

Table 2.
L36 orthogonal array (OA) for experiments the parameters setting.
According to the Taguchi optimization and the S/N ratio to quantify the output quality, the S/N ratio was selected as the optimization criterion. The best experimental results obtained by the calculation of average S/N ratio for each factor and the optimal level of the process parameters were indicated by the level with the largest S/N ratio. We analyzed the S/N ratio using three different performance characteristics: larger-is-better, nominal-is-better, and smaller-is-better []. In this study, ‘larger-is-better’ performance characteristics were preferred.
where n is the total number of replications of per test run and yi is the accuracy of the 2D CNN in the replication experiment for lung cancer recognition i executed under the same experimental conditions per test run.
We used analysis of variance (ANOVA) on the experimental data from the Taguchi method to determine the significance of process parameters statistically affecting extraction and the measurements [].
where T represents the sum of all observations, N is the total number of experiments, SST is the sum of squares of total change, SSA is the sum of squares due to factor A, KA is number of levels of factor A, Ai is the sum of all observations of level i of factor A, is the number of all observations at level i of factor A, DF is the degrees of freedom, MSA is the variance of the factor, and FA represents the F ratio of the factor.
2.2. Materials
The images used in this study were collected from lung cancer International Society for Optics and Photonics with the support of the American Association of Physicists in Medicine (SPIE-AAPM) Lung CT Challenge and Lung Image Database Consortium and Infectious Disease Research Institute (LIDC-IDRI) datasets, consisting of 22,489 CT and 244,617 CT scans 512 × 512 pixels in size, respectively [,]. The sample benign and malignant CT images of lungs from the LIDC-IDRI dataset for 2D CNN model training are shown in Figure 2 and Figure 3, and sample CT images of lungs from the SPIE-AAPM Lung CT Challenge dataset are shown in Figure 4. We used 70% of the CT image cases to train the 2D CNN model, and the other 30% for validation.
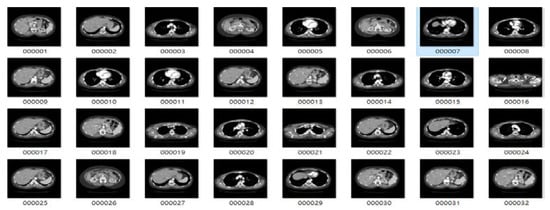
Figure 2.
Sample benign tumor computed tomography (CT) images from the Lung Image Database Consortium and Infectious Disease Research Institute (LIDC-IDRI) dataset.
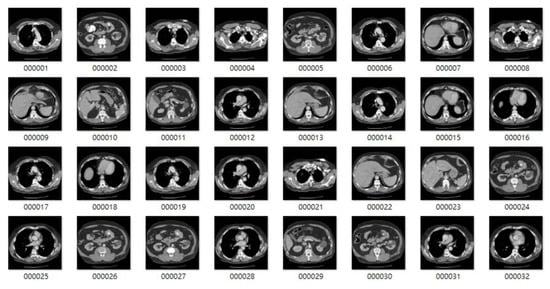
Figure 3.
Sample malignant tumor CT images from the LIDC-IDRI dataset.
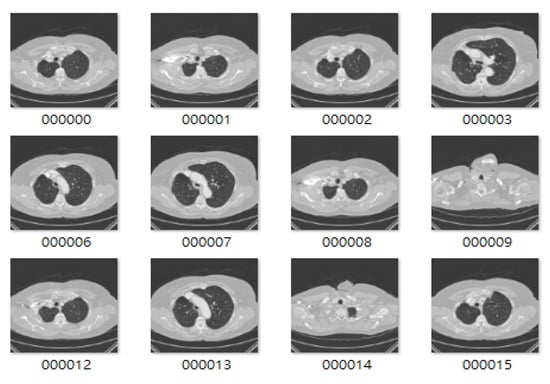
Figure 4.
Sample CT images of lungs from the International Society for Optics and Photonics with the support of the American Association of Physicists in Medicine (SPIE-AAPM) dataset.
2.3. 2D CNN Model
The 2D CNN designs the correct model of single views by reducing the bias of a single network, thereby effectively and accurately reducing the false positive rate of lung nodule []. This study proposes a 2D CNN architecture includes of two convolutional, two pooling and two fully-connected layers. The input image sizes 50 × 50 × 3, and the convolutional layer is based on the process of circulating a particular filter over the entire image. Kernel size, stride, padding, and filters can be different sizes, and the proposed 2D CNN architecture is described in Table 3.

Table 3.
The proposed 2D convolutional neural network (CNN) architecture.
We statistically analyzed the classification performance of the 2D CNN in terms of the Taguchi parametric optimization, accuracy, sensitivity, specificity, and false positive rate (FPR) derived from the confusion matrix. Accuracy is the correct prediction from the whole dataset, sensitivity is the ability of the test to correctly identify patients with the disease, and the specificity of a clinical test refers to the ability of the test to correctly identify patients without the disease. The following commonly used validation measurements are shown [].
where true positive (TP) is malignant correctly identified as malignant, false positive (FP) is benign incorrectly identified as malignant, true negative (TN) is benign correctly identified as benign, and false negative (FN) is malignancy incorrectly identified as benign.
3. Experimental Results
For implementing the proposed framework, we trained and validated the 2D CNN model with Taguchi parametric optimization both on LIDC-IDRI [] and SPIE-AAPM [] separately. The architecture of the proposed CAD system is shown in Figure 5.
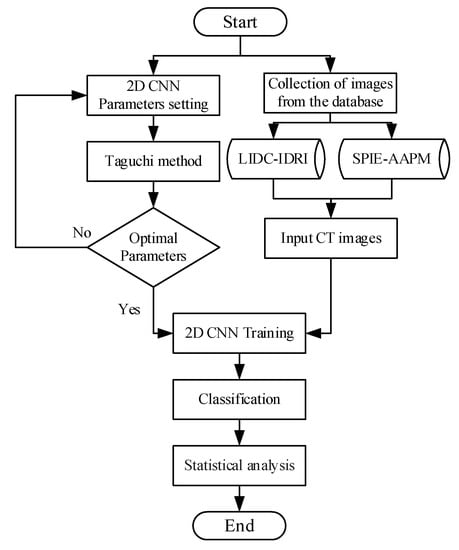
Figure 5.
Architecture of the proposed CAD system.
3.1. LIDC-IDRI
LIDC-IDRI [] consists of 1018 CT scans case, and 244,617 CT images. Pulmonary nodule diameters range from 3 to 30 mm, including only 4702 images of small nodules (<3 mm). For differential diagnostic imaging features, the larger the size (maximum diameter) of the nodule, the higher its malignancy: ≤8 mm is mostly benign and >8 mm is more likely malignant. Other types are very faint with smaller lesions called ground-glass opacity (GGO): ≤10 mm is mostly benign; >10 mm is more malignant. We performed experiments according to the combination of experiments in the OA table to improve the reliability of the experimental results. We obtained observations by combining experiments with three replications. The values of the parameters, the measured accuracies of the 2D CNN for lung cancer recognition, and S/N ratios in each run are listed in Table 4. The S/N ratio values of the accuracy were calculated by implementing of the-higher-the-better concept. A larger S/N ratio means less quality loss, that is, higher quality. Based on the results of the 36 runs, the maximum mean accuracy of 98.48% was achieved with Run #31, whereas the minimum mean accuracy of 78.29% occurred for Run #30.

Table 4.
The signal-to-noise (S/N) ratios of accuracy for different combinations of factors and levels.
The mean responses for the S/N ratios and the ANOVA table are shown in Table 5 and Table 6. Table 6 shows that four factors–C1_S, C2_S, C1_KS, and C2_F–have a significant impact. Table 5 and Figure 6 show that the factors affecting the S/N ratios based on yield extraction method are affected for each level of the parameters. The most significant factors can be determined by the larger difference in the S/N ratio. The experimental results indicate that factor C2_F has noticeable delta value of 0.845%, meaning that factor C2_F has the big significant parameter on model response. The importance of parameter was ranked from C2_F > C1_KS > C2_S > C2_P > C1_S > C2_KS > C1_F > C1_P. Hence, the best levels are A1 (level 1 for C1_S), B1 (level 1 for C1_P), C1 (level 1 for C2_S), D2 (level 2 for C2_P), E1 (level 1 for C1_KS), F1 (level 1 for C1_F), G3 (level 3 for C2_KS) and H3 (level 3 for C2_F). Optimum parameter C1_S is 1, C1_P is 0, C2_S is 1, C2_P is 1, C1_KS is 3, C1_F is 4, C2_KS is 7 and C2_F is 32.

Table 5.
Mean responses of the S/N ratios for each level and optimal parameter for the LIDC-IDRI dataset.

Table 6.
ANOVA for the S/N ratios of the accuracy on the LIDC-IDRI dataset.
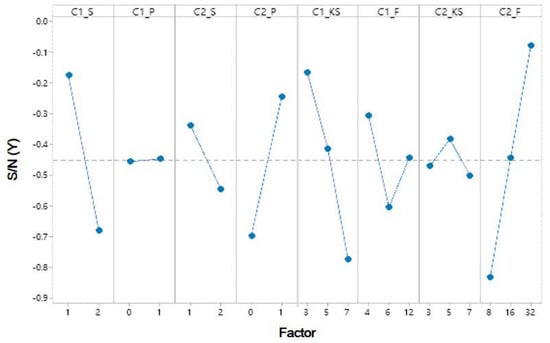
Figure 6.
Response graph of S/N ratio values of factor levels on the LIDC-IDRI dataset.
To compare our 2D CNN performance with Taguchi parametric optimization with the other neural networks such as deep fully convolutional neural network (DFCNet) [] and deep features + k-nearest neighbor (kNN) + minimum redundancy maximum relevance (mRMR) [] based on LIDC-IDRI dataset. The results are shown in Table 7. The best accuracy was 99.51% for the Deep features + kNN + mRMR, but the best sensitivity was produced by 2D CNN with Taguchi parametric optimization with a sensitivity of 99.97%, the best specificity of 99.93%, and FPR of 0.06%.

Table 7.
Recognition results of different techniques on the LIDC-IDRI dataset.
3.2. SPIE-AAPM
SPIE-AAPM [] consists of 22,489 CT images for 68 samples, including 31 benign and 37 malignant samples. The mean responses for the S/N ratios and ANOVA are shown in Table 8 and Table 9, respectively. Table 9 shows that four factors—C1_S, C2_P, C1_KS, and C2_F—have a significant impact. From Table 8 and Figure 7, the factors affecting the S/N ratios based on the yield extraction method are affected at each level of the parameters. The most significant factors can be determined by the larger differences in the S/N ratio. The experimental results indicated that factor C2_F has a noticeable delta value of 0.76%, meaning that factor C2_F is the most significant parameter in the model response. In terms of the importance, the parameters were ranked: C2_F > C1_KS > C1_S > C2_P > C1_F > C2_S > C2_KS > C1_P. Hence, the best levels are A1 (level 1 for C1_S), B2 (level 2 for C1_P), C1 (level 1 for C2_S), D2 (level 2 for C2_P), E1 (level 1 for C1_KS), F1 (level 1 for C1_F), G2 (level 2 for C2_KS) and H3 (level 3 for C2_F). Optimum parameter C1_S is 1, C1_P is 1, C2_S is 1, C2_P is 1, C1_KS is 3, C1_F is 4, C2_KS is 5 and C2_F is 32.

Table 8.
Mean responses for S/N ratios for each level and optimal parameter on the SPIE-AAPM dataset.

Table 9.
ANOVA for the S/N ratios of the accuracy on the SPIE-AAPM dataset.
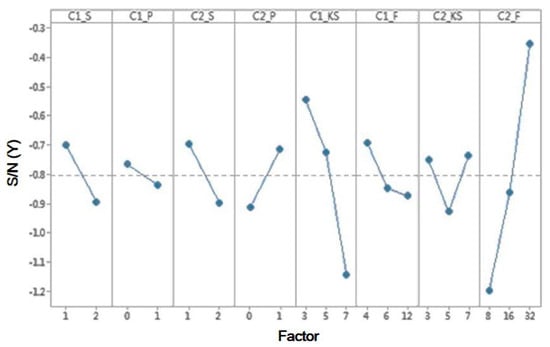
Figure 7.
Response graph of S/N ratio values of factor levels on the SPIE-AAPM dataset.
To evaluate the classification performance of the proposed 2D CNN with Taguchi parametric optimization, we selected DFCNet [] and back propagation neural network-variational inertia weight (BPNN-VIW) [] for comparison based on the SPIE-AAPM dataset. The results are provided in Table 10. The best sensitivity was 100% for BPNN-VIW, but the best accuracy was recorded for 2D CNN with Taguchi parametric optimization with an accuracy of 99.97%, the best specificity of 99.94%, and an FPR of 0.06%.

Table 10.
Recognition results of different techniques on the SPIE-AAPM dataset.
3.3. Discussion
We compared the performance of the proposed 2D CNN with Taguchi parametric optimization with the original 2D CNN model on the LIDC-IDRI and SPIE-AAPM datasets. We optimized the convolutional neural network model with new data on the LIDC-IDRI and SPIE-AAPM datasets using the best parameter configuration to improve accuracy. The comparison with the original accuracy, and parameters settings are shown in Table 11 and Table 12, respectively. On the LIDC-IDRI dataset, the original 2D CNN model attained an accuracy of 91.97%. After three more experiments with the best combination of parameters, the average accuracy rate was 98.83% (98.04%, 98.91%, and 99.53%). On the SPIE-AAPM dataset for the original 2D CNN model the accuracy was 94.68%. After three more experiments with the best combination of parameters, the average accuracy rate was 99.97% (100%, 99.90%, and 100%). Therefore, compared with the original model, the accuracy significant improved 6.86% and 5.29% for the two datasets, respectively. The results showed that the design model for 2D CNN with parametric optimization is necessary.

Table 11.
2D CNNs architecture parameters setting comparison on the LIDC-IDRI dataset.

Table 12.
2D CNNs architecture parameters setting comparison on the SPIE-AAPM dataset.
4. Conclusions
In this study, we constructed a 2D CNN Taguchi parametric optimization method for recognizing lung cancer from CT images. We successfully applied the Taguchi method to determine optimal 2D CNN architecture to increase the classification accuracy of lung cancer recognition, which was verified by running an eight-factor mixed-level experimental design. Additionally, our results identified the significant factors affecting the extraction yield. In this study, the LIDC-IDRI and SPIE-AAPM datasets were used to verify the proposed method. In the LIDC-IDRI dataset experimental results, the deep features + kNN + mRMR method for lung nodule type classification in a CAD system, the accuracy was 99.51%. Although this is better than the 98.83% result for the proposed method, the sensitivity, specificity, and FPR for the proposed method are superior to their results. In the SPIE-AAPM dataset experimental results, the best accuracy of the BPNN-VIW method in classifying lung CT was 97.2%. The proposed method had an accuracy of 99.97%, so it is better than the previously proposed CAD system. The experimental findings showed the proposed method provides increased classification accuracy, the essential model design with parametric optimization is necessary.
The classification accuracy of lung cancer using the original 2D CNN was 91.97% on the LIDC-IDRI dataset; the lung cancer classification accuracies for the three experiments with the proposed method were 98.04%, 98.91%, and 99.53%. Hence, the average accuracy rate of three observations of the proposed method was 98.83%. In addition, the lung cancer classification accuracy of the original 2D CNN was 94.68% on the SPIE-AAPM dataset; the accuracies were 100%, 99.90%, and 100% for the proposed method. Hence, the average accuracy rate of three observations of the proposed method was 99.97%. The accuracies of the proposed method were 6.86% and 5.29% higher than the original 2D CNN on the LIDC-IDRI and SPIE-AAPM datasets, respectively. The findings show that the proposed 2D CNN with Taguchi parametric optimization is more accurate for this task.
The limitation of this study is the depth of layers. However, the 2D CNN with Taguchi parametric optimization shows potential feature learning and is robust to variable sizes of training and testing datasets. Future studies should focus on the optimal size of the input patch for the deep learning algorithms development of new architectures to improve the computational efficiency and diagnosis of human diseases.
Author Contributions
C.-J.L. and S.-Y.J. conceived and designed the experiments; M.-K.C. performed the experiments; C.-J.L., S.-Y.J. and M.-K.C. analyzed the data; C.-J.L. and S.-Y.J. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Technology of the Republic of China, grant number MOST 108-2221-E-167 -026.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Togaçar, M.; Ergen, B.; Cömert, Z. Detection of lung cancer on chest CT images using minimum redundancy maximum relevance feature selection method with convolutional neural networks. Biocybern. Biomed. Eng. 2020, 40, 23–39. [Google Scholar] [CrossRef]
- Zhanga, S.; Hanb, F.; Lianga, Z.; Tane, J.; Caoa, W.; Gaoa, Y.; Pomeroyc, M.; Ng, K.; Hou, W. An investigation of CNN models for differentiating malignant from benign lesions using small pathologically proven datasets. Comput. Med Imaging Graph. 2019, 77. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, S.; Jiang, X.; Zhang, T.; Huang, H.; Ge, F.; Zhao, L.; Li, X.; Hu, X.; Han, J.; et al. The cerebral cortex is bisectionally segregated into two fundamentally different functional units of gyri and sulci. Cereb. Cortex 2019, 29, 4238–4252. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, H.; Huang, H.; Zhao, Y.; Jiang, X.; Bowers, B.; Guo, L.; Hu, X.; Sanchez, M.; Liu, T. Deep learning models unveiled functional difference between cortical gyri and sulci. IEEE Trans. Biomed. Eng. 2019, 66, 1297–1308. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Gu, Y.; Lv, X. Image classification toward lung cancer recognition by learning deep quality model. J. Vis. Commun. Image Represent. 2019, 78. [Google Scholar] [CrossRef]
- Bonavita, I.; Rafael-Palou, X.; Ceresa, M.; Piella, G.; Ribas, V.; Ballester, M.A.G. Integration of convolutional neural networks for pulmonary nodule malignancy assessment in a lung cancer classification pipeline. Comput. Methods Programs Biomed. 2020, 185. [Google Scholar] [CrossRef]
- Dolz, J.; Desrosiers, C.; Wang, L.; Yuan, J.; Shen, D.; Ayed, I.B. Deep CNN ensembles and suggestive annotations for infant brain MRI segmentation. Comput. Med Imaging Graph. 2020, 79. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated classification of lung Cancer types from cytological images using deep convolutional neural networks. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef]
- Trigueros, D.S.; Meng, L.; Hartnett, M. Enhancing convolutional neural networks for face recognition with occlusion maps and batch triplet loss. Image Vis. Comput. 2018, 79, 99–108. [Google Scholar] [CrossRef]
- Giménez, M.; Palanca, J.; Botti, V. Semantic-based padding in convolutional neural networks for improving the performance in natural language processing. A case of study in sentiment analysis. Neurocomputing 2020, 378, 315–323. [Google Scholar] [CrossRef]
- Suresh, S.; Mohan, S. NROI based feature learning for automated tumor stage classification of pulmonary lung nodules using deep convolutional neural networks. J. King Saud Univ. Comput. Inf. Sci. 2019. [Google Scholar] [CrossRef]
- Yunus, M.; Alsoufi, M.S. A Statistical Analysis of Joint Strength of dissimilar Aluminium Alloys Formed by Friction Stir Welding using Taguchi Design Approach, ANOVA for the Optimization of Process Parameters. IMPACT Int. J. Res. Eng.Technol. 2015, 3, 63–70. [Google Scholar]
- Canel, T.; Zeren, M.; Sınmazçelik, T. Laser parameters optimization of surface treating of Al 6082-T6 with Taguchi method. Opt. Laser Technol. 2019, 120. [Google Scholar] [CrossRef]
- Özakın, A.N.; Kaya, F. Experimental thermodynamic analysis of air-based PVT system using fins indifferent materials: Optimization of control parameters by Taguchi method and ANOVA. Sol. Energy 2020, 197, 199–211. [Google Scholar] [CrossRef]
- Özel, S.; Vural, E.; Binici, M. Optimization of the effect of thermal barrier coating (TBC) on diesel engine performance by Taguchi method. Fuel 2020, 263. [Google Scholar] [CrossRef]
- Rezania, A.; Atouei, S.A.; Rosendahl, L. Critical parameters in integration of thermoelectric generators and phase change materials by numerical and Taguchi methods. Mater. Today Energy 2020, 16. [Google Scholar] [CrossRef]
- Idris, F.N.; Nadzir, M.M.; Shukor, S.R.A. Optimization of solvent-free microwave extraction of Centella asiatica using Taguchi method. J. Environ. Chem. Eng. 2020. [Google Scholar] [CrossRef]
- Armato, S.G., III; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Clarke, L.P. Data from LIDC-IDRI. Cancer Imaging Arch. 2015. [Google Scholar] [CrossRef]
- Armato, S.G., III; Hadjiiski, L.; Tourassi, G.D.; Drukker, K.; Giger, M.L.; Li, F.; Redmond, G.; Farahani, K.; Kirby, J.S.; Clarke, L.P. SPIE-AAPM-NCI Lung Nodule Classification Challenge Dataset. Cancer Imaging Arch. 2015. [Google Scholar] [CrossRef]
- Eun, H.; Kim, D.; Jung, C.; Kim, C. Single-view 2D CNNs with fully automatic non-nodule categorization for false positive reduction in pulmonary nodule detection. Comput. Methods Programs Biomed. 2018, 165, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Masood, A.; Sheng, B.; Li, P.; Hou, X.; Wei, X.; Qin, J.; Feng, D. Computer-Assisted Decision Support System in Pulmonary Cancer detection and stage classification on CT images. J. Biomed. Inform. 2018, 79, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Nithila, E.E.; Kumar, S.S. Automatic detection of solitary pulmonary nodules using swarm intelligence optimized neural networks on CT images. Eng. Sci. Technol. Int. J. 2017, 20, 1192–1202. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).