Simple Summary
This study developed and validated a rapid, accurate, and cost-effective genetic sex identification method for the freshwater fish Opsariichthys bidens. Using whole-genome resequencing and the chromosome quotient approach, 45 male-specific genomic regions were identified. Among 50 designed primer pairs, the Mar28 primer pair consistently distinguished males from females across multiple populations, amplifying two bands in males and only one band in females. This marker provides a practical tool for monosex breeding and aids further research on sex determination in O. bidens.
Abstract
Sex-specific markers are important basic tools for the sex-controlled breeding of farmed fish. Here, we aimed to develop a rapid yet accurate, cost-effective method for determining the genetic sex of the Chinese hooksnout carp (Opsariichthys bidens), a freshwater fish. Using whole-genome resequencing technology, along with bulked segregant analysis (BSA) and chromosome quotient (CQ) methods, sex-specific regions were screened, and corresponding primers were designed to validate the screening results. A total of 45 sex-specific regions were successfully screened through BSA sequencing and CQ analysis, and 50 pairs of primers were designed for use in the screening verification. The Mar28 primer pair showed stable sex specificity in multiple populations of O. bidens, accurately distinguishing male from female individuals. This primer pair amplified two bands (509 and 814 bp) in males, but only one band (509 bp) in females. The genetic sex identification method established here provides a theoretical basis for studying the mechanism of sex determination in O. bidens, has implications for the monosex culture and molecular breeding of O. bidens, and has significant scientific and practical value.
1. Introduction
Fish species have complex modes of sex determination, namely genetic sex determination, environmental sex determination, or a combination of both [1,2]. The fish sex chromosome system is intricate and diverse, harboring almost all the vertebrate sex chromosome types, the two most common being male heterozygosity (XX/XY) and female heterozygosity (ZW/ZZ) [3]. Many fish species are sexually dimorphic, with males and females differing markedly in growth rate. For example, males of Mastacembelus armatus [4] and Channa argus [5] grow faster than females. Conversely, females of Siniperca chuatsi [6] and Siniperca scherzeri [7] grow faster than conspecific males. Hence, understanding how sex is determined in fish is critically important in aquaculture because it enables the production of all-male (or all-female) juveniles (monosex culture), which increases the yield per unit of culture. Sex control and single-sex culture have long been key topics in fisheries and aquaculture worldwide, and sex-specific molecular markers have always been at the forefront of fish breeding research. The international community has long sought sex-specific molecular markers and reliable genetic sex determination methods for different fish species [8].
Whole-genome resequencing involves the resequencing of the entire genome of a species at the chromosome level. Comparison with the reference genome permits inherent variation in the whole genome to be detected and yields molecular genetic information for individuals or populations [9]. Compared with de novo genome assembly, whole-genome resequencing has various advantages, including low sequencing costs and the simplicity of bioinformatics analyses; it has thus become the main approach for the development of sex identification markers [9,10]. This method has yielded sex-specific markers for various aquaculture fish species, such as Megalobrama amblycephala, Platichthys stellatus, and Mastacembelus armatus [9,10,11,12,13]. Bulked segregant analysis (BSA) is based on genome-wide resequencing technology and can quickly identify specific genes as well as regions of variation within the genome, in addition to screening out markers associated with certain phenotypes; hence, it is a simple and efficient trait mapping technique [14]. Sex identification markers have been developed successfully for fish such as Micropterus salmoides and Trachinotus ovatus using BSA [15,16].
The Chinese hooksnout carp (Opsariichthys bidens) in the order Cypriniformes, family Cyprinidae, is named because of the gap and protrusions in its upper and lower jaw, which resemble a horse’s mouth. This freshwater fish is widely distributed in East Asia and is often found in clear, fast-flowing shoals, sandy bottom streams, and river tributaries, where it feeds on small fish, shrimp, and aquatic insects [17]. Owing to its rapid growth and plump, tender flesh, O. bidens is an economically important aquaculture species with high aquaculture value. This species shows sexual dimorphism, with males growing faster than females, and males have a blue–green pattern during the breeding period with high ornamental value; females have an ordinary body color [18,19]. Therefore, the male-only culture of O. bidens could significantly improve both its yield and economic benefits. However, because males and females are not readily distinguishable prior to adulthood, rapid and efficient sex identification markers are needed. Earlier work reported that RAPD primers derived from gonadal DNA could amplify bands distinguishing the sex of O. bidens, but this requires gonadal tissue, which is difficult to extract from young fish [20]. The complete genome of O. bidens has been sequenced [18,21], and its XX/XY sex determination system [22] is conducive to the development of new sex determination techniques. Recently, genome-wide resequencing technology was used in a comparative study of the male and female genomes of O. bidens. A male-specific insertion fragment was detected, from which a PCR-based sex identification method was then constructed; the marker was amplified in males only, not in females [23]. A sex-linked SNP marker in O. bidens has been reported, which is heterozygous in males and homozygous in females, and is located within an exonic region [24]. Nevertheless, more sex identification markers are needed to aid sex-controlled breeding and facilitate further research on the mechanisms of sex determination and differentiation in O. bidens.
In the present study, mixed pool sequencing was performed on male and female O. bidens based on whole-genome resequencing. The chromosome quotient (CQ) method was used to screen for regions with sex-specific sequences. The CQ method maps resequencing reads to a reference genome and then slides a window across the alignment to count the number of mapped reads in each bin. Multiple pairs of primers were designed for each region, and their sex specificity in the O. bidens population was verified through PCR amplification. This yielded sex identification markers for O. bidens, which amplified two bands in males but just one band in females. Our findings provide a theoretical reference for future research on sex determination mechanisms and the sex-controlled breeding of O. bidens and have practical implications for enhancing breeding management.
2. Materials and Methods
2.1. Sample and DNA Extraction
The samples used for resequencing and primer screening were derived from an artificially bred population with a known genetic background in Jiangxi Province. In June 2023 (breeding season: June–September), 25 males (body length 13.02 ± 0.82 cm, body weight 24.94 ± 4.69 g) and 25 females (body length 12.24 ± 0.54 cm, body weight 19.39 ± 1.69 g) were selected for sequencing; males and females were distinguished via morphology and inspection of their gonads after dissection. The tail fin of each fish was immediately flash-frozen and then stored at ultralow temperatures until the DNA was extracted. This DNA was subsequently used to create a mixed pool for BSA sequencing. The fish populations used for primer validation were collected from Jiangxi, Zhejiang, and Fujian during the breeding season (20 male and 20 female adults per population). To extract the genomic DNA of O. bidens, the BayBiopure magnetic bead animal tissue genomic DNA extraction kit was used (BayBio, Guangzhou, China). The amount of genomic DNA was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and its quality was assessed via 0.7% agarose gel electrophoresis. Care of animals was in compliance with the guidelines of the Animal Experiment Committee, Zhejiang Institute of Freshwater Fishery (ZJIFF20230603).
2.2. Library Construction and BSA Sequencing
The qualified genomic DNA samples of female and male O. bidens were mixed in equal amounts, resulting in two genomic DNA pools for females and males, respectively. Their DNA samples were broken into short fragments (200–400 bp each), using a Covaris ultrasonic breaker (50 hertz, 20 min) (Covaris, Woburn, MA, USA). After undergoing terminal repair, poly(A) tails and sequencing adapters were attached, and this was followed by purification and PCR amplification; the resulting PCR products were then cyclized to complete the library construction. Next, the DNA concentration was determined using a Qubit 3.0 fluorometer (Life Technologies, Carlsbad, CA, USA), and the library was quantified. The library’s insert size was detected using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) to confirm that the size of the inserted fragment was as expected. The precise effective concentration of the library was subsequently determined using qPCR to ensure the overall quality of the library. Finally, each pooled library was sequenced on the DNBSEQ platform, with each pool producing approximately 20 GB of data.
2.3. Initial Data Processing and Quality Assessment
Initial image data files were converted into raw sequencing reads via base calling, and clean reads were generated using fastp (v0.20.1) for quality control with six filtering criteria [25]. The obtained clean reads from all samples were aligned to the O. bidens reference genome using BWA software (v0.7.17) [26]. The alignment results were converted from SAM files into sorted BAM files, from which any duplicates were removed using SAMtools software (v1.19) [27]. The alignment rate and coverage were then calculated using a Python (v3.12) script.
2.4. Screening of Sex-Specific Regions
Based on the resulting data (BAM file) generated by BSA and the reference genome (GWHBJYU00000000, https://ngdc.cncb.ac.cn/gwh/Assembly/26062/show, accessed on 30 September 2023), the chromosome quotient (CQ) was used to screen the male-specific sequence regions. The genome of male O. bidens was screened (window size = 500 bp, step = 1000 bp), and the CQ value of each sliding window was calculated as follows: CQ = Hf/Hm (where Hm is the number of reads in the reference genome of the male mixed-pool comparison; Hf is the number of reads in the reference genome of the female mixed-pool comparison). The male-specific regions with a CQ < 0.2 and an Hm > 30 were retained. These thresholds were based on the references from published literature [28]. Overlapping windows identified during screening were merged. Integrative Genomic Viewer (v2.16.2) was used to compare reads in the selected regions and further confirm male-specific reads.
2.5. Design of Primers for Sex-Specific Regions and Their Screening Validation
Flanking sequences (1 kb upstream and downstream) of each sex-specific region were extracted, and common PCR amplification primers were designed using Primer Premier software (v6.0). These primers were screened via two rounds of PCR amplification. First, the genomic DNA of three males and three females of O. bidens served as the template for the preliminary screening of primers. For those primers with either unclear amplification bands or no target bands, the annealing gradient was adjusted to find the optimal annealing temperature until clear and complete target bands were obtained. For primers that met the expected specificity for the target region, sex specificity was further validated using the O. bidens populations from Jiangxi, Zhejiang, and Fujian (20 males and 20 females per population). Every PCR amplification was performed in a 25 μL reaction system, consisting of 12.5 μL of 2 × Rapid Taq Master Mix, 1.0 μL of each F/R primer (10 μmol/L), 1.5 μL of DNA template, and 9.0 μL of ddH2O. A negative control using ddH2O as the template was also included. The PCR amplification program was as follows: initial denaturation at 94 °C for 5 min; 30 cycles of denaturation at 94 °C for 30 s, annealing at 53–59 °C for 30 s, followed by an extension at 72 °C for 2 min; and a final extension at 72 °C for 10 min. Samples were then stored at 4 °C. The primers’ specificity was confirmed by 1% agarose gel electrophoresis of the PCR products. The PCR products of primers that targeted the expected sex-specific region and yielded sex-differentiating bands were purified and ligated into the PMD-18 vector for cloning. The ligation mixtures were transformed into DH5α Escherichia coli competent cells, putative positive colonies were selected, and these were sequenced by Shengong Bioengineering (Shanghai, China) Co., Ltd. Male and female sequences of O. bidens were analyzed in SnapGene software (v6.0).
3. Results
3.1. Sample Information
The genetic background, specifications, and age information of the male and female fish populations used for resequencing and primer validation are shown in Table 1. Photographs clearly illustrating the differences in sexual dimorphism between the sexes are shown in Figure 1.

Table 1.
Information on the populations sampled in this study.

Figure 1.
Male and female Opsariichthys bidens.
The genomic DNA of 25 male and 25 female fish was detected using a NanoDrop spectrophotometer. Their average DNA concentration was 275.92 ng/μL, with an OD260/OD280 between 1.8 and 2.0. The results of 1% agarose gel electrophoresis showed clear DNA bands and no serious degradation. This indicated that the collected genomic DNA of O. bidens was of high quality and suitable for BSA-seq library preparation.
3.2. Sequencing Data Statistics
From the BSA sequencing, raw data totaling 21.54 GB and 28.20 GB were generated for the mixed pools of male and female O. bidens, respectively. After undergoing data filtering and quality control, effective data amounting to 20.73 GB and 26.98 GB were obtained for males and females, respectively. Their Q20 was 98.96% and 99.03%, their Q30 was 95.64% and 95.92%, and their GC content was 38.95% and 38.94%, respectively (Table 2). After duplicate reads were removed, 142,668,885 and 186,590,651 sequences were obtained from the male and female mixed pools, respectively. Of those, 141,763,631 and 185,468,646 sequences were aligned to the O. bidens reference genome, yielding alignment rates of 99.37% and 99.40%, genome coverage of 96.33% and 96.44%, and an average sequencing depth of 24.22× and 31.46×.

Table 2.
BSA sequencing data statistics.
3.3. Screening and Validation of Sex-Specific Markers
After the IGV examination, 45 sex-specific regions (CQ < 0.2) were found (Figure 2). For these, 50 pairs of primers were designed according to their specific regional flanking sequences (Table 3). Three males and three females were randomly selected to screen those 50 primer pairs in the first PCR amplification round: the amplification bands of four pairs (the primers Mar26, Mar28, Mar37, and Mar38) showed pronounced differences between males and females (Figure 3). Hence, these four pairs of primers were subjected to further screening in a second PCR amplification round; that is, 40 samples (20 male and 20 female adult fish per population) from each of the Jiangxi, Zhejiang, and Fujian populations were used for verification. These results revealed that Mar28, located on chromosome 8, amplified two bands in males but only a single band in females in all three O. bidens populations (Figure 4 and Table 3). In contrast, the amplified bands of the other three primers failed to yield a difference between the sexes in all individuals. Further sequencing of the Mar28 primer amplification products showed that Mar28 amplified two respective bands of 509 and 814 bp in males, while a single band of 509 bp was amplified in females (Figure 5).
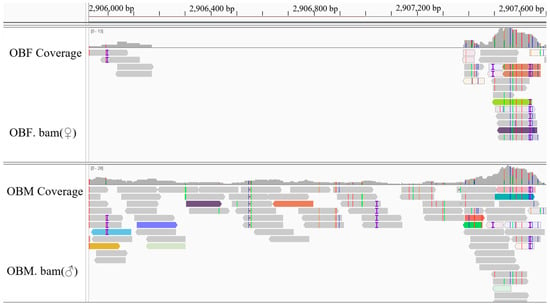
Figure 2.
Visualization of the sex-specific region for the primer Mar38. The target region was located on chromosome 20 (genomic coordinates: chr20: 2,906,224–2,907,256), spanning 1032 bp. The analysis was extended to include 300 bp of the flanking sequence on both sides of this core interval.

Table 3.
Primer sequences designed for the sex-specific regions of Opsariichthys bidens. TM, annealing temperature.
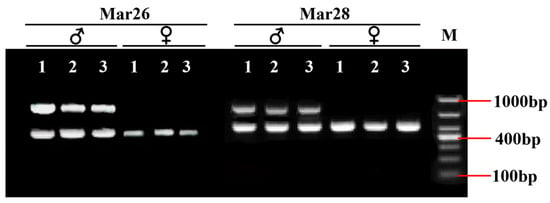
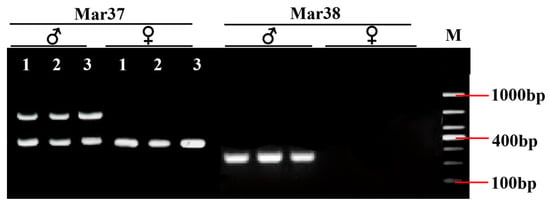
Figure 3.
PCR amplification electrophoretogram of four pairs of primers in six Opsariichthys bidens individuals.
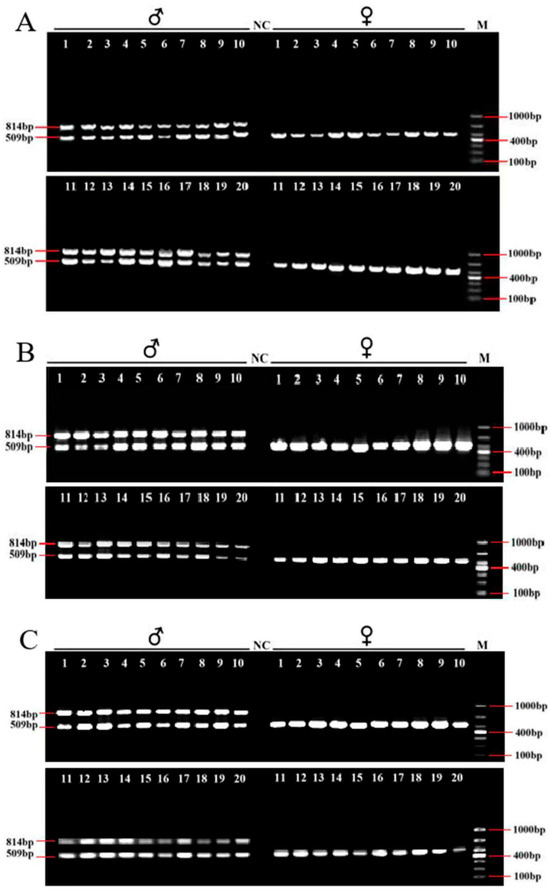
Figure 4.
Amplified PCR electrophoretogram of Mar28 in 40 Opsariichthys bidens individuals from Jiangxi (A), Zhejiang (B), and Fujian (C). NC, the negative control. M, the DL1000 Marker.
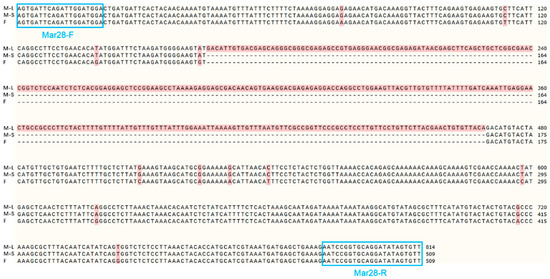
Figure 5.
Sequence alignment of the Mar28 amplified fragments in male and female Opsariichthys bidens. M-L denotes the long sequence in males, while M-S denotes the short sequence in males; F refers to the female sequence, with “—” indicating a missing sequence. Highlighted in red are the base differences between the male and female sequences; the blue box shows the position of the primer pair.
4. Discussion
Fish are the mainstay of aquaculture, and fish farming is an important industry for ensuring China’s national food security. China is home to a wide variety of farmed fish species, and the development of sex-specific molecular markers can greatly aid the aquaculture industry. Research on sex control and monosex breeding techniques is critically important, given that many cultured fish exhibit sexual differences. O. bidens is an omnivorous species, although it clearly prefers consuming animals and will only consume plants when prey is scarce. Males grow faster than females and display striking coloration during the breeding period. The breeding of all-male fish can thus aid O. bidens breeding programs. Determining the genetic sex of O. bidens is crucial for achieving a monosex culture. Sex-specific markers are also important for studying the mechanisms of sex determination and differentiation in aquaculture and fisheries.
RAPD, SSRs, AFLPs, and other methods have been successfully used to develop sex identification markers for a variety of fish [29,30,31]. However, these molecular markers have several drawbacks, such as their low efficiency and poor repeatability [32]. In our study, we utilized BSA to address these limitations. BSA allows for the pooling of DNA from individuals with extreme phenotypes, which significantly enhances the signal-to-noise ratio in genetic mapping [33]. This approach enabled us to focus on the most informative genetic markers, thereby increasing the efficiency of identifying trait-associated loci. Furthermore, by using a larger number of individuals per bulk, we improved the repeatability of our results, as the genetic noise from individual variation was minimized. This concrete application of BSA in our study demonstrates its effectiveness in overcoming the challenges posed by traditional methods. Sex identification markers developed via the BSA technique are also being increasingly applied to fish species [34]. A pair of sex markers was selected for Channa argus via SSR and BSA, and differential bands could be amplified in most females, but not males [35]. Combining BSA analysis with the resequencing of multiple individuals can eliminate individual differences, provide greater genome coverage of genome-wide information, and reduce the probability of false positives [32]. Using this approach, six male-specific markers were recently developed in Spinibarbus hollandi [36]. Some researchers have used the combination of BSA and CQ methods to screen 279 sex-specific regions in Oryzias curvinotus, from which two sex markers were developed [28].
The Mar28 marker in our study was obtained by comparing and analyzing genomic resequencing data of female and male O. bidens derived from divergent alleles of the same locus, and this marker flanked a Y-specific insertion within a pseudoautosomal region. The Y chromosome has an additional 305-bp fragment, but its function requires further study. Insertion mutations are a common type of mutation characterized by the incorporation of extra nucleotides or DNA segments into existing sequences, which modifies gene sequences and expression patterns [37]. When such insertions occur within intronic regions, they can disrupt or create novel splice sites, which can affect protein structure and functionality [38]. Similar insertional events have been documented in Oplegnathus punctatus [39]. Whereas a previous GWAS by Xu et al. [23] mapped all sex-associated loci in O. bidens to chromosome 24, the marker developed here was fully embedded within the Y-chromosome insertion and therefore produced a single, male-specific band. In contrast, our newly developed marker resided on chromosome 8 and spanned the insertion junction; consequently, males yielded two bands, whereas females yielded one.
Mining sex-determining or candidate genes via transcriptome sequencing to develop sex-specific molecular markers is also a highly efficient strategy. De novo transcriptome analysis of the testis and ovary of O. bidens revealed that cyp19a was highly expressed only during the early stage of oocyte development and was not expressed in the testis, while vasa was expressed at significantly different levels in the testis and ovary [40]. Transcriptome sequencing analysis with reads mapped to the genome and RT-qPCR verification revealed that zp3, cyp19a, hsd17b1, and gdf9 were female-biased genes, while msh4, dmrt1, rspo2, and kif23 were male-biased genes [41]. Transcriptome sequencing of the testis with genome-mapped reads was performed during four developmental stages of male O. bidens to determine the genes and pathways related to testis and sperm formation in O. bidens [42]. The discovery of these genes will aid the development of sex-specific markers for O. bidens. In this study, no association analysis was conducted between the molecular markers obtained and the sex-specific genes. This will be a goal of our future work.
5. Conclusions
In this study, sex-specific regions were selected by comparing the resequencing data of mixed pools of females and males to the reference genome of O. bidens. Unique primers were designed for these regions, which were then confirmed and validated through PCR, to obtain a robust sex marker. We developed a rapid, accurate, and cost-effective approach for the genetic sex identification of O. bidens. Our empirical results will facilitate the production of monosex populations of O. bidens and provide a theoretical basis for future investigations of its sex determination and differentiation mechanisms.
Author Contributions
Conceptualization, X.W. and C.Z.; methodology, R.Z., Y.W. and R.W.; formal analysis, R.Z. and N.L.; investigation, M.M. and N.L.; resources, M.M. and D.D.; data curation, R.Z. and S.O.; writing—original draft preparation, C.Z. and R.W.; writing—review and editing, C.Z. and F.L.; visualization, R.Z. and D.D.; supervision, S.O. and X.W.; project administration, Y.W.; funding acquisition, F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the major project of Science and Technology of Zhejiang Province (2023C02039), the Joint Breeding Project of the Fishery Seed Industry in Jiangxi Province (2023YYZYGG-08), and the Special Fund of Zhejiang Province (2025CZZX01).
Institutional Review Board Statement
The animal study protocol was approved by the Animal Experiment Committee, Zhejiang Institute of Freshwater Fishery (ZJIFF20230603, 12 June 2023).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in the present study can be downloaded from the NCBI database under project number PRJNA1286929, https://www.ncbi.nlm.nih.gov/nuccore/?term=+PRJNA1286929 (accessed on 20 May 2025).
Acknowledgments
We would like to express our gratitude to Lingzhan Xue from the Fujian Institute of Freshwater Aquatic Research for providing the samples of the Fujian population for this research. We thank Chongguang Chen, Hongfei Hu, and Yi Zhou at Nanchang University, China, for their help with sample collection. We also thank Yuting Dai at Nanchang University, China, for performing the data analysis.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to have influenced the work reported in this paper.
References
- Stelkens, R.B.; Wedekind, C. Environmental sex reversal, Trojan sex genes, and sex ratio adjustment: Conditions and population consequences. Mol. Ecol. 2010, 19, 627–646. [Google Scholar] [CrossRef]
- Anastasiadi, D.; Vandeputte, M.; Sánchez-Baizán, N.; Allal, F.; Piferrer, F. Dynamic epimarks in sex-related genes predict gonad phenotype in the European sea bass, a fish with mixed genetic and environmental sex determination. Epigenetics 2018, 13, 988–1011. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Bian, C.; Jiao, K.; Zhang, L.; Huang, Y.; Yang, W.; Li, Y.; Shi, G.; Huang, Y.; et al. Allelic variation and duplication of the dmrt1 were associated with sex chromosome turnover in three representative Scatophagidae fish species. Commun. Biol. 2025, 8, 627. [Google Scholar] [CrossRef]
- Xue, L.; Guo, X.; Zhou, Y.; Wang, Z.; Fan, H.; Li, D.; Gui, J. Screening and characterization of sex-specific markers by 2b-RAD sequencing in zig-zag eel (Mastacembelus armatus) with implication of XY sex determination system. Aquaculture 2020, 528, 735550. [Google Scholar] [CrossRef]
- Wang, L.; Xie, N.; Shen, Y.; Ye, B.; Yue, G.; Feng, X. Constructing high-density genetic maps and developing sexing markers in northern snakehead (Channa argus). Mar. Biotechnol. 2019, 21, 348–358. [Google Scholar] [CrossRef]
- Han, C.; Zhu, Q.; Lu, H.; Wang, C.; Zhou, X.; Peng, C.; Tang, L.; Han, L.; Chen, J.; Li, S.; et al. Screening and characterization of sex-specific markers developed by a simple NGS method in mandarin fish (Siniperca chuatsi). Aquaculture 2020, 527, 735495. [Google Scholar] [CrossRef]
- Li, Y.; Cui, Y.; Ye, X.; Wang, M.; Xu, H.; Wu, Y.; Jin, J.; Huang, H.; Lai, M.; Wang, Z.; et al. Identification of the sex-linked region of Siniperca scherzeri and development of sex-specific markers. Aquaculture 2025, 600, 742231. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S. Fish sex-specific molecular marker screening and marker-assisted sex-controlled breeding: History, progress and prospect. J. Fish. China 2023, 47, 34–43. [Google Scholar]
- Qin, W.; Han, C.; Yang, J.; Yu, Z.; Feng, Y.; Wu, Y.; Lu, B.; Cui, M.; Shu, H. Development of genetic sex markers of zig-zag eel (Mastacembelus armatus) by a NGS method. Aquaculture 2023, 571, 739498. [Google Scholar] [CrossRef]
- Bentley, D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006, 16, 545–552. [Google Scholar] [CrossRef]
- Wen, M.; Wang, S.; Zhu, C.; Zhang, Y.; Liu, Z.; Wu, C.; Wang, S.; Wang, Y.; Ren, L.; Tao, M.; et al. Identification of sex locus and a male-specific marker in blunt-snout bream (Megalobrama amblycephala) using a whole genome resequencing method. Aquaculture 2024, 582, 740559. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, L.; Wang, S.; Yu, Y.; Wang, Y.; Gao, Z. Identification of sex-specific markers using genome re-sequencing in the blunt snout bream (Megalobrama amblycephala). BMC Genom. 2024, 25, 963. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Li, S.; Liu, Y.; Li, C.; Han, S.; Liu, K.; Doretto, L.B.; Liu, B.; Huang, H.; et al. Investigation of sex determination in starry flounder (Platichthys stellatus) reveals sex chromosome evolution in Pleuronectiformes and identifies a sex-specific marker. Zool. Res. 2024, 45, 1347–1356. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zhang, Y.; Wang, S.; Hu, F.; Tang, C.; Li, Q.; Qin, Q.; Tao, M.; Zhang, C.; Zhao, R.; et al. Sex locus and sex markers identification using whole genome pool-sequencing approach in the largemouth bass (Micropterus salmoides L.). Aquaculture 2022, 559, 738375. [Google Scholar] [CrossRef]
- He, P.; Wei, P.; Ma, Y.; Hu, S.; Yao, J.; Jiang, X.; Xu, Y.; Zhu, P.; Wei, M.; Jiang, W.; et al. Candidate sex-associated gene identification in Trachinotus ovatus (Carangidae) using an integrated SLAF-seq and bulked segregant analysis approach. Gene 2022, 809, 146026. [Google Scholar] [CrossRef]
- Liu, S.; Lian, Q.; Jia, Y.; Chi, M.; Li, F.; Jiang, J.; Liu, Y.; Zheng, J.; Cheng, S.; Gu, Z. Genetic diversity analysis of three Opsariichthys bidens populations in Zhejiang Province based on mitochondrial Cyt b gene sequences. Acta Agric. Zhejiangensis 2023, 35, 293–300. [Google Scholar]
- Xu, X.; Guan, W.; Niu, B.; Guo, D.; Xie, Q.; Zhan, W.; Yi, S.; Luo, B. Chromosome-level assembly of the Chinese hooksnout carp (Opsariichthys bidens) genome using PacBio sequencing and hi-C technology. Front. Genet. 2022, 12, 788547. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, Q.; Wang, W.; He, B.; Hou, Y.; Lin, Y.; Ye, J.; Ren, S.; Qin, Y.; Xiao, A.; et al. Transcriptomic analysis of colour dimorphism of Opsariichthys bidens provides insights into the mechanism of male colour. Aquac. Rep. 2023, 33, 101756. [Google Scholar] [CrossRef]
- Qiu, Q.; Luo, W. Apply RAPD to screen out sex differentiation related molecular marker in Opsariichthys bidens. J. Aquacult. 2012, 33, 38–42. [Google Scholar]
- Liu, D.; Gui, L.; Zhu, Y.; Xu, C.; Zhou, W.; Li, M. Chromosome-level assembly of male Opsariichthys bidens genome provides insights into the regulation of the GnRH signaling pathway and genome evolution. Biology 2022, 11, 1500. [Google Scholar] [CrossRef]
- Guan, W.; Xu, X.; Zhan, W.; Niu, B.; Luo, B. Induction of gynogenesis with ultra-violet irradiated Koi carp (Cyprinus carpio haematopterus) sperm demonstrates the XX/XY sex determination system in Opsariichthys bidens. Aquac. Rep. 2022, 26, 101286. [Google Scholar] [CrossRef]
- Xu, X.; Yu, J.; Ge, J.; Yi, S.; Weng, X.; Guan, W.; Niu, B.; Zhang, X.; Luo, B. A male-specific insert of Opsariichthys bidens identified based on genome-wide association analyses and comparative genomics. Aquac. Rep. 2024, 35, 101982. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, R.; Deng, Y.; Wu, X.; Ouyang, S.; Lin, F.; Zhou, C. Development of SNP markers for sex identification of Opsariichthys bidens. J. Nanchang Univ. Nat. Sci. 2025, 49, 214–221. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Wang, Z.; He, C.; Yao, Z.; Li, J.; Chen, Z.; Deng, A.; Lai, Z.; Li, Y.; Dong, Z.; Guo, Y. Screening of Sex Genetic Markers in the Sanya Population of Oryzias curvinotus by Whole-Genome Resequencing. J. Guangdong Ocean Univ. 2023, 43, 69–75. [Google Scholar]
- Akkaya, M.S.; Bhagwat, A.A.; Cregan, P.B. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics 1992, 132, 1131–1139. [Google Scholar] [CrossRef]
- Waldbieser, G.C.; Bosworth, B.G.; Nonneman, D.J.; Wolters, W.R. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics 2001, 158, 727–734. [Google Scholar] [CrossRef]
- Felip, A.; Young, W.P.; Wheeler, P.A.; Thorgaard, G.H. An AFLP-based approach for the identification of sex-linked markers in rainbow trout (Oncorhynchus mykiss). Aquaculture 2001, 247, 35–43. [Google Scholar] [CrossRef]
- Han, C.; Zhou, X.; Lu, H.; Zhu, Q.; Han, L.; Li, S.; Lin, H.; Zhang, Y. A simple PCR-based genetic sex identification method in the blotched snakehead (Channa maculata) developed by high-throughput sequencing. Aquaculture 2021, 538, 736579. [Google Scholar] [CrossRef]
- Best, N.B.; McSteen, P. Mapping Maize Mutants Using Bulked-Segregant Analysis and Next-Generation Sequencing. Curr. Protoc. 2022, 2, e591. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, J.; Liu, C.; Yang, Y.; Wang, Y.; Mu, X. Identification of two effective sex-specific DNA markers in silver arowana (Osteoglossum bicirrhosum). Aquaculture 2025, 596, 741748. [Google Scholar] [CrossRef]
- Liu, G.; Chen, K.; Zheng, G.; Zhu, X.; Zhao, J.; Xu, P.; Sun, X. Screening and identification of female-specific DNA fragments in Channa argus using SSR-BSA. J. Fish. China 2011, 35, 170–175. [Google Scholar]
- Huang, W.; Lai, J.; Liang, W.; Ye, S.; Li, J.; Zhou, J.; Zhang, Y.; Peng, S.; Zhan, H.; Zheng, P.; et al. Identification of sex-specific DNA markers in the army fish (Spinibarbus hollandi) by whole genome re-sequencing method. Aquaculture 2024, 583, 740605. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Lupski, J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016, 17, 224–238. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xiao, Y.; Xiao, Z.; Li, J. Development of DNA insertion-specific markers based on the intergenic region of Oplegnathus punctatus Cdkn1/srsf3 for sex identification. Mar. Biotechnol. 2024, 26, 687–695. [Google Scholar] [CrossRef]
- Tang, R.; Zhu, Y.; Gan, W.; Zhang, Y.; Yao, Z.; Ren, J.; Li, M. De novo transcriptome analysis of gonads reveals the sex-associated genes in Chinese hook snout carp Opsariichthys bidens. Aquac. Rep. 2022, 23, 101068. [Google Scholar] [CrossRef]
- Zhou, C.; Lian, X.; Wang, R.; Wu, X.; Lin, F.; Ouyang, S.; Jian, S.; Hua, Q. Gonadal transcriptome analysis of Opsariichthys bidens reveals sex-associated genes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 54, 101379. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Tang, D.; Zhang, Y.; Gao, X.; Du, C.; Shen, W.; Jin, S.; Zhu, J. Transcriptomes of testes at different developmental stages in the Opsariichthys bidens predict key genes for testis development and spermatogenesis. Mar. Biotechnol. 2023, 25, 123–139. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).