Genomic Characterization of the Kazakh Fat-Tailed Coarse-Wool Sheep Breed Using ROH Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Genotyping
2.3. ROH Detection
2.4. Estimation of Genomic Inbreeding and Correlation Analysis
2.5. Gene Annotation and Functional Enrichment Analysis
3. Results
3.1. ROH Analysis Results
3.2. Genomic Inbreeding Estimates and Correlation Between FROH and FGRM
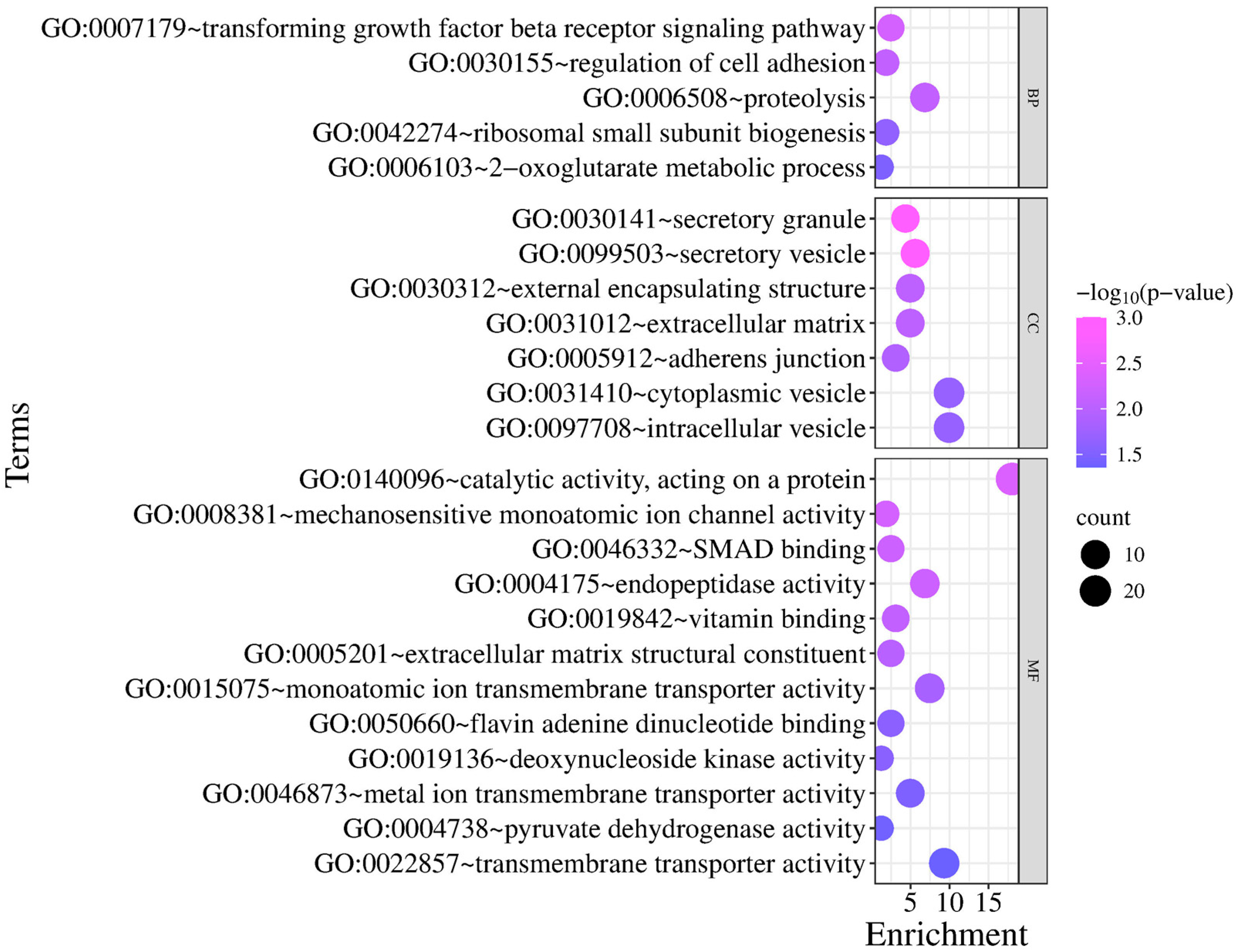
3.3. Gene Annotation and Functional Clustering
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ROH | Runs of homozygosity |
| FROH | Inbreeding coefficient based on ROH |
| FGRM | Inbreeding coefficient based on genomic relationship matrix |
| MAS | Marker-assisted selection |
| SNP | Single-nucleotide polymorphism |
References
- Bureau of National Statistics of the Agency for Strategic Planning and Reforms of the Republic of Kazakhstan. Available online: https://stat.gov.kz/ru/industries/business-statistics/stat-forrest-village-hunt-fish/dynamic-tables/ (accessed on 15 April 2025).
- Katasheva, A.; Iskakov, K.; Kulataev, B.; Аbdrаmаnоv, А.; Sattorov, S. Improving the efficiency of the production of mutton of kazakh fat-tailed coarse-wool sheep. Izdenister Natigeler 2024, 2, 55–62. (In Russian) [Google Scholar] [CrossRef]
- Malmakov, N.; Kulataev, B.; Iskakov, K.; Tastaganov, M. Features of effective breeding methods to improve the reproduction ability of Kazakh coarse-wooled sheep. Izdenister Natigeler 2023, 3, 5–10. (In Russian) [Google Scholar]
- Zhumadila, K.; Zhumadilaev, N.K.; Esengaliev, K.; Burambaeva, N.B. Recommendations for Breeding Work with Kazakh Fat-Tailed Coarse-Wool Sheep: Mynbayevo, Kazakhstan. 2009, p. 28, ISBN 978-601-7226-02-2. Available online: http://rpo-kaz.kz/learn/ (accessed on 21 August 2025). (In Russian).
- Ma, R.; Liu, J.; Ma, X.; Yang, J. Genome-wide runs of homozygosity reveal inbreeding levels and trait-associated candidate genes in diverse sheep breeds. Genes 2025, 16, 316. [Google Scholar]
- Deng, T.X.; Ma, X.Y.; Duan, A.Q.; Lu, X.R.; Abdel-Shafy, H. Genomic insights into selection signatures and candidate genes for milk production traits in buffalo population. Animal 2025, 19, 101427. [Google Scholar] [CrossRef]
- Ma, K.; Li, X.; Ma, S.; Zhang, M.; Wang, D.; Xu, L.; Chen, H.; Wang, X.; Qi, A.; Ren, Y.; et al. Analysis of population structure and selective signatures for milk production traits in Xinjiang Brown cattle and Chinese Simmental cattle. Int. J. Mol. Sci. 2025, 26, 2003. [Google Scholar] [CrossRef]
- Nandolo, W.; Mészáros, G.; Banda, L.J.; Gondwe, T.N.; Lamuno, D.; Mulindwa, H.A.; Nakimbugwe, H.N.; Wurzinger, M.; Utsunomiya, Y.T.; Woodward-Greene, M.J.; et al. Timing and Extent of inbreeding in African goats. Front. Genet. 2019, 10, 537. [Google Scholar] [CrossRef]
- Silva, G.A.A.; Harder, A.M.; Kirksey, K.B.; Mathur, S.; Willoughby, J.R. Detectability of runs of homozygosity is influenced by analysis parameters and population-specific demographic history. PLoS Comput. Biol. 2024, 20, e1012566. [Google Scholar] [PubMed]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Meyermans, R.; Gorssen, W.; Aerts, N.; Hooyberghs, K.; Bhaskaran, B.C.; Chapard, L.; Buys, N.; Janssens, S. Genomic characterisation and diversity assessment of eight endangered Belgian sheep breeds. Animal 2024, 18, 101315. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, F.C.; Joshi, P.K.; Clark, D.W.; Ramsay, M.; Wilson, J.F. Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 2018, 19, 220–234. [Google Scholar] [CrossRef]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef]
- Ferenčaković, M.; Hamzić, E.; Gredler, B.; Solberg, T.R.; Klemetsdal, G.; Curik, I.; Sölkner, J. Estimates of autozygosity derived from runs of homozygosity: Empirical evidence from selected cattle populations. J. Anim. Breed. Genet. 2013, 130, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Halvoník, A.; Moravčíková, N.; Chalupková, M.; Kasarda, R. Commonly used genomic estimators of individual inbreeding in livestock. Czech. J. Anim. Sci. 2024, 69, 269–279. [Google Scholar] [CrossRef]
- Howrigan, D.P.; Simonson, M.A.; Keller, M.C. Detecting autozygosity through runs of homozygosity: A comparison of three autozygosity detection algorithms. BMC Genom. 2011, 12, 460. [Google Scholar] [CrossRef]
- Dadousis, C.; Ablondi, M.; Cipolat-Gotet, C.; van Kaam, J.T.; Marusi, M.; Cassandro, M.; Sabbioni, A.; Summer, A. Genomic inbreeding coefficients using imputed genotypes: Assessing different estimators in Holstein-Friesian dairy cows. J. Dairy Sci. 2022, 105, 5926–5945. [Google Scholar] [CrossRef]
- Illa, S.K.; Mumtaz, S.; Nath, S.; Mukherjee, S.; Mukherjee, A. Characterization of runs of homozygosity revealed genomic inbreeding and patterns of selection in indigenous Sahiwal cattle. J. Appl. Genet. 2024, 65, 167–180. [Google Scholar] [CrossRef]
- Amandykova, M.; Akhatayeva, Z.; Kozhakhmet, A.; Kapassuly, T.; Orazymbetova, Z.; Yergali, K.; Khamzin, K.; Iskakov, K.; Dossybayev, K. Distribution of runs of homozygosity and their relationship with candidate genes for productivity in Kazakh meat–wool sheep breed. Genes 2023, 14, 1988. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; He, X.; Wang, X.; Di, R.; Chu, M. Litter size of sheep (Ovis aries): Inbreeding depression and homozygous regions. Genes 2021, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Dzomba, E.F.; Chimonyo, M.; Pierneef, R.; Muchadeyi, F.C. Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genom. 2021, 22, 7. [Google Scholar] [CrossRef]
- Selli, A.; Ventura, R.V.; Fonseca, P.A.S.; Buzanskas, M.E.; Andrietta, L.T.; Balieiro, J.C.C.; Brito, L.F. Detection and visualization of heterozygosity-rich regions and runs of homozygosity in worldwide sheep populations. Animals 2021, 11, 2696. [Google Scholar] [CrossRef]
- Islam, R.; Li, Y.; Liu, X.; Berihulay, H.; Abied, A.; Gebreselassie, G.; Ma, Q.; Ma, Y. Genome-wide runs of homozygosity, effective population size, and detection of positive selection signatures in six Chinese goat breeds. Genes 2019, 10, 938. [Google Scholar] [CrossRef] [PubMed]
- An, Z.X.; Shi, L.G.; Hou, G.Y.; Zhou, H.L.; Xun, W.J. Genetic diversity and selection signatures in Hainan black goats revealed by whole-genome sequencing data. Animal 2024, 18, 101147. [Google Scholar] [CrossRef]
- Kim, E.S.; Cole, J.B.; Huson, H.; Wiggans, G.R.; Van Tassell, C.P.; Crooker, B.A.; Liu, G.; Da, Y.; Sonstegard, T.S. Effect of artificial selection on runs of homozygosity in U.S. Holstein cattle. PLoS ONE 2013, 8, e80813. [Google Scholar] [CrossRef]
- Mulim, H.A.; Brito, L.F.; Pinto, L.F.B.; Ferraz, J.B.S.; Grigoletto, L.; Silva, M.R.; Pedrosa, V.B. Characterization of runs of homozygosity, heterozygosity-enriched regions, and population structure in cattle populations selected for different breeding goals. BMC Genom. 2022, 23, 209. [Google Scholar] [CrossRef]
- Abdoli, R.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Zamani, P.; Moradi, M.H.; Ferdosi, M.H.; Sargolzaei, M.; Gondro, C. Runs of homozygosity and cross-generational inbreeding of Iranian fat-tailed sheep. Heredity 2023, 130, 358–367. [Google Scholar] [CrossRef]
- Kusza, S.; Badaoui, B.; Wanjala, G. Insights into the genomic homogeneity of Moroccan indigenous sheep breeds though the lens of runs of homozygosity. Sci. Rep. 2024, 14, 16515. [Google Scholar] [CrossRef]
- Liu, J.; Shi, L.; Li, Y.; Chen, L.; Garrick, D.; Wang, L.; Zhao, F. Estimates of genomic inbreeding and identification of candidate regions that differ between Chinese indigenous sheep breeds. J. Anim. Sci. Biotechnol. 2021, 12, 95. [Google Scholar] [CrossRef]
- Bao, J.; Xiong, J.; Huang, J.; Yang, P.; Shang, M.; Zhang, L. Genetic diversity, selection signatures, and genome-wide association study identify candidate genes related to litter size in Hu sheep. Int. J. Mol. Sci. 2024, 25, 9397. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Zerehdaran, S.; Javadmanesh, A. Genetic diversity in some domestic and wild sheep and goats in Iran. Small Rumin. Res. 2022, 209, 106641. [Google Scholar] [CrossRef]
- Zhao, C.; Raza, S.H.A.; Khan, R.; Sabek, A.; Khan, S.; Ullah, I.; Memon, S.; El-Aziz, A.H.A.; Shah, M.A.; Shijun, L.; et al. Genetic variants in MYF5 affected growth traits and beef quality traits in Chinese Qinchuan cattle. Genomics 2020, 112, 2804–2812. [Google Scholar] [CrossRef]
- Nosrati, M.; Asadollahpour Nanaei, H.; Javanmard, A.; Esmailizadeh, A. The pattern of runs of homozygosity and genomic inbreeding in world-wide sheep populations. Genomics 2021, 113, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Bayrak, B.; Şen, U.; Gökçek, D.; Şirin, E. Differential Myf5 and Myf6 expression and muscle fiber traits in Angora, Hair, Honamlı, and Kilis goats. Trop. Anim. Health Prod. 2025, 57, 36. [Google Scholar] [CrossRef]
- Fadhil, M.; Zülkadir, U. Association between polymorphismsof Myf5, MSTN and CAST genes and fattening performance in Brown Swiss and Holstein cattle breeds. Anim. Biotechnol. 2021, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Hong, J.H. Chloride/multiple anion exchanger SLC26A family: Systemic roles of SLC26A4 in various organs. Int. J. Mol. Sci. 2024, 25, 4190. [Google Scholar] [CrossRef] [PubMed]
- John, S.; Thiebach, L.; Frie, C.; Mokkapati, S.; Bechtel, M.; Nischt, R.; Rosser-Davies, S.; Paulsson, M.; Smyth, N. Epidermal transglutaminase (TGase 3) is required for proper hair development, but not the formation of the epidermal barrier. PLoS ONE 2012, 7, e34252. [Google Scholar] [CrossRef]
- Sun, R.; Li, M.; He, N.; Wen, X.; Zhang, J. Molecular characterization, expression profiles of SMAD4, SMAD5 and SMAD7 genes and lack of association with litter size in Tibetan sheep. Animals 2022, 12, 2232. [Google Scholar] [CrossRef]
- Chen, N.N.; An, X.J.; Li, T.T.; Wang, H.H.; Xu, S.R.; Man, Y.H.; Ma, Y.J. Expression and localization of BMP6/SMAD5 in Tibetan sheep testis. Prat. Sci. 2022, 39, 2434–2441. [Google Scholar]
- Zhong, Y.; Di, R.; Yang, Y.; Liu, Q.; Chu, M. Transcriptome analysis of neuroendocrine regulation of ovine hypothalamus-pituitary-ovary axis during ovine anestrus and the breeding season. Genes 2021, 12, 1861. [Google Scholar] [CrossRef]

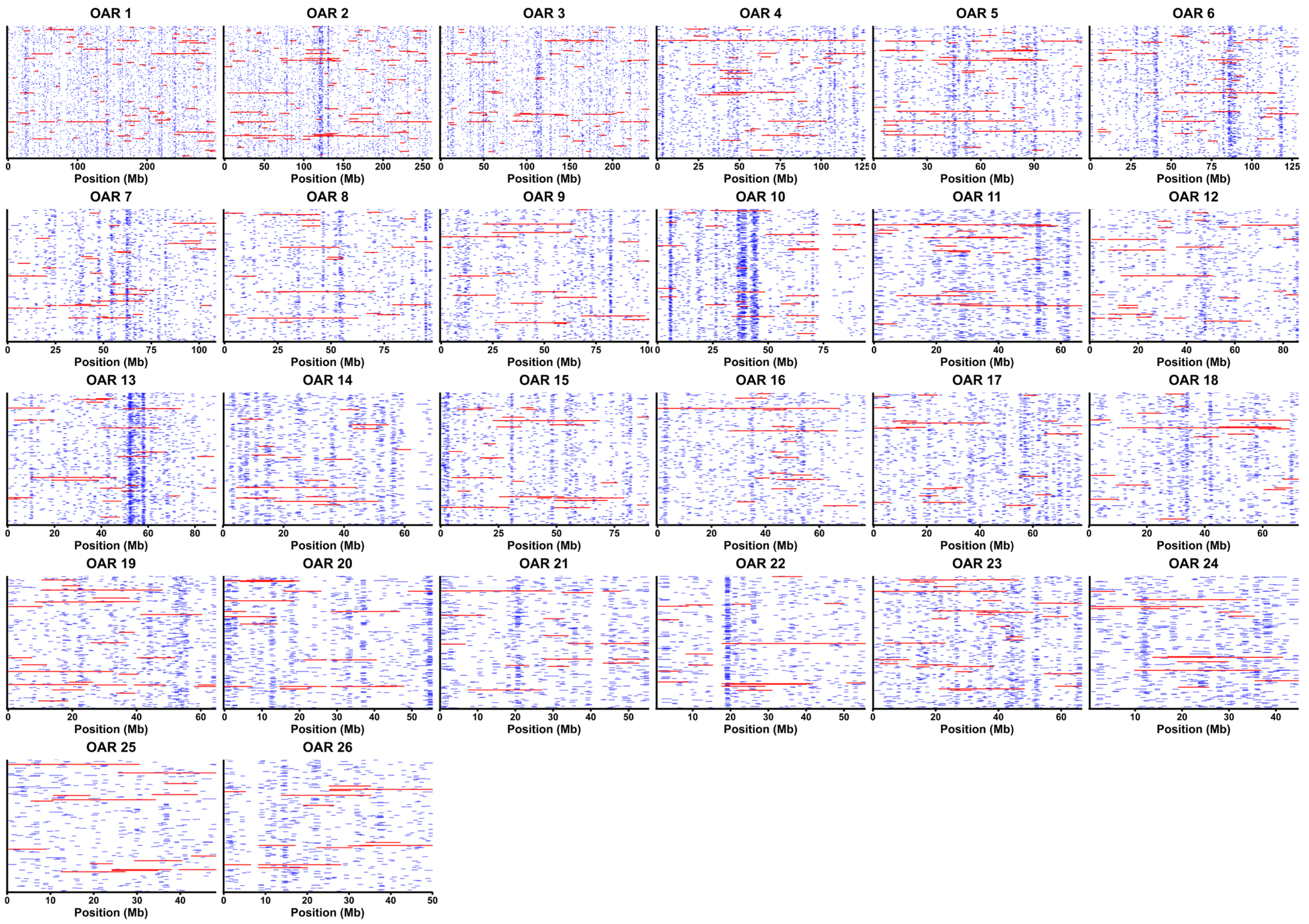

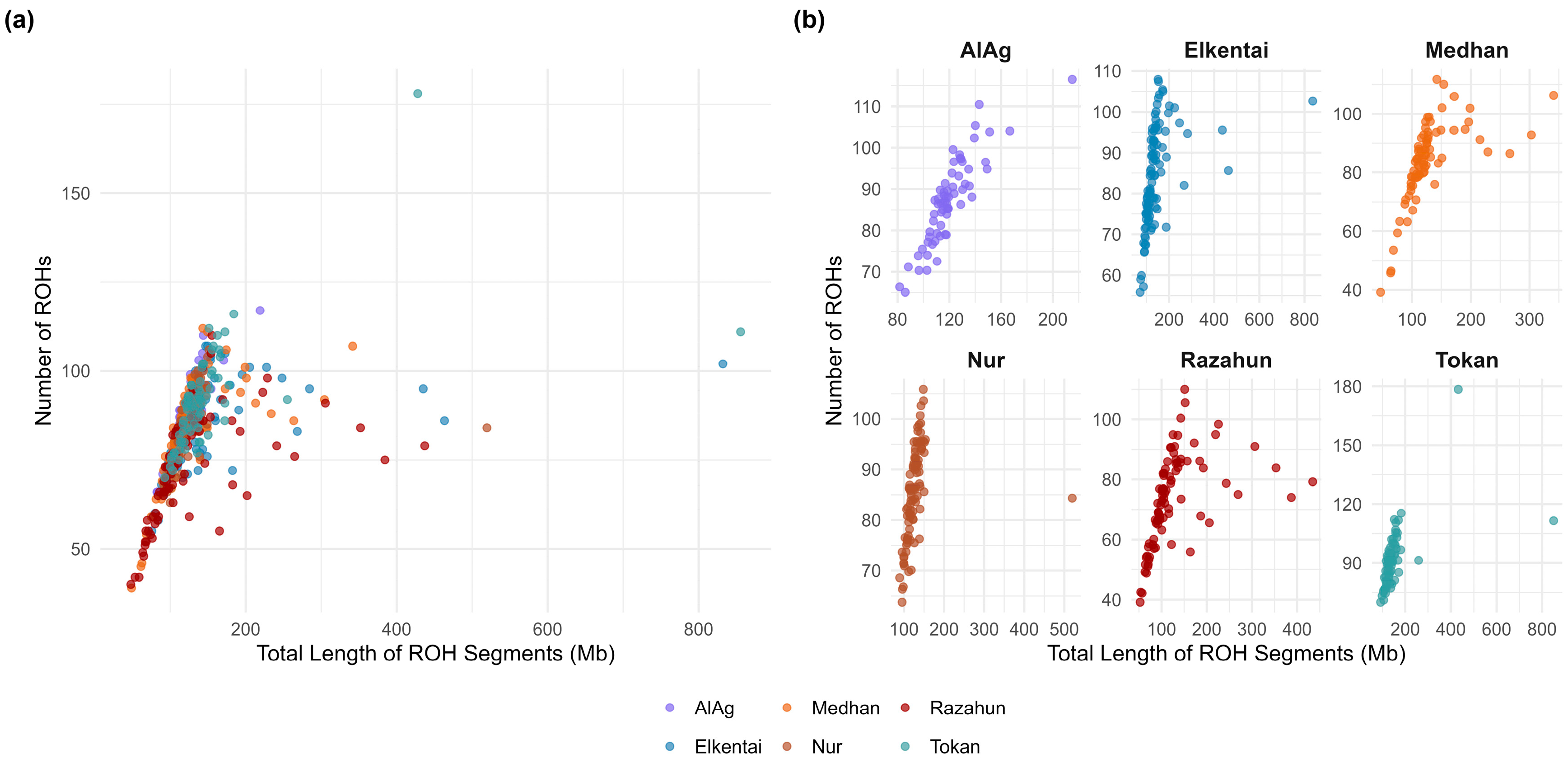
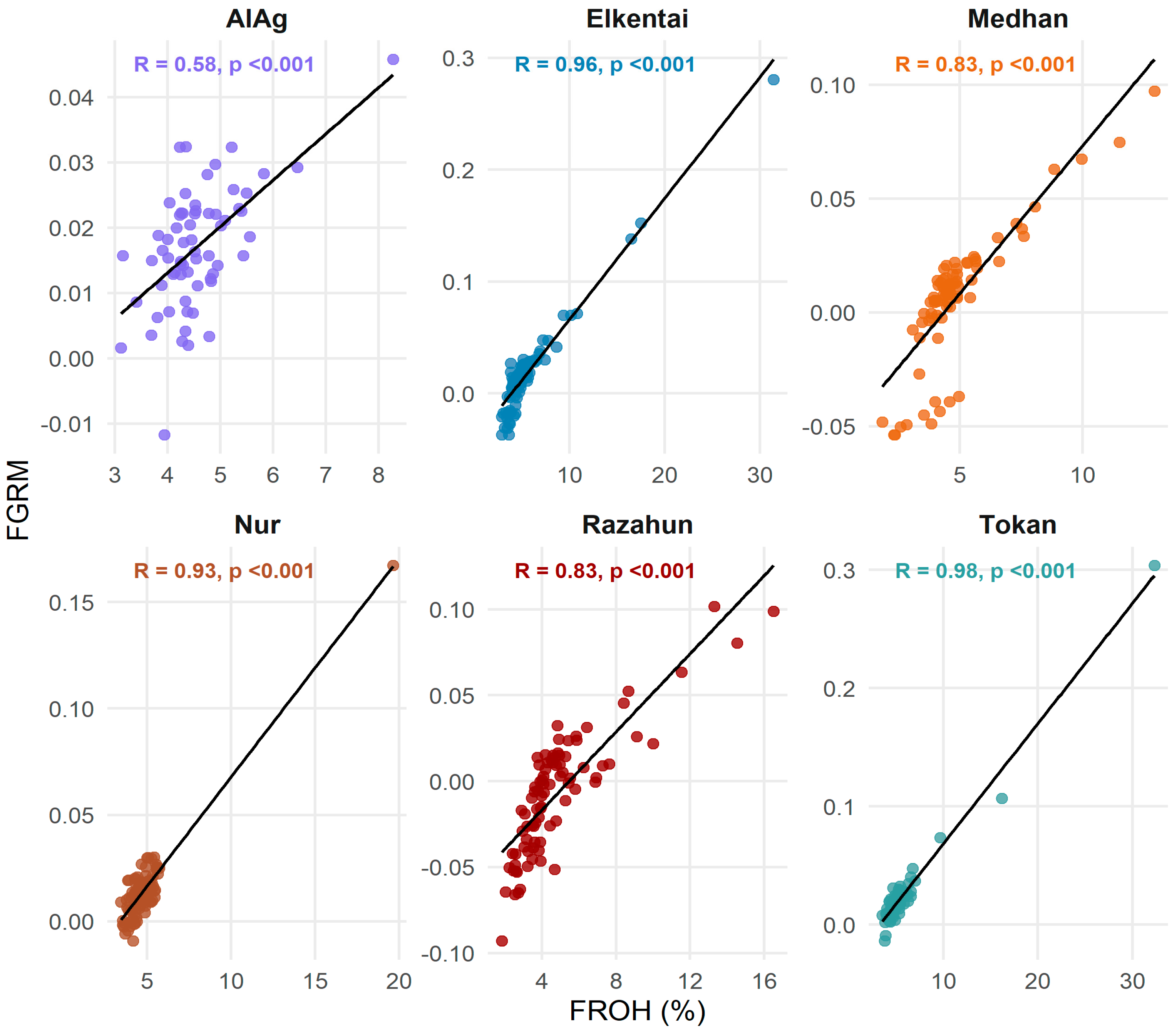
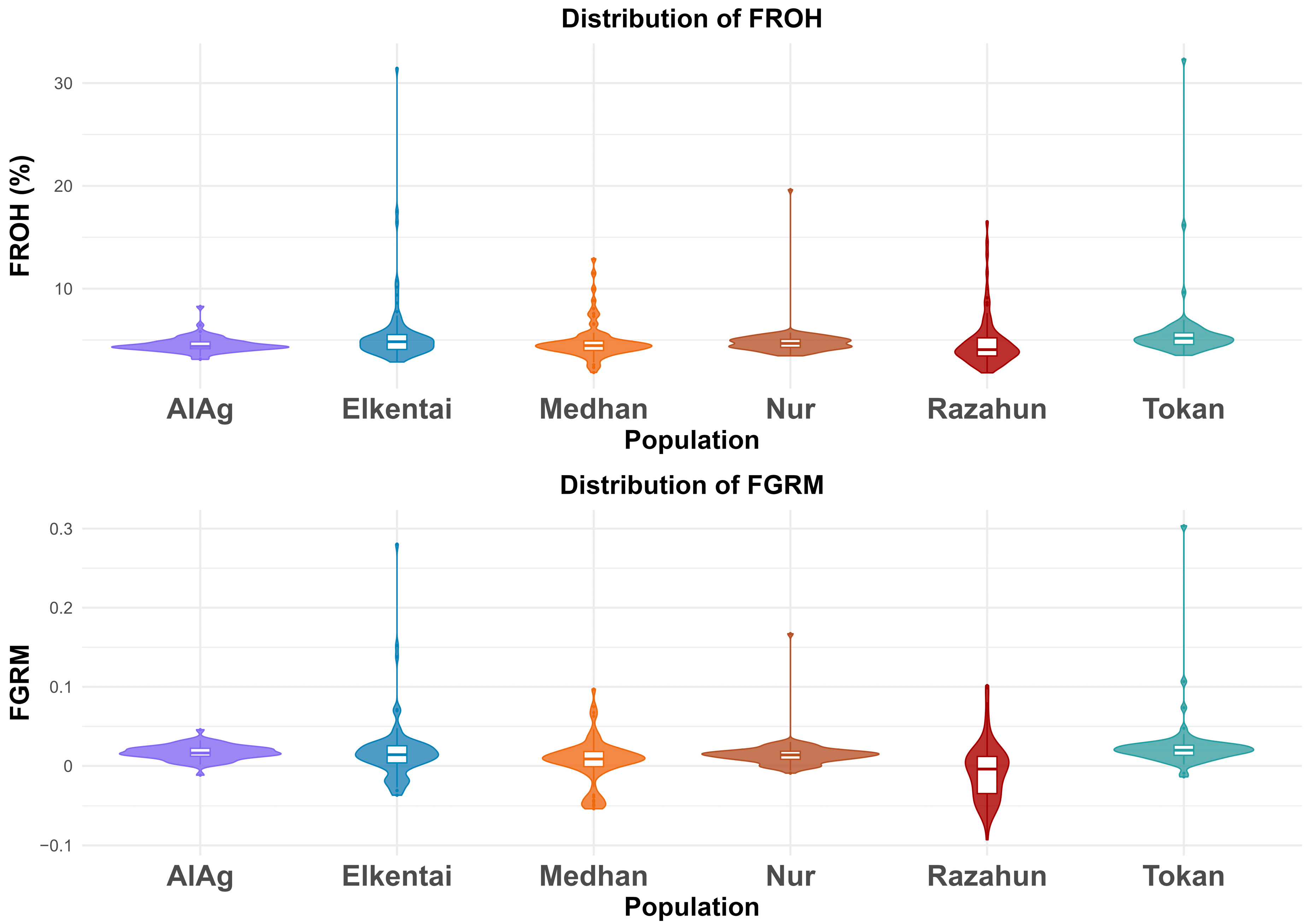
| Population | Total Number of ROH (n) | Mean ROH Length (mb) |
|---|---|---|
| AlAg | 5656 | 1.381 |
| Elkentai | 8267 | 1.721 |
| Medhan | 6613 | 1.530 |
| Nur | 8374 | 1.490 |
| Razahun | 6321 | 1.735 |
| Tokan-1 | 6497 | 1.645 |
| Total | 41,728 | 1.589 |
| FID | ROH_1-2 Mb | ROH_2-4 Mb | ROH_4-8 Mb | ROH_8-16 Mb | ROH_>16 Mb | Total ROH |
|---|---|---|---|---|---|---|
| AlAg | 81 ± 9.3 | 5.4 ± 2.6 | 1.5 ± 0.9 | 1.7 ± 1.5 | NA | 87 ± 10.2 |
| Elkentai | 75.7 ± 11 | 6 ± 2.8 | 2.2 ± 1.7 | 2.3 ± 1.9 | 3 ± 3.9 | 84.4 ± 12.2 |
| Medhan | 77 ± 12.3 | 5.2 ± 2.2 | 2.3 ± 2.2 | 2.4 ± 2.1 | 1.9 ± 1.2 | 83.7 ± 13.6 |
| Nur | 78.1 ± 8.7 | 5.9 ± 2.4 | 1.6 ± 0.8 | 1.3 ± 1.2 | 3 ± 4 | 85.4 ± 9.2 |
| Razahun | 66.2 ± 13.7 | 5 ± 2.8 | 2.3 ± 1.6 | 2.4 ± 1.9 | 3.2 ± 2.6 | 73.5 ± 14.9 |
| Tokan | 81.9 ± 10.3 | 6.8 ± 4.5 | 2.4 ± 3.1 | 1.7 ± 1.5 | 2.7 ± 4.3 | 91.5 ± 14.8 |
| Population | FROH 1–4 Mb | FROH 4–8 Mb | FROH 8–16 Mb | FROH >16 Mb | Total | FROH-FGRM (Mean ± CI) |
|---|---|---|---|---|---|---|
| AlAg | 0.045 | 0.001 | 0.001 | 0.000 | 0.046 | 0.017 (−0.012–0.046) |
| Elkentai | 0.043 | 0.003 | 0.003 | 0.007 | 0.056 | 0.017 (−0.037–0.281) |
| Medhan | 0.042 | 0.002 | 0.002 | 0.003 | 0.049 | 0.006 (−0.054–0.097) |
| Nur | 0.044 | 0.002 | 0.001 | 0.002 | 0.049 | 0.015 (−0.009–0.167) |
| Razahun | 0.037 | 0.002 | 0.004 | 0.006 | 0.049 | −0.007 (−0.093–0.102) |
| Tokan | 0.047 | 0.004 | 0.002 | 0.005 | 0.058 | 0.025 (−0.014–0.304) |
| Population | r (>1 Mb) | r (>4 Mb) | r (>8 Mb) | r (>16 Mb) | r (Total) |
|---|---|---|---|---|---|
| AlAg | 0.451 | 0.397 | 0.486 | NA | 0.583 |
| Elkentai | 0.233 | 0.698 | 0.706 | 0.894 | 0.959 |
| Medhan | 0.576 | 0.483 | 0.554 | 0.649 | 0.831 |
| Nur | 0.103 | 0.181 | 0.819 | 0.878 | 0.932 |
| Razahun | 0.604 | 0.639 | 0.488 | 0.654 | 0.832 |
| Tokan | 0.361 | 0.514 | 0.752 | 0.918 | 0.980 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozhakhmet, A.; Akhatayeva, Z.; Dossybayev, K.; Yermekova, M.; Kapassuly, T.; Yergali, K.; Torekhanov, A.; Bissenov, U.; Lan, X.; Kulataev, B. Genomic Characterization of the Kazakh Fat-Tailed Coarse-Wool Sheep Breed Using ROH Analysis. Animals 2025, 15, 2714. https://doi.org/10.3390/ani15182714
Kozhakhmet A, Akhatayeva Z, Dossybayev K, Yermekova M, Kapassuly T, Yergali K, Torekhanov A, Bissenov U, Lan X, Kulataev B. Genomic Characterization of the Kazakh Fat-Tailed Coarse-Wool Sheep Breed Using ROH Analysis. Animals. 2025; 15(18):2714. https://doi.org/10.3390/ani15182714
Chicago/Turabian StyleKozhakhmet, Altynay, Zhanerke Akhatayeva, Kairat Dossybayev, Marina Yermekova, Tilek Kapassuly, Kanagat Yergali, Aibyn Torekhanov, Utepbergen Bissenov, Xianyong Lan, and Beibit Kulataev. 2025. "Genomic Characterization of the Kazakh Fat-Tailed Coarse-Wool Sheep Breed Using ROH Analysis" Animals 15, no. 18: 2714. https://doi.org/10.3390/ani15182714
APA StyleKozhakhmet, A., Akhatayeva, Z., Dossybayev, K., Yermekova, M., Kapassuly, T., Yergali, K., Torekhanov, A., Bissenov, U., Lan, X., & Kulataev, B. (2025). Genomic Characterization of the Kazakh Fat-Tailed Coarse-Wool Sheep Breed Using ROH Analysis. Animals, 15(18), 2714. https://doi.org/10.3390/ani15182714





