Featured Application
The molecular methods here identified as the best performing tools to type K. humilis, S. cerevisiae and L. sanfranciscensis isolates could be further utilized for the selection of sourdough yeasts and LAB to be used as starters for the production of functional baked goods.
Abstract
The diverse metabolites, positively affecting the nutritional, organoleptic and technological traits of leavened baked goods, are produced by different sourdough lactic acid bacteria (LAB) and yeast strains, as the result of their genetic intraspecific diversity. Therefore, the molecular and functional strain-level characterization of sourdough microbiota is crucial to valorize traditional or origin protected baked end-products, develop innovative starter cultures and design functional cereal-based foods. To this aim, the genetic intraspecific diversity of 96 Lactobacillus sanfranciscensis, 65 Kazachstania humilis and three Saccharomyces cerevisiae characterizing Protected Designation of Origin (PDO) Tuscan bread sourdough, was investigated, using P4, P7 and M13 random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR), (GTG)5 repetitive element sequence-based (rep)-PCR and inter-delta region analyses, respectively. Regarding LAB, the combination of P4, P7 and M13 RAPD-PCR analyses revealed a huge degree of intraspecific variability, discriminating 43 biotypes out of 96 isolates of L. sanfranciscensis. (GTG)5 rep-PCR showed a discriminatory index of 0.95, grouping the 65 K. humilis isolated from PDO Tuscan bread sourdough in 9 biotypes. The high polymorphism among both LAB and yeast isolates of PDO Tuscan bread sourdough outlines a highly complex microbial community structure, whose relative composition and specific physiological characteristics could be responsible for the peculiar organoleptic, rheological, nutritional and potentially nutraceutical features of PDO Tuscan bread.
1. Introduction
Sourdough is a mixture of cereal flour and water, fermented by a complex biological ecosystem, consisting of different genera, species and strains of lactic acid bacteria (LAB) and yeasts, which interact among themselves, often establishing stable associations. During fermentation, yeasts are primarily responsible for the leavening, LAB for the acidification of the dough, and both produce a wide variety of compounds (organic acids, alcohols, aldehydes, esters, ketones, bacteriocins) that act in a synergistic way to prevent or eliminate deleterious microbial contaminations, improving the shelf-life of baked goods. Moreover, microbial compounds contribute to unique flavours and textures of the diverse fermented bread, enhancing also their nutritional and nutraceutical quality [1]. The production of bioactive metabolites varies significantly among the different LAB and yeast strains occurring in each sourdough [2,3], as the result of their genetic intraspecific diversity. Therefore, the molecular and functional strain-level characterization of sourdough microbiota is crucial to give value to traditional or origin protected baked end-products, to develop innovative starter cultures and to design functional cereal-based foods. To this aim, molecular tools for the intraspecific characterization of sourdough microbiota are fundamental, though there is no universally applicable typing method to discriminate among isolates from the same ecological niche, harbouring different microbial communities well adapted to the environment and often closely related among them [4].
Lattanzi et al. (2013) revealed the composition of the LAB microbiota of 18 Italian traditional sourdoughs, using random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) technique with P4, P7 and M13 primers [5]. Multiplex RAPD-PCR with two singly (P4 and MV1) and two combined (OPL-05 + RD1 and P1 + RD1) primers was utilized by Venturi et al. (2012) for typing Lactobacillus sanfranciscensis isolates from traditional Italian sourdoughs [6]. Another technique, which has been found particularly useful for differentiating sourdough LAB at the strain level, is repetitive element sequence-based (rep)–PCR using (GTG)5 primer. Such a method was applied by Liu et al. (2016) to analyse a total of 246 LAB strains isolated from 15 different samples of Chinese traditional sourdoughs [7].
Many studies have focused on the intraspecific characterization of LAB, while only a few investigated sourdough yeasts. Among DNA-fingerprinting methods, yeast intraspecific heterogeneity has been studied using RAPD- and rep-PCR. The former has been successfully used, with M13 and RF2 primers, to differentiate three distinct biotypes out of 58 Saccharomyces cerevisiae strains from southern Italian sourdoughs [8]. Recently, Palla et al. (2019) revealed a high polymorphism among the 77 S. cerevisiae isolates from Tuscan sourdoughs, using inter-delta region analysis [9]. Rep-PCR with (GACA)4 primer was utilized to reveal the composition of the yeast microbiota of 18 traditional Italian sweet sourdoughs and 19 typical Italian bread sourdoughs, respectively [5,10]. Vigentini et al. (2014), using intron splice site dispersion (ISS-PCR) and (GTG)5 rep-PCR analyses, discriminated different strains within the species Candida milleri (now reclassified as Kazachstania humilis) [11].
Considering that a high genetic intraspecific diversity of sourdough microbial communities may provide differential functional features affecting nutritional, organoleptic and technological properties of leavened baked goods, here, for the first time, we investigated the molecular diversity of Protected Designation of Origin (PDO) Tuscan bread sourdough microbiota at the strain level. To this aim, we assessed the diversity of 96 Lactobacillus sanfranciscensis, 65 Kazachstania humilis and 3 Saccharomyces cerevisiae, using P4, P7 and M13 RAPD-PCR, (GTG)5 rep-PCR and inter-delta region analyses, respectively, as the best discriminating molecular techniques.
2. Materials and Methods
2.1. Microorganisms
The 96 L. sanfranciscensis, 65 K. humilis and 3 S. cerevisiae isolates used in this study belong to the sourdough germoplasm bank of the PDO Tuscan bread (Official Journal of the European Union C 235 of 14 August 2013), maintained in the International Microbial Archives (IMA) of the Microbiology Labs of the Department of Agriculture, Food and Environment (DAFE) of the University of Pisa (Table 1) [12]. LAB were maintained in the modified De Man, Rogosa and Sharpe (mMRS) broth medium [13] obtained by adding 2% maltose and 5% fresh yeast extract and adjusted to pH 5.6, and yeasts were maintained in the Wallerstein Laboratory Nutrient (WLN) broth medium (Oxoid, Basingstoke, UK), both supplemented with 20% (w/v) glycerol, at −80 °C.

Table 1.
Lactic acid bacteria and yeast strains used in this study.
2.2. Molecular Intraspecific Diversity of LAB Isolates Characterizing the PDO Tuscan Bread Sourdough
In order to discriminate the L. sanfranciscensis isolates at the strain level, a preliminary identification of the best performing molecular technique was carried out on 12 isolates (IMA 2LAB, 13, 15, 23, 24, 27, 59, 63, 83, 87 93, 98), which were previously selected on the basis of their variability to solubilize phytate and of their polymorphism within 16S rRNA gene [12]. LAB isolates were analysed using different fingerprinting techniques: (GTG)5 rep-PCR and RAPD-PCR using different sets of primers. The best discriminating technique was then used to study the intraspecific diversity of the 96 L. sanfranciscensis isolates, along with the reference strain L. sanfranciscensis DSMZ 20451T.
The rep-PCR analysis was carried out using 160 ng of DNA, as described by Liu et al. (2016) [7]. The reaction conditions were: 94 °C initial denaturation for 5 min, 40 amplification cycles of 15 s at 94 °C, 30 s at 55 °C, 90 s at 72 °C, final extension at 72 °C for 4 min. The first RAPD-PCR tested was carried out using two single reactions with P4 (5’-CCG CAG CGT T- 3’) and MV1 (5’-GGA CGC TTC TG-3’) primers, and two multiplex reactions with the combined OPL-05 (5’-ACG CAG GCA-3’) + RD1 (5’-GCT TAA GGA GGT GAT CCA GCC-3’) and P1 (5’-ACG CGC CCT-3’) + RD1 primers, as described by Venturi et al. (2012) [6]. The second RAPD-PCR tested, was performed using three single reaction with P4 (5’-CCG CAG CGT T- 3’), P7 (5’-AGC AGC GTG G-3’) and M13 (5’-GAG GGT GGC GGT TCT-3’) primers, as described by Lattanzi et al. (2013), except for the primer concentration (5 μM) and for the PCR conditions: 94 °C initial denaturation for 1 min, 40 amplification cycles of 1 min at 94 °C, 20 s at 40 °C, 2 min at 72 °C, final extension at 72 °C for 5 min [5]. All PCR amplifications were carried out with an iCycler-iQ Multicolor Real-Time PCR Detection System (Bio-Rad, Milan, Italy), using Taq DyNAzyme II DNA polymerase and 10X DyNAzyme Buffer Mg2+ free (Finnzymes, Thermofisher, Milan, Italy). All primers were bought at Eurofins Genomics, Ebersberg, Germany. The presence of amplicons was confirmed by electrophoresis in 1.8% (w/v) agarose gel stained with 0.5 µg/mL REALSAFE Nucleic Acid Staining Solution (20000x, REAL, Durviz S.L., Valencia, Spain), run in 1X TBE at 65 V for 2 h. A 100 bp DNA ladder (New England BioLabs) was added on each side and in the centre of agarose gels, in order to better align patterns from different gels. All gels were visualized by UV and captured as TIFF format files by the UVI 1D v. 16.11a program for FIRE READER V4 gel documentation systems (Uvitec Cambridge, Eppendorf).
LAB profiles were digitally processed and analysed with BioNumerics software ver. 7.6 (Applied Maths, St-Martens-Latem, Belgium). Profiles were compared using the band matching tool with a position tolerance and optimization of 1%. For cluster analysis, the similarity was calculated using Dice’s coefficient and unpaired group method with arithmetic average (UPGMA) trees were constructed. In particular, RAPD profiles were processed under a unique dendrogram, by combining the patterns from the different PCR amplification, while the other profiles were analyzed individually. The cophenetic correlation coefficient was used as a statistical method to estimate the error associated with dendrogram branches, while the Cluster Cutoff method was applied to define the most reliable cluster.
The reproducibility of fingerprints was assessed by comparing the combined fingerprinting patterns, obtained using DNA prepared from two separate cultures of the same ten randomly selected strains (L. sanfranciscensis IMA 3LAB, 18, 27, 45, 55, 56, 76, 94, 98 and DSMZ 20451T). All pairs of duplicates were amplified in different PCR reactions and the average similarity coefficient was calculated as described by Eriksson et al. (2005) [14].
To estimate the typing efficiency of the three different molecular techniques, a numerical index of discrimination (D) was calculated as described by Hunter and Gaston (1988) [15].
2.3. Molecular Intraspecific Diversity of Yeast Isolates Characterizing the PDO Tuscan Bread Sourdough
In order to discriminate yeast isolates at the strain level, a preliminary identification of the best performing molecular technique was carried out on nine isolates (K. humilis IMA 1Y, 8, 13, 33, 37, 122, S. cerevisiae IMA 19Y, 36 and 105) along with K. humilis DBVPG 6753T, which were previously selected on the basis of functional analysis results [12]. Such yeasts were analysed using the following fingerprinting techniques: restriction fragment length polymorphism analysis of mitochondrial DNA (RFLP mtDNA), mitochondrial COX1 gene introns amplification, inter-delta regions amplification, RAPD-PCR and two different rep-PCR analyses. The best discriminating techniques were then used to study the intraspecific diversity of the 65 K. humilis and of the 3 S. cerevisiae isolates, separately.
For the analysis of RFLP mtDNA, mitochondrial DNA from yeast isolates was extracted and digested using HinfI and RsaI endonucleases (New England BioLabs), as described by Agnolucci et al. (2009) [16]. Restriction fragments were separated in 0.8% (w/v) agarose gel electrophoresis in 1X TBE buffer at 100 V for 3 h and then stained in a water bath containing 0.5 µg/mL of REALSAFE Nucleic Acid Staining Solution (20000x, REAL, Durviz S.L., Valencia, Spain). The multiplex amplification reaction of mitochondrial COX1 gene introns was carried out using 100 ng of the mitochondrial DNA as described by Lopez et al. (2003) [17]. Inter-delta regions amplification was performed using 160 ng of DNA following the method reported by Palla et al. (2019) [9]. RAPD-PCR was carried out by two single reactions with M13 (5’-GAG GGT GGC GGT TCT-3’) and RF2 (5’-CGG CCC CTG T- 3’) primers, using the same conditions described by Succi et al. (2003) [8]. For the first rep-PCR, the primer (GTG)5 was used, and the amplification reaction was performed as described by Vigentini et al. (2014) [11]. The second one was performed with (GACA)4 primer, according to Andrade et al. (2006) PCR protocol [18].
All PCR amplifications were carried out as previously reported. The presence of amplicons was confirmed by electrophoresis in 1.8% (w/v) agarose gel stained with 0.5 µg/mL REALSAFE Nucleic Acid Staining Solution (20000x, REAL, Durviz S.L., Valencia, Spain), run in 1X TBE at 65 V for 2 h, using a 100 bp DNA ladder (New England BioLabs) as molecular marker. Finally, all gels were visualized and digitally processed by BioNumerics software ver. 7.6 (Applied Maths, St-Martens-Latem, Belgium), as previously described.
The reproducibility of fingerprints was assessed by comparing the fingerprinting patterns, obtained using DNA prepared from two separate cultures of the same five randomly selected strains (K. humilis IMA 22Y, 24, 27, 51 and 104). All pairs of duplicates were amplified in different PCR reactions and run in different electrophoresis gels. The average similarity coefficient from duplicates was calculated as described by Eriksson et al. (2005) [14].
In order to estimate the typing efficiency of the different molecular techniques, Discriminatory index (D) was calculated.
3. Results
3.1. Molecular Intraspecific Diversity of LAB Isolates Characterizing the PDO Tuscan Bread Sourdough
The preliminary test performed using 3 different molecular techniques was carried out on 12 L. sanfranciscensis isolates. Profiles obtained using rep-PCR with (GTG)5 primer were digitally processed. The resulted dendrogram grouped the isolates into two main clusters with a similarity level of 50% (Figure 1), showing a discriminatory index of 0.82 and was able to discriminate 6 different biotypes. RAPD-PCR profiles obtained using two single reactions with P4 and MV1 primers, and two multiplex reactions with the combined OPL-05 + RD1 and P1 + RD1 primers were compared and analysed. The dendrogram, created by combining the four different RAPD profiles, grouped the isolates into two main clusters with a similarity level of 63% (Figure 1), showing a discriminatory index of 0.98 and was able to discriminate 11 different biotypes. Finally, RAPD-PCR profiles, obtained using three single reactions with P4, P7 and M13 primers, were combined to create a dendrogram, which grouped the isolates into two main clusters with a similarity level of 79%, and clustered L. sanfranciscensis IMA 23LAB separately, at a similarity of 65% (Figure 1). Such a method showed a discriminatory index of 0.95 differentiating 10 different biotypes.
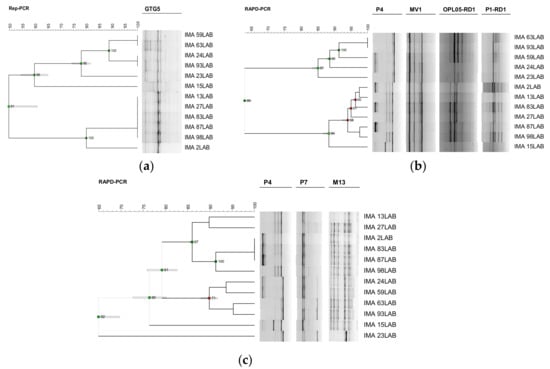
Figure 1.
Screening of the best discriminating tool carried out on 12 selected L. sanfranciscensis isolates characterizing Protected Designation of Origin (PDO) Tuscan bread sourdough. Dendrograms obtained from unpaired group method with arithmetic average (UPGMA) analysis, using Dice’s coefficient, of (GTG)5 repetitive element sequence based-polymerase chain reaction (rep-PCR) profiles (a), of combined random amplified polymorphic DNA (RAPD)-PCR profiles, obtained using P4, MV1, OPL-05 + RD1 and P1 + RD1 primers (b) and using P4, P7 and M13 primers (c). The scale indicates the similarity level. Cophenetic correlation is shown at each branch by numbers and coloured dots, ranging between green-yellow-orange-red, according to decreasing values.
Comparing the discriminatory indices obtained from the three molecular techniques, both RAPD-PCR methods showed high discrimination performance. Although RAPD-PCR with P4, MV1, OPL-05 + RD1 and P1 + RD1 primers was able to discriminate one biotype more than RAPD-PCR with P4, P7 and M13 primers, the latter clustered the isolates in accordance with the polymorphism differences observed within 16S rRNA gene [12] and was used to analyse the intraspecific diversity of the 96 L. sanfranciscensis isolates. Such isolates, along with the reference strain L. sanfranciscensis DSMZ 20451T, were analysed combining RAPD profiles obtained with each primer and considering 81.47% as the similarity value for separation of biotypes. Such a method was able to discriminate 43 biotypes out of the 96 LAB isolates, revealing high biodiversity among L. sanfranciscensis strains characterizing PDO Tuscan bread sourdough (Figure 2).
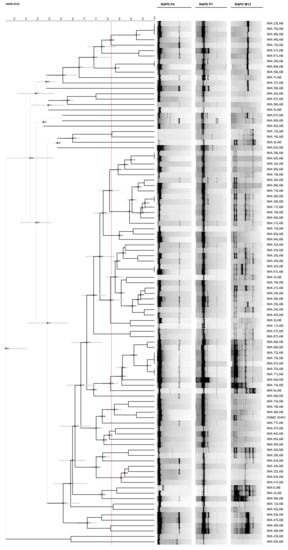
Figure 2.
Dendrogram obtained from UPGMA analysis, using Dice’s coefficient, based on P4, P7 and M13 random amplified polymorphic DNA (RAPD) profiles, of the 96 L. sanfranciscensis isolates characterizing PDO Tuscan bread sourdough and one reference strain. The red line indicates the similarity value (81.47%) for separation of biotypes. Standard deviation is shown at each node by a grey bar. Cophenetic correlation is shown at each branch by numbers and coloured dots, ranging between green-yellow-orange-red, according to decreasing values.
3.2. Molecular Intraspecific Diversity of Yeast Isolates Characterizing the PDO Tuscan Bread Sourdough
A preliminary test performed using 6 different molecular techniques was carried out on 7 selected K. humilis and on the only 3 S. cerevisiae strains isolated. COX1 gene introns amplification and RFLP mtDNA analyses were unable to discriminate neither among K. humilis nor among S. cerevisiae isolates (data not shown). Profiles obtained using inter-delta regions amplification analysis originated the dendrogram reported in Figure 3, discriminating S. cerevisiae from K. humilis isolates, at a similarity level of 43%. Such a method discriminated 5 different biotypes among the 7 K. humilis isolates, showing a discriminatory index of 0.90. The dendrogram, created by combining the two RAPD profiles obtained using M13 and RF2 primers, grouped K. humilis isolates into two main clusters with a similarity level of 93%, discriminating only 3 different biotypes (Figure 3). The discriminatory index resulted in 0.67. The cluster analysis of profiles obtained using rep-PCR with (GTG)5 primer grouped the K. humilis isolates at a similarity level of 78% with K. humilis DBVPG 6753T (Figure 3). Such a technique showed a discriminatory index of 0.95 and allowed the detection of 6 different biotypes, revealing a high ability of discrimination among K. humilis isolates. The cluster analysis of profiles obtained using rep-PCR with (GACA)4 primer grouped K. humilis isolates in two main clusters at a similarity level of 46%, discriminating 4 different biotypes. The discriminatory index resulted in 0.81 (Figure 3).
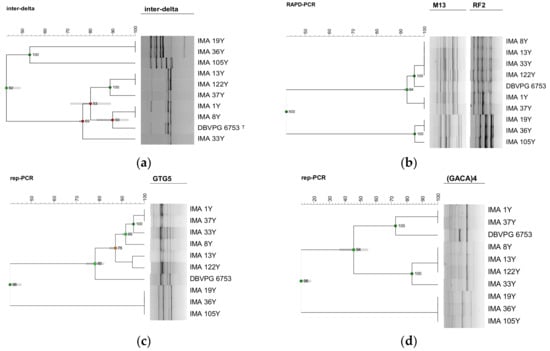
Figure 3.
Screening of the best discriminating tool carried out on 10 selected yeast isolates characterizing PDO Tuscan bread sourdough. Dendrogram obtained from UPGMA analysis, using Dice’s coefficient, of inter-delta profiles (a), RAPD-PCR profiles, obtained using M13 and RF2 primers (b), (GTG)5 (c) and (GACA)4 (d) rep-PCR profiles. The scale indicates the similarity level. Cophenetic correlation is shown at each branch by numbers and coloured dots, ranging between green-yellow-orange-red, according to decreasing values.
Comparing the discriminatory index obtained by the 6 molecular techniques, rep-PCR with (GTG)5 primer resulted in the best differentiation tool for the typization of K. humilis isolates and was used to characterize at strain level the 65 K. humilis isolates, considering 93.78% as the similarity value for separation of biotypes. Results showed the occurrence of 9 biotypes out of the 65 K. humilis isolates characterizing PDO Tuscan bread sourdough (Figure 4).
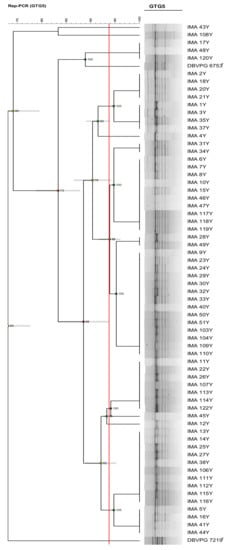
Figure 4.
Dendrogram obtained from UPGMA analysis, using Dice’s coefficient, based on (GTG)5 rep-PCR profiles, of the 65 K. humilis isolates characterizing PDO Tuscan bread sourdough and two reference strains. The red line indicates the similarity value (93.78%) for separation of biotypes. Standard deviation is shown at each node by a grey bar. Cophenetic correlation is shown at each branch by numbers and coloured dots, ranging between green-yellow-orange-red, according to decreasing values.
Among the 6 typing methods screened, only the inter-delta regions amplification (Figure 3) and the M13 and RF2 RAPD-PCR (Figure 3) techniques differentiated two biotypes out of the three S. cerevisiae isolates, showing that S. cerevisiae IMA 19Y and IMA 36Y belonged to the same biotype, and S. cerevisiae IMA 105Y was a different one. As inter-delta regions amplification differentiated the two biotypes at a lower similarity level (53%) than RAPD-PCR (96%), the former technique resulted in the best tool for the typization of our S. cerevisiae isolates.
4. Discussion
Here, the molecular characterization at the strain level of K. humilis, S. cerevisiae and L. sanfranciscensis isolates occurring in PDO Tuscan bread sourdough was performed. Rep–PCR and inter-delta region analyses revealed the highest intraspecific diversity in K. humilis and S. cerevisiae, respectively, while the combination of three RAPD-PCR analyses was the best discriminating method for L. sanfranciscensis.
The DNA fingerprinting methods rep-PCR with (GTG)5 primer and RAPD-PCR with two different sets of primers, used to characterize L. sanfranciscensis isolates, showed a high discriminatory power, particularly the two multiplex RAPD-PCR techniques (D ≥ 0.95). RAPD-PCR with P4, P7 and M13 primers clustered the isolates in accordance with the polymorphism observed within 16S rRNA gene and revealed a large degree of intraspecific variability among L. sanfranciscensis strains isolated from PDO Tuscan bread sourdough. Actually, the occurrence of only one LAB species, L. sanfranciscensis, may have boosted a marked intraspecific differentiation. Similar findings were found by Kitahara et al. (2005), who, studying the LAB communities of 5 different sourdoughs, found L. sanfranciscensis intraspecific diversity only in the sourdough where such microorganism was the only LAB species [19].
Concerning the yeast community of PDO Tuscan bread sourdough, the intraspecific characterization has been carried out comparing 6 different molecular methods. Inter-delta regions amplification resulted in the best discrimination tool for the typization of S. cerevisiae, differentiating two biotypes among the three S. cerevisiae isolates. Similar results were obtained by Osimani et al. (2009), who used this method to investigate the intraspecific diversity of the S. cerevisiae isolates characterizing the sourdough from the Marche region [20]. Also, Pulvirenti et al. (2004) found a high degree of polymorphism among S. cerevisiae isolated from Sicilian sourdoughs, using inter-delta region analysis [21]. Conversely, Venturi et al. (2012), studying S. cerevisiae strains isolated from Legaccio and Panettone mother sponges, revealed only one inter-delta profile for each sourdough [22].
The rep-PCR with (GTG)5 primer method showed the best discriminatory index (0.95) for K. humilis. Cluster analyses grouped the 65 K. humilis in 9 biotypes, revealing a high degree of intraspecific polymorphism. Accordingly, a high genetic diversity at the strain level within C. milleri and Candida humilis (reclassified as K. humilis) isolated from traditional Italian sourdoughs, using (GTG)5 microsatellite analyses was reported [11]. By contrast, a very low polymorphism among C. milleri isolates of traditional sourdoughs was detected using MV1-RAPD PCR [22].
In conclusion, our work showed a high degree of genetic diversity among LAB and yeast strains of PDO Tuscan bread sourdough. Such highly complex microbial communities could be further studied for their functional traits conferring peculiar organoleptic, rheological and nutritional properties to PDO Tuscan bread. Actually, preliminary tests carried out on these isolates revealed the ability to solubilize phytate in 18 out of the 96 L. sanfranciscensis strains, in the 3 S. cerevisiae and in 3 out of 65 K. humilis isolates, 50% of which showed also protease activity [12]. The molecular and functional characterization of PDO Tuscan bread sourdough microbiota will allow the selection of autochthonous Tuscan sourdough yeasts and LAB to be used as starters in the years to come.
Author Contributions
Conceptualization, M.P., M.G. and M.A.; methodology, C.C.; software, M.P. and C.C.; validation, M.P. and M.A.; writing—original draft preparation, M.P.; writing—review and editing, M.A. and M.G.; supervision, M.A. and M.G.; funding acquisition, M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Italian Ministry of Education, University and Research (MIUR) programme PRIN 2015 (grant number 2015SSEKFL) “Processing for healthy cereal foods”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hutkins, R. Fermented vegetables. In Microbiology and Technology of Fermented Foods, 1st ed.; Hutkins, R., Ed.; IFT Press Blackwell Publishing: Oxford, UK, 2006; Volume 22, pp. 223–259. [Google Scholar]
- Dolci, P.; Alessandria, V.; Rantsiou, K.; Cocolin, L. Advanced methods for the identification, enumeration, and characterization of microorganisms in fermented foods. In Advances in fermented foods and beverages: Improving quality, technologies and health benefits; Holzapfel, W., Ed.; Woodhead Publishing Series in Food Science; Technology and Nutrition: Cambridge, UK, 2015; Volume 265, pp. 157–176. [Google Scholar]
- Gänzle, M.G.; Vermeulen, N.; Vogel, R.F. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 2007, 24, 128–138. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Tassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 2007, 13, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Lattanzi, A.; Minervini, F.; Di Cagno, R.; Diviccaro, A.; Antonielli, L.; Cardinali, G.; Cappelle, S.; De Angelis, M.; Gobbetti, M. The lactic acid bacteria and yeast microbiota of eighteen sourdoughs used for the manufacture of traditional Italian sweet leavened baked goods. Int. J. Food Microbiol. 2013, 163, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Venturi, M.; Guerrini, S.; Granchi, L.; Vincenzini, M. Typing of Lactobacillus sanfranciscensis isolates from traditional sourdoughs by combining conventional and multiplex RAPD-PCR profiles. Int. J. Food Microbiol. 2012, 156, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Chen, J.; Sadiq, F.A.; Zhang, G.; Li, Y.; He, G. Prevalence and diversity of lactic acid bacteria in Chinese traditional sourdough revealed by culture dependent and pyrosequencing approaches. LWT Food Sci. Technol. 2016, 68, 91–97. [Google Scholar] [CrossRef]
- Succi, M.; Reale, A.; Andrighetto, C.; Lombardi, A.; Sorrentino, E.; Coppola, R. Presence of yeasts in Southern Italian sourdough from Triticum aestivum flour. FEMS Microbiol. Lett. 2003, 225, 143–148. [Google Scholar] [CrossRef][Green Version]
- Palla, M.; Agnolucci, M.; Calzone, A.; Giovannetti, M.; Di Cagno, R.; Gobbetti, M.; Rizzello, C.G.; Pontonio, E. Exploitation of autochthonous Tuscan sourdough yeasts as potential starters. Int. J. Food Microbiol. 2019, 302, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Di Cagno, R.; Lattanzi, A.; De Angelis, M.; Antonielli, L.; Cardinali, G.; Cappelle, S.; Gobbetti, M. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 2012, 78, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Vigentini, I.; Antoniani, D.; Roscini, L.; Comasio, A.; Galafassi, S.; Picozzi, C.; Corte, L.; Compagno, C.; Dal Bello, F.; Cardinali, G.; et al. Candida milleri species reveals intraspecific genetic and metabolic polymorphisms. Food Microbiol. 2014, 42, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Identification and characterization of lactic acid bacteria and yeasts of PDO Tuscan bread sourdough by culture dependent and independent methods. Int. J. Food Microbiol. 2017, 250, 19–26. [Google Scholar] [CrossRef] [PubMed]
- De Man, J.C.; Rogosa, D.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Eriksson, J.; Löfströma, C.; Aspán, A.; Gunnarsson, A.; Karlsson, I.; Borch, E.; De Jonge, B.; Rådström, P. Comparison of genotyping methods by application to Salmonella livingstone strains associated with an outbreak of human salmonellosis. Int. J. Food Microbiol. 2005, 104, 93–103. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
- Agnolucci, M.; Vigentini, I.; Capurso, G.; Merico, A.; Tirelli, A.; Compagno, C.; Foschino, R.; Nuti, M. Genetic diversity and physiological traits of Brettanomyces bruxellensis strains isolated from Tuscan Sangiovese wines. Int. J. Food Microbiol. 2009, 130, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Lopez, V.; Fernández-Espinar, M.T.; Barrio, E.; Ramón, D.; Querol, A. A new PCR-based method for monitoring inoculated wine fermentations. Int. J. Food Microbiol. 2003, 81, 63–71. [Google Scholar] [CrossRef]
- Andrade, M.J.; Rodriguez, M.; Sánchez, B.; Aranda, E.; Córdoba, J.J. DNA typing methods for differentiation of yeasts related to dry-cured meat products. Int. J. Food Microbiol. 2006, 107, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, M.; Sakata, S.; Benno, Y. Biodiversity of Lactobacillus sanfranciscensis strains isolated from five sourdoughs. Lett. Appl. Microbiol. 2005, 40, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Osimani, A.; Zannini, E.; Aquilanti, L.; Mannazzu, I.M.; Comitini, F.; Clementi, F. Lactic acid bacteria and yeasts from wheat sourdoughs of the Marche region. Ital. J. Food Sci. 2009, 21, 269–286. [Google Scholar]
- Pulvirenti, A.; Solieri, L.; Gullo, M.; De Vero, L.; Giudici, P. Occurrence and dominance of yeast species in sourdough. Lett. Appl. Microbiol. 2004, 38, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Venturi, M.; Guerrini, S.; Vincenzini, M. Stable and non-competitive association of Saccharomyces cerevisiae, Candida milleri and Lactobacillus sanfranciscensis during manufacture of two traditional sourdough baked goods. Food Microbiol. 2012, 31, 107–115. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).