Abstract
Despite the increasing role played by artificial intelligence methods (AI) in pharmaceutical sciences, model deployment remains an issue, which only can be addressed with great difficulty. This leads to a marked discrepancy between the number of published predictive studies based on AI methods and the models, which can be used for new predictions by everyone. On these grounds, the present paper describes the Tree2C tool which automatically translates a tree-based predictive model into a source code with a view to easily generating applications which can run as a standalone software or can be inserted into an online web service. Moreover, the Tree2C tool is implemented within the VEGA environment and the generated program can include the source code to calculate the required attributes/descriptors. Tree2C supports various programming languages (i.e., C/C++, Fortran 90, Java, JavaScript, JScript, Lua, PHP, Python, REBOL and VBScript and C-Script). Along with a detailed description of the major features of this tool, the paper also describes two examples which are aimed to predict the blood–brain barrier (BBB) permeation as well as the mutagenicity. They permit a clear evaluation of the potentials of Tree2C and of its related features as implemented by the VEGA suite of programs. The Tree2C tool is available for free.
1. Introduction
The increasing amount of available digital data has been paralleled by the increasing demand for effective methods for exploring them. Consequently, artificial intelligence (AI) methods (mostly based on machine (ML) and deep learning (DL) approaches) are finding countless applications not only in scientific contexts but in everyday life, ranging from business automation to self-driving cars, as well (e.g., [1,2]).
The remarkable success that AI methods have enjoyed in the scientific community has been fostered by the high number of available tools to develop predictive models based on ML or DL approaches. These tools comprise standalone programs (e.g., Weka [3]), which allow the generation of AI-based predictive models in a rather easy (and assisted) way without requiring a clear mastery of their founding principles, as well as programming libraries (e.g., Keras or TensorFlow in Python [4]), which allow more skilled users to generate and apply predictive models with a more accurate control of the parameters influencing their performances. In particular, medicinal chemistry has experienced a huge increase in the available chemical and biological data, mostly arising from high-throughput screening (HTS) campaigns and omics disciplines, and has strongly benefitted from the AI approaches [5,6], which find successful applications in activity prediction, compound characterization, de novo drug design and absorption, distribution, metabolism, excretion (ADME) profiling [7,8].
Similarly to what was observed in many other fields, the countless number of highly performing models, which are routinely published in the specialized scientific literature, is not paralleled by the (markedly more limited) number of models which can be easily utilized for new predictions: relatively few models are indeed available as online webservers, and even fewer models are delivered as standalone applications. As a matter of fact, the transformation of a predictive model into an application that everyone can use (the so-called model deployment) is always a problematic step [9]. Such difficulty can be explained by considering different factors, among which the most common cause can be ascribed to professional computer science skills which are required to develop an application which should be stable, portable and easy to use to permit an extensive use by the scientific community. Within a multidisciplinary environment, one may imagine that the developed model can be handed out to a specialized team for its deployment, while the model dissemination often remains an unmanageable task in more limited contexts. As a consequence of this scenario, many published models cannot be used by the readers and are exploited only within the scientific group which has developed them or for retrospective analyses.
On these grounds, the study reports the Tree2C program, which converts tree-based models into source codes which require no or very limited modifications to be compiled and then used for new predictions. In particular, Tree2C reads the input classification models as described by a formatted scheme which is based on that produced by the Weka program [3], but can be easily generated also by other predictive tools. Next, Tree2C generates the corresponding source code supporting several programming languages (e.g., C, C++, Fortran 90, Java, JavaScript, JScript, Lua, PHP, Python, REBOL and VBScript). When utilized within the VEGA environment [10], Tree2C recognizes the molecular attributes/descriptors which can be directly calculated, and adds the code required to calculate them.
Together with the key features of the Tree2C program, the paper reports its application in two exemplificative ML-based predictive studies, which are aimed to predict the blood–brain barrier (BBB) permeation and the mutagenicity as based on Li’s and Bursi’s datasets, respectively [11,12]. Along with the availability of suitable databases and reference studies, these two examples were chosen by considering the remarkable medicinal interest in predicting both the capacity of a given molecule to penetrate the CNS as well as its potential toxicity concerns as exemplified by mutagenicity. In more detail, the capacity to predict the BBB permeation based on the physicochemical properties of a given permeant is of crucial relevance to assist the rational design of molecules developed for neurological disorders. To this end, different approaches have been applied including standard linear regressions [13], machine learning methods [14] or tailored algorithms (as seen with the recent BBB score [15]). Notably, some of the developed predictive models are freely available as online services [16].
Similarly, the prediction of potential toxicity concerns relating to novel molecules is of crucial relevance to minimize the health risks during drug development [17], as well as to assure the overall safety of the industrial chemicals [18]. In detail, in silico mutagenicity prediction has gained a key role in the last few years, since this can also be used for regulatory purposes, provided that the developed models assure appropriate robustness and predictive power with a well-defined applicability domain and considered endpoint [19]. Hence, the remarkable number of predictive studies recently published comes as no surprise: the applied computational techniques include classic QSAR methods, rule-based methods, AI algorithms and consensus approaches [20].
By describing the entire process required to develop and to deploy the corresponding models, these predictive exercises offer a meaningful exemplification of the potentials of Tree2C and of the related features implemented by the VEGA suite of programs.
2. Materials and Methods
2.1. The Tree2C Implementation
In this section, the implementation and the main features of the here-proposed Tree2C tool are described in depth. The section is subdivided into three parts which parallel the three major modules of the program, by discussing: (1) the features of the required input tree model, (2) the way by which Tree2C generates the code, and (3) the key characteristics of the produced source code.
2.1.1. Tree2C: The Input Model
As explained above, the input file required by the Tree2C program is based on the output generated by the Weka software when performing a tree-based classifier (e.g., random tree, random forest) [21]. This input format was chosen because it is a text file and thus can be quite easily (and even manually) generated and/or modified when using other statistical programs. In detail, the required input file can be subdivided into two parts: (1) a header with the information about the attributes and the learning approach (Run information section); (2) one or more than one decision tree (Classifier model section). With regard to the first Run information section, Tree2C requires only two tags, namely Scheme and Attributes. The former is used by the program to recognize the file and to detect the model type. In these tags, Weka also reports the type of the tree and the parameters used for its generation, but Tree2C does not require these data. The Attribute tag reports the number of attributes and optionally their list. Since Tree2C can directly extract the attributes from the next section, the list is not necessary. The second section begins with the “Classifier model” label and comprises one or more than one decision tree. The “RandomTree” label indicates the beginning of each tree. The tree is drawn from left to right and the splitting nodes are not indicated but are placed virtually in the middle of the segment built by multiple pipe characters (“|”). At the end of two branches of the fork, there is a leaf represented by an attribute and a condition, which can be either a threshold or a Boolean value.
2.1.2. Tree2C: How It Works
The program code is organized in three logical modules: (1) input modules; (2) processing module; (3) output modules. In detail, there are two input modules: the ARFF loader and the tree decoder. The former loads the data from the ARFF file [22] used to develop the model and which Tree2C uses to generate the code to check the applicability domain (see below). The latter reads the tree output file and decodes the decision trees, the number of which depends on the method used to build the model. Moreover, this module finds the attributes, which are directly involved in the model and are a subset of those included in the ARFF file. Each decoded tree is stored as a list of nodes hierarchically ordered according to the distance from the root of the tree. Each node includes the logical condition, defined by (1) an attribute, (2) a logical operator, and (3) a threshold value. The condition must be fulfilled to split the branch and to move to the next node or to yield the final prediction if no branch is further available. Next, the processing module checks if all the tree attributes are comprised within the ARFF file, renames them, removing the characters which are not compatible with some programming languages (e.g., spaces, dashes, punctuation marks, etc.), and calculates the corresponding applicability domain. Moreover, the processing module sets the correct output module according to the selected programming language which can be object-oriented (C++, Fortran 90, Java, JavaScript, JScript, Lua, PHP, Python, REBOL and VBScript), non-object-oriented (C) and C-Script. Finally, all the output modules translate the node lists of the decision trees into a set of IF THAN ELSE conditions and (if requested) include the source code required to determine the applicability domain according to the syntax of the selected programming language. Moreover, the output module for the C-Script, which is the scripting language implemented by the VEGA environment, can add the code required to calculate the attributes by the host program and eventually by MOPAC7 or MOPAC2016 [23]. Since C-Scripts are ready-to-run and can be directly used within the VEGA environment to perform predictions, the output module can also install the generated scripts in the VEGA environment to be easily and promptly used for new predictions.
2.1.3. Tree2C: The Output Source Code
This section provides a brief description of the output source code developed by Tree2C to deploy the input model. For the sake of simplicity and as an example, the code description is focused on the C language, while the corresponding details for the other supported languages can be found in the online Tree2C manual [24]. All constants, data structures and functions are named according to the rule by which each object has the “T2C_MODEL_NAME_” prefix, where “MODEL_NAME” is derived by capitalizing the name of the used tree file. Along with some constants used to initialize the required calculations, the only data type defined in the source code is that required as the input for the functions, which perform the prediction and the analysis of the applicability domain (see below). This is named T2C_MODEL_NAME_INPUT and can include Boolean and discrete attributes, which are defined as integer numbers, while the other parameters are defined as single precision floating point numbers. Two main functions are included in the generated source code and represent the core of the program: T2C_MODEL_NAME_Classify (T2C_MODEL_NAME_INPUT *Input) and T2C_MODEL_NAME_DomCheck (T2C_SUPER_INPUT *Input). The former is always present in the code generated by Tree2C and performs the classification based on the required input attributes included in T2C_MODEL_NAME_INPUT structure, which have to be pre-calculated. This function returns the predicted class as an integer number (usually 0 and 1) according to the given input data. The second function is optional and checks if the attributes used for the prediction are in the same domain of those used to build the model. The parameters are the same as those of the previous function and the function returns the number of domain violations. It ranges from 0 (= no violations) to the total number of the attributes included in the classification model.
2.2. Computational Details
As mentioned in the Introduction, two applications are here included to better illustrate the potentials of Tree2C. In both cases, the utilized datasets are retrieved as lists of SMILES strings from the reference papers [11,12]. By using the features implemented in the VEGA environment [25], all SMILES strings were automatically converted into 3D structures and then optimized in their neutral form by PM7 semi-empirical calculations [26], as implemented in MOPAC2016 (keywords: PM7 PRECISE GEO-OK SUPER). Based on the prepared molecules, 129 properties/attributes were calculated by both VEGA and MOPAC2016 and their values were exported into the ARFF file format. By using the Weka program, the most significant attributes were selected according to both the BestFirst search algorithm (direction = Forward; lookupCacheSize = 1; searchTermination = 5) and the WrapperSubsetEval attribute evaluator (classifier = RandomForest with default settings; doNotCheckCapabilities = False; evaluationMeasure = accuracy, RMSE; folds = 5; seed = 1; threshold = 0.01).
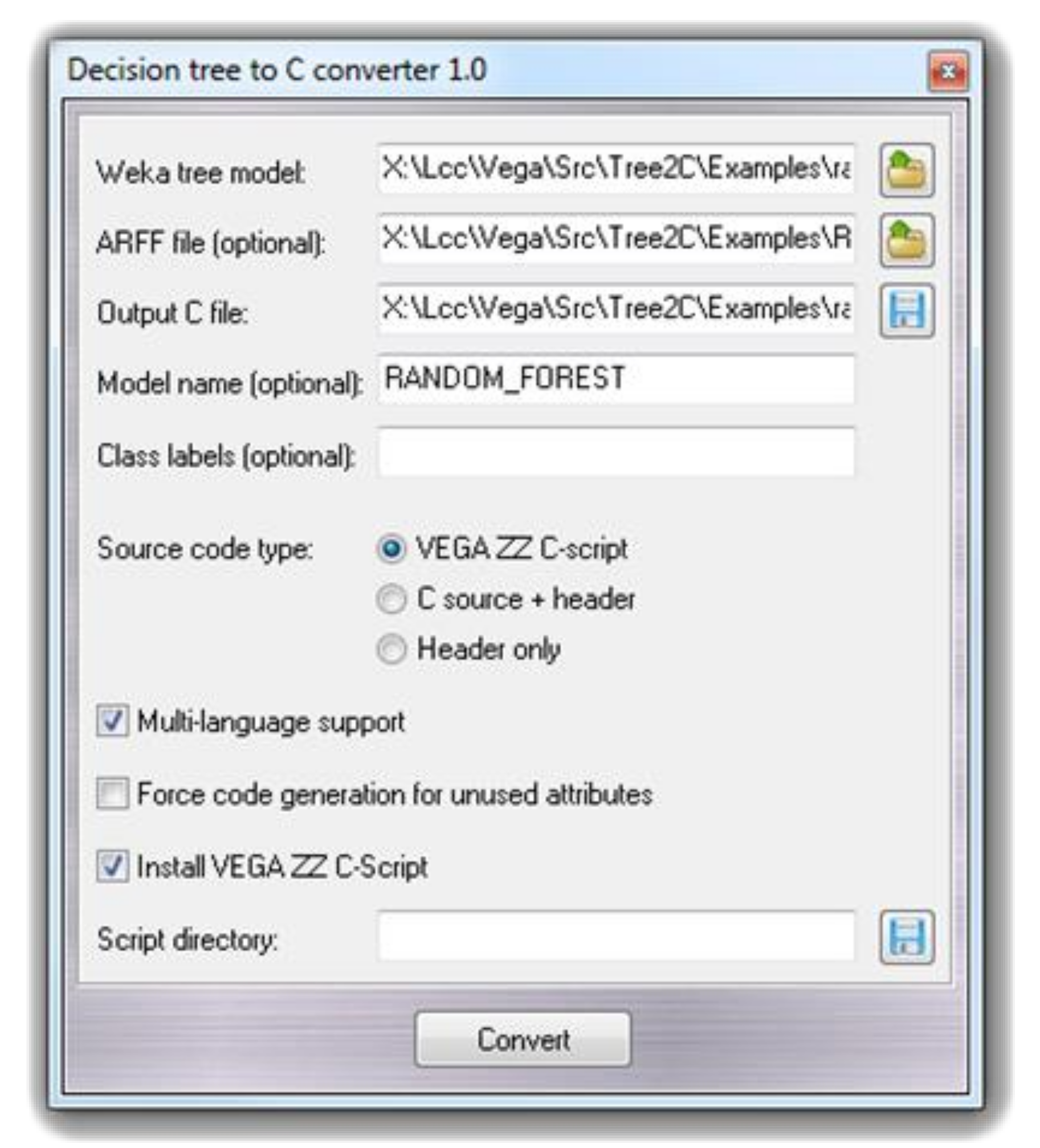
The final model was developed by the random forest machine learning algorithm as implemented in Weka with default parameters (bagging with 100 iterations and base learner) and performing a 10-fold cross-validation. The Weka output files along with the input ARFF files are then utilized by the Tree2C tool to generate the source code which deploys the developed models to be used for new predictions. Tree2C can be run either outside the VEGA environment by command line according to a set of options, which can be seen in the manual, or within the VEGA environment by using a specific graphical interface. To better illustrate this second option, Figure 1 shows the Tree2C graphical interface as implemented in the VEGA environment. The user has to define the required input files (the ARFF file is required only to determine the applicability domain) and the output source code file. The user can assign a name to the model and can select if the output code should be a VEGA C-script or should be compiled as a standalone tool.
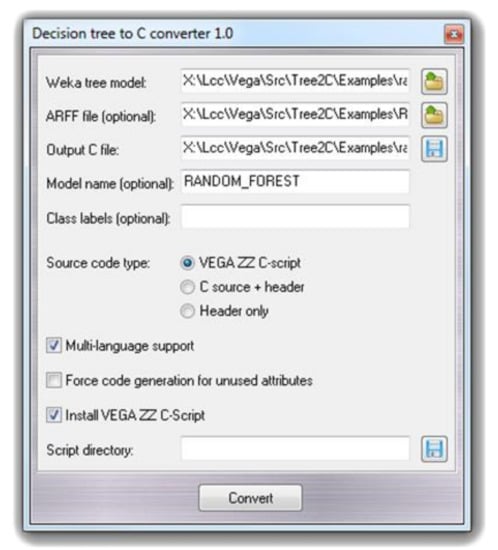
Figure 1.
The VEGA ZZ graphical user interface for Tree2C.
3. Results
3.1. Prediction of the BBB Permeation
The first example concerns the prediction of the BBB permeation based on Li’s dataset, which includes 415 compounds, 276 permeants and 139 non-permeants [11]. Among the 129 computed descriptors, the variable selection collected only nine attributes: they include size/shape descriptors (such as mass, or volume), the log P as computed by the Molecular Lipophilicity Potential (MLP) approach [27], three Kier–Hall E-state descriptors [28] and the total charge, which encodes for the quaternary ammonic molecules included in the dataset. The selected descriptors are in agreement with the above-mentioned BBB score [15] and confirms that molecular size and polarity play a crucial role. The final model obtained by the random forest algorithm shows satisfactory results as confirmed by the 83.4% of correctly predicted molecules with a Matthews correlation coefficient (MCC) equal to 0.620 and an area under the curve (AUC) equal to 0.865. Table 1 summarizes the performances for each class, where 0 and 1 indicate non-permeant and permeant molecules, respectively.

Table 1.
Predictive performances reached by the model for the blood–brain barrier (BBB) permeation (0 and 1 indicate non-permeant and permeant molecules, respectively).
The obtained results are in line with those reported by Li in the reference paper. In detail, Li developed the predictive model by support vector machine (SVM) with recursive feature elimination (RFE) [29] reaching an 83.7% of correctly predicted molecules with an MCC value of 0.645. The major difference between the two models is that the here-proposed model includes only descriptors, while that of Li required 35 attributes. In detail, the here-proposed model does not include quantum mechanical parameters, thus markedly reducing the computational costs without affecting the overall predictive power. Finally, both models comprise Kier–Hall E-state descriptors, but the here-proposed model includes only three descriptors, while the SVM + RFE model requires 17 Kier-Hall E-state descriptors.
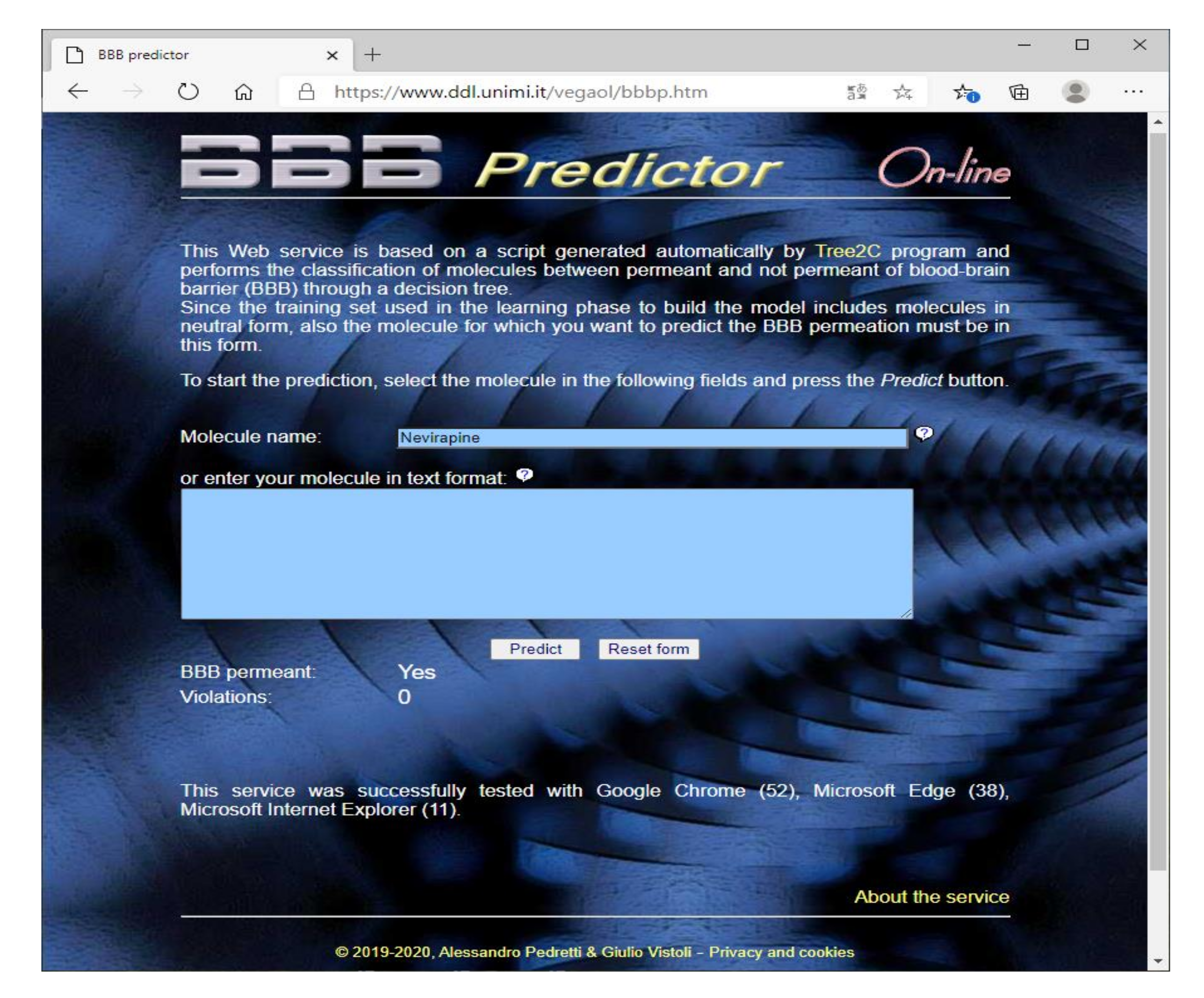
Along with the source codes/script routinely generated by Tree2C, the predictive model was also used to develop a web service based on a PHP script generated by Tree2C (https://www.ddl.unimi.it/vegaol/bbbp.htm) in order to predict the BBB permeation. As depicted by Figure 2, this requires as input the molecular structure in one of the file formats supported by VEGA, or only the molecule name. In this last case, the structure is directly downloaded from PubChem by its PUG REST interface [30]. The VEGA engine then calculates the molecular descriptors required to perform the prediction by the decision tree encoded by the PHP code.
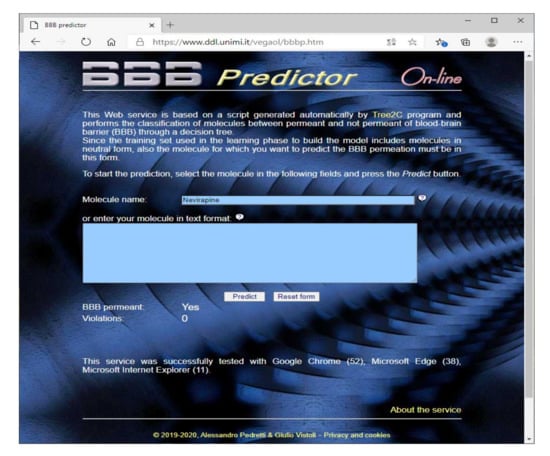
Figure 2.
The graphic interface of the web service based on the code generated by Tree2C to predict the BBB permeation.
3.2. Prediction of Mutagenicity
The second example aimed to predict the mutagenicity and involved the dataset collected by Bursi and co-workers including 4337 molecular structures subdivided into 2401 mutagens and 1936 non-mutagens based on the corresponding Ames test data [12]. Here, the variable selection kept 24 attributes which include (1) stereo-electronic parameters (such as LUMO Energy and Parr and Pople absolute hardness [31]) which encode for the reactivity/electrophilicity of the molecule; (2) constitutive descriptors (such as number of unsaturated systems, number of H-bonding groups) which may indirectly encode for the presence of reactive moieties; (3) 13 Kier–Hall E-state descriptors which reasonably play the same role as described for constitutive attributes [28].
The final model obtained by the random forest algorithm reaches satisfactory results, as confirmed by an MCC equal to 0.644 and an AUC equal to 0.896. Table 2 summarizes the performances for each class, where 0 and 1 indicate non-permeant and permeant molecules, respectively, and shows very similar performances for both classes. In detail, the model correctly predicted 3575 molecules out of 4337, with an overall accuracy equal to 82.43%, truly superimposable to that obtained by the original study (82.5%). For this second example, Tree2C was utilized to generate the corresponding C-script embedded within the VEGA environment by which new predictions can be easily performed. Notice that since the predictive model comprises stereo-electronic descriptors, the corresponding script also includes the source code required to calculate them by linking the script with the MOPAC2016 program. The interested reader can find the corresponding source code in the Supporting Material (File S1).

Table 2.
Predictive performances reached by the model for the mutagenicity (0 and 1 indicate non-permeant and permeant molecules, respectively).
4. Conclusions
In summary, we here present a tool which supports the deployment of predictive models based on tree algorithms by transforming the modelling output into a source code which can either be utilized within the VEGA environment or as part of a web service or a standalone program. By exploiting the VEGA features, the generated source code can automatically calculate a rich arsenal of molecular descriptors. Nevertheless, and with limited modifications, the source code can be rendered independent by the VEGA functions and can become a versatile tool in all the contexts, which require a rapid deployment and dissemination of the developed predictive models. This is not to mention that some supported languages are particularly effective in developing web services, thus making Tree2C a fruitful tool to develop online resources. The Tree2C tool is freely available at www.vegazz.net.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/21/7704/s1, File S1: C source code of the mutagenicity predictor.
Author Contributions
A.P. conceived the study and wrote the source code of Tree2C, S.G. and A.M. performed the included calculations, G.V. coordinated the study and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abduljabbar, R.; Dia, H.; Liyanage, S.; Bagloee, S.A. Applications of Artificial Intelligence in Transport: An Overview. Sustainability 2019, 11, 189. [Google Scholar]
- Nemitz, P. Constitutional democracy and technology in the age of artificial intelligence. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2018, 376, 20180089. [Google Scholar]
- Smith, T.C.; Frank, E. Introducing Machine Learning Concepts with WEKA. Methods Mol. Biol. 2016, 1418, 353–378. [Google Scholar]
- Rampasek, L.; Goldenberg, A. TensorFlow: Biology’s Gateway to Deep Learning? Cell Syst. 2016, 27, 12–14. [Google Scholar]
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A., Jr.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020, 19, 353–364. [Google Scholar]
- Yang, X.; Wang, Y.; Byrne, R.; Schneider, G.; Yang, S. Concepts of Artificial Intelligence for Computer-Assisted Drug Discovery. Chem. Rev. 2019, 119, 10520–10594. [Google Scholar]
- Mazzolari, A.; Afzal, A.M.; Pedretti, A.; Testa, B.; Vistoli, G.; Bender, A. Prediction of UGT-mediated Metabolism Using the Manually Curated MetaQSAR Database. ACS Med. Chem. Lett. 2019, 10, 633–638. [Google Scholar]
- Šícho, M.; Stork, C.; Mazzolari, A.; de Bruyn Kops, C.; Pedretti, A.; Testa, B.; Vistoli, G.; Svozil, D.; Kirchmair, J. FAME 3: Predicting the Sites of Metabolism in Synthetic Compounds and Natural Products for Phase 1 and Phase 2 Metabolic Enzymes. J. Chem. Inf. Model. 2019, 59, 3400–3412. [Google Scholar]
- Coiera, E. The Last Mile: Where Artificial Intelligence Meets Reality. J. Med. Internet Res. 2019, 21, e16323. [Google Scholar]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA—An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 2004, 18, 167–173. [Google Scholar]
- Li, H.; Yap, C.W.; Ung, C.Y.; Xue, Y.; Cao, Z.W.; Chen, Y.Z. Effect of selection of molecular descriptors on the prediction of blood-brain barrier penetrating and nonpenetrating agents by statistical learning methods. J. Chem. Inf. Model. 2005, 45, 1376–1384. [Google Scholar]
- Kazius, J.; McGuire, R.; Bursi, R. Derivation and validation of toxicophores for mutagenicity prediction. J. Med. Chem. 2005, 48, 312–320. [Google Scholar]
- Morales, J.F.; Montoto, S.S.; Fagiolino, P.; Ruiz, M.E. Current State and Future Perspectives in QSAR Models to Predict Blood-Brain Barrier Penetration in Central Nervous System Drug R&D. Mini Rev. Med. Chem. 2017, 17, 247–257. [Google Scholar]
- Saxena, D.; Sharma, A.; Siddiqui, M.H.; Kumar, R. Blood Brain Barrier Permeability Prediction Using Machine Learning Techniques: An Update. Curr. Pharm. Biotechnol. 2019, 20, 1163–1171. [Google Scholar]
- Gupta, M.; Lee, H.J.; Barden, C.J.; Weaver, D.F. The Blood-Brain Barrier (BBB) Score. J. Med. Chem. 2019, 62, 9824–9836. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar]
- Wu, F.; Zhou, Y.; Li, L.; Shen, X.; Chen, G.; Wang, X.; Liang, X.; Tan, M.; Huang, Z. Computational Approaches in Preclinical Studies on Drug Discovery and Development. Front. Chem. 2020, 8, 726. [Google Scholar]
- Rim, K.T. In silico prediction of toxicity and its applications for chemicals at work. Toxicol. Environ. Health Sci. 2020, 12, 191–202. [Google Scholar]
- Honma, M. An assessment of mutagenicity of chemical substances by (quantitative) structure-activity relationship. Genes Environ. 2020, 42, 23. [Google Scholar]
- Benigni, R.; Bossa, C. Data-based review of QSARs for predicting genotoxicity: The state of the art. Mutagenesis 2019, 34, 17–23. [Google Scholar]
- Frank, E.; Hall, M.A.; Witten, I.H. The WEKA Workbench. Online Appendix for Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Burlington, MA, USA, 2016; Available online: https://www.cs.waikato.ac.nz/ml/weka/Witten_et_al_2016_appendix.pdf (accessed on 30 October 2020).
- ARFF Format. Available online: https://waikato.github.io/weka-wiki/formats_and_processing/arff/ (accessed on 30 October 2020).
- James, J.P. MOPAC2016, Stewart, Stewart Computational Chemistry, Colorado Springs, CO, USA. Available online: http://OpenMOPAC.net (accessed on 30 October 2020).
- Tree2C. Classification Tree to Code Converter. Available online: https://www.ddl.unimi.it/manual/utilities/tree2c.htm (accessed on 30 October 2020).
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA: A versatile program to convert, handle and visualize molecular structure on Windows-based PCs. J. Mol. Graph. Model. 2002, 21, 47–49. [Google Scholar]
- Stewart, J.J. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013, 19, 1–32. [Google Scholar]
- Gaillard, P.; Carrupt, P.A.; Testa, B.; Boudon, A. Molecular lipophilicity potential, a tool in 3D QSAR: Method and applications. J. Comput. Aided Mol. Des. 1994, 8, 83–96. [Google Scholar]
- Hall, L.H.; Mohney, B.; Kier, L.B. The electrotopological state: Structure information at the atomic level for molecular graphs. J. Chem. Inf. Comput. Sci. 1991, 31, 76–82. [Google Scholar]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar]
- Parr, R.G.; Chattaraj, P.K. Principle of maximum hardness. J. Am. Chem. Soc. 1991, 113, 1854–1855. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).