Genomic Characterization of Peruvian Creole Goats: Insights into Population Structure and Runs of Homozygosity
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
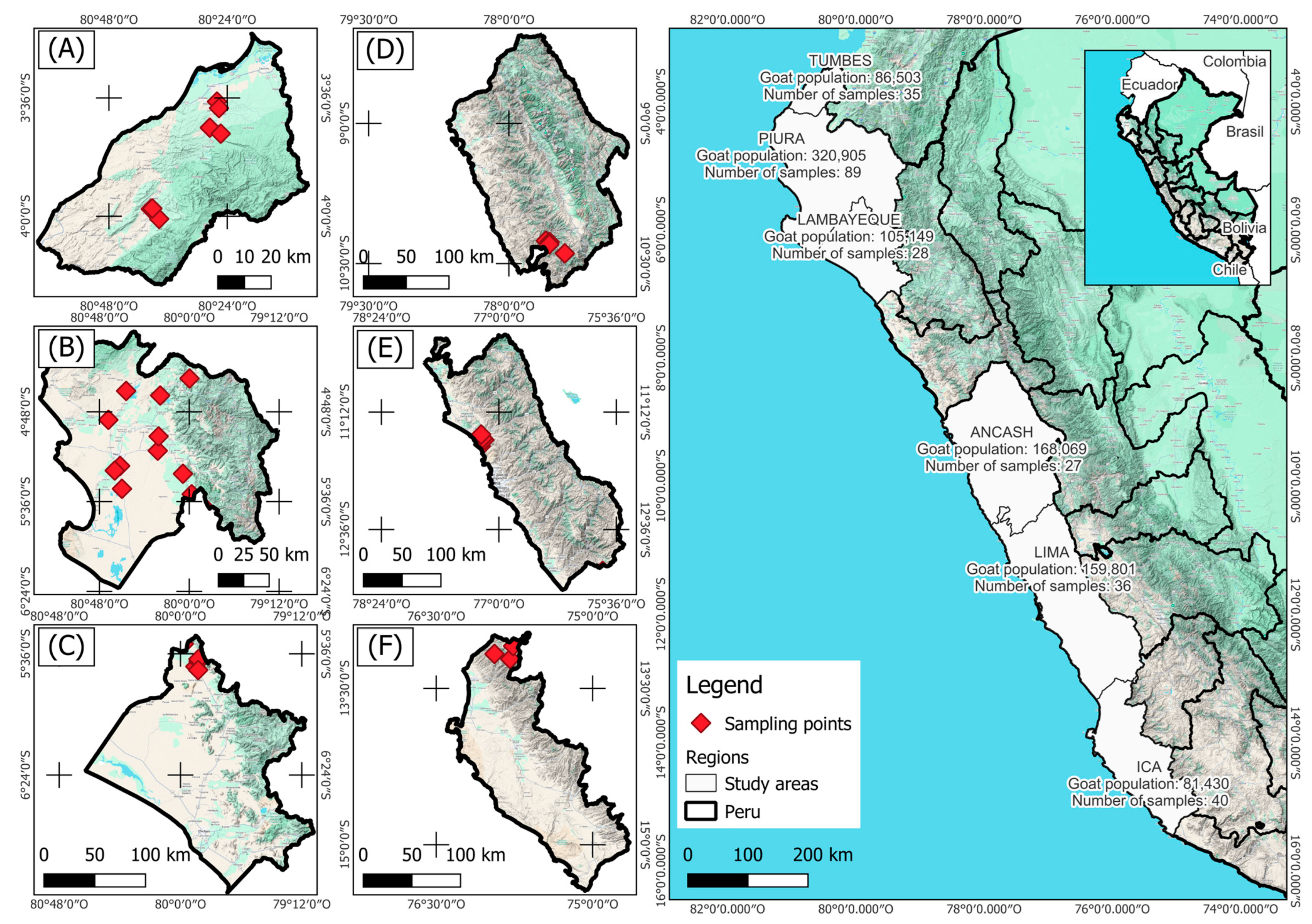
2.2. Animals and Genotyping Data
2.3. Quality Control and Data Filtering
2.4. Population Structure and Runs of Homozygosity
2.5. Population Metrics
3. Results
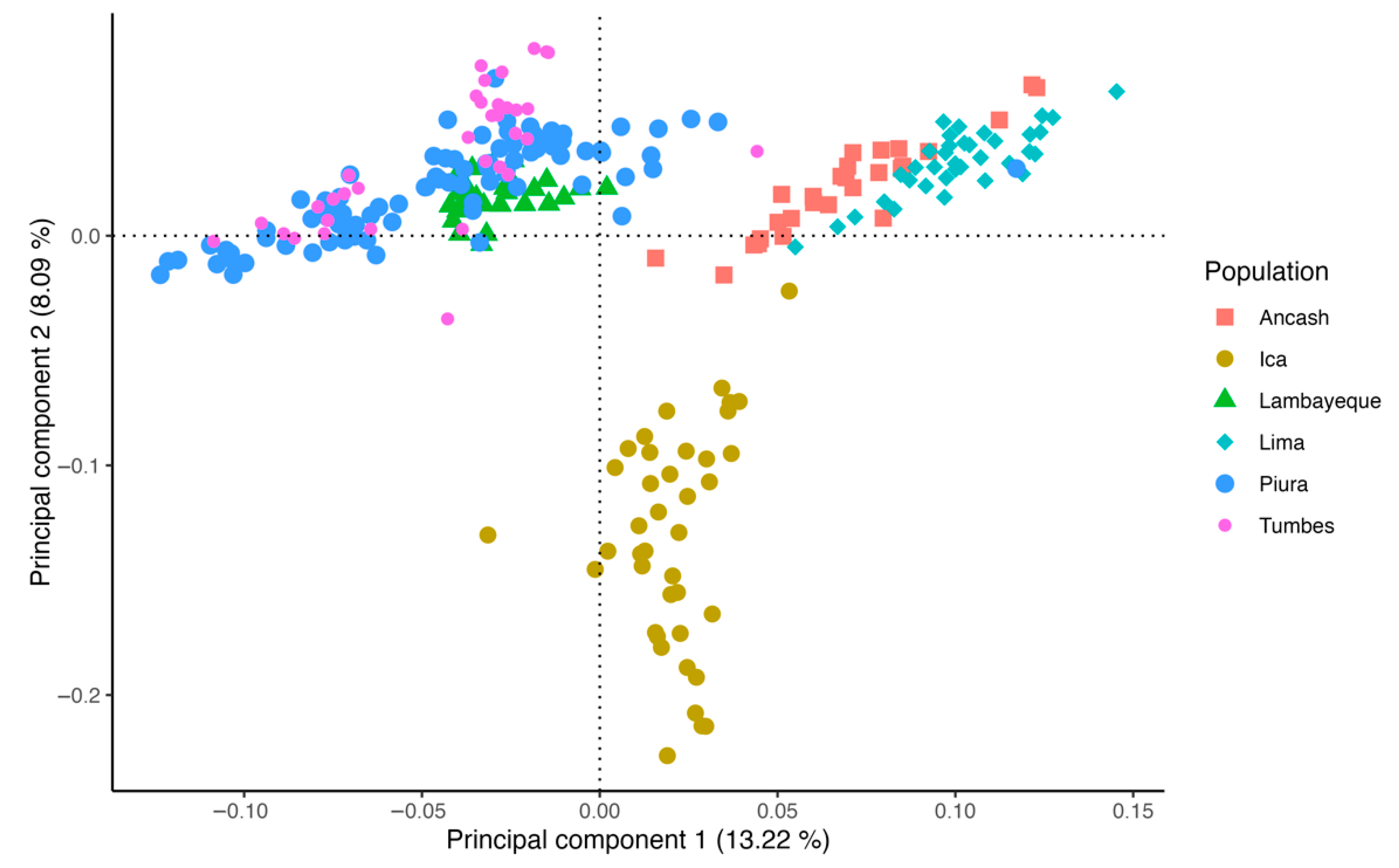
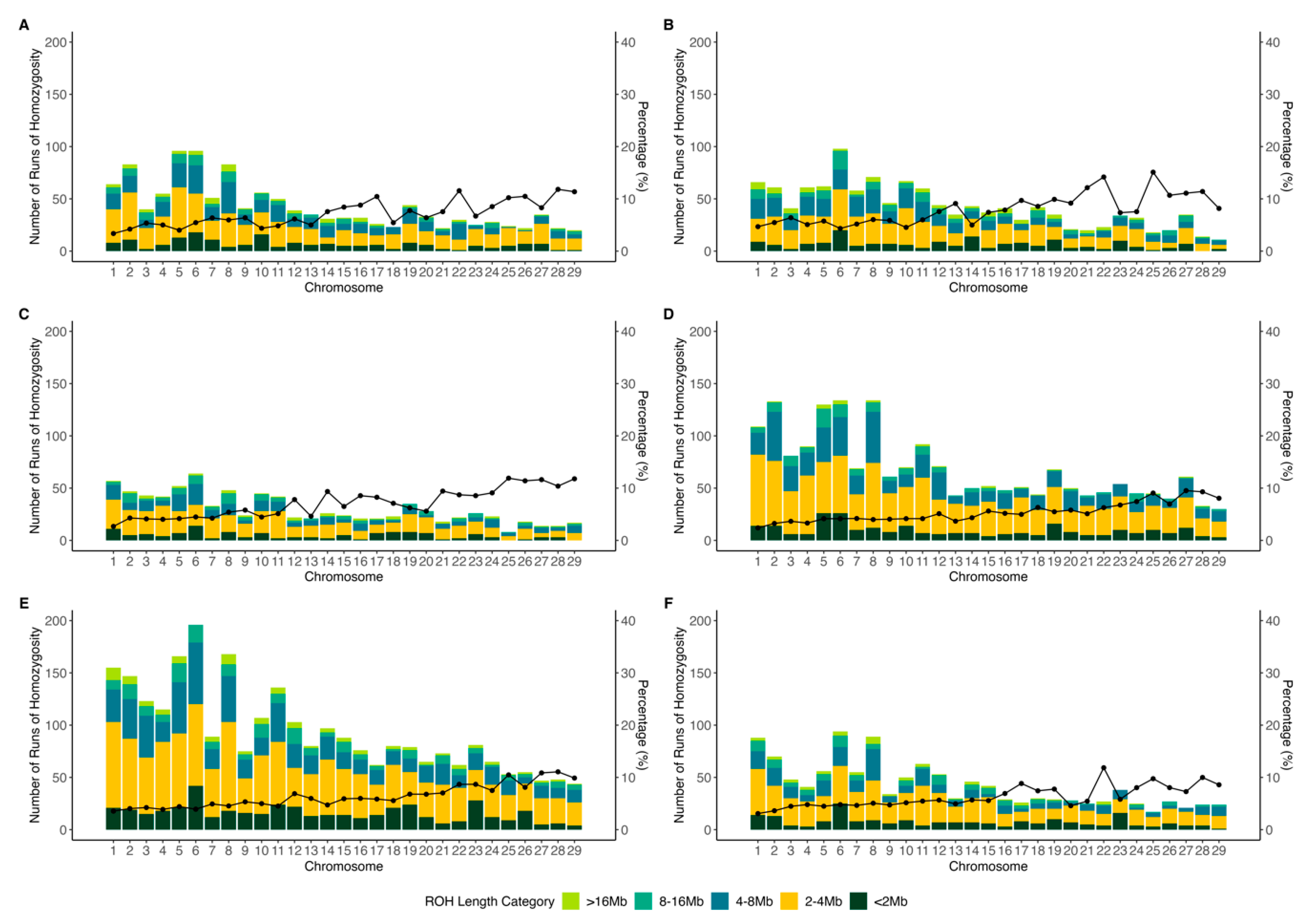
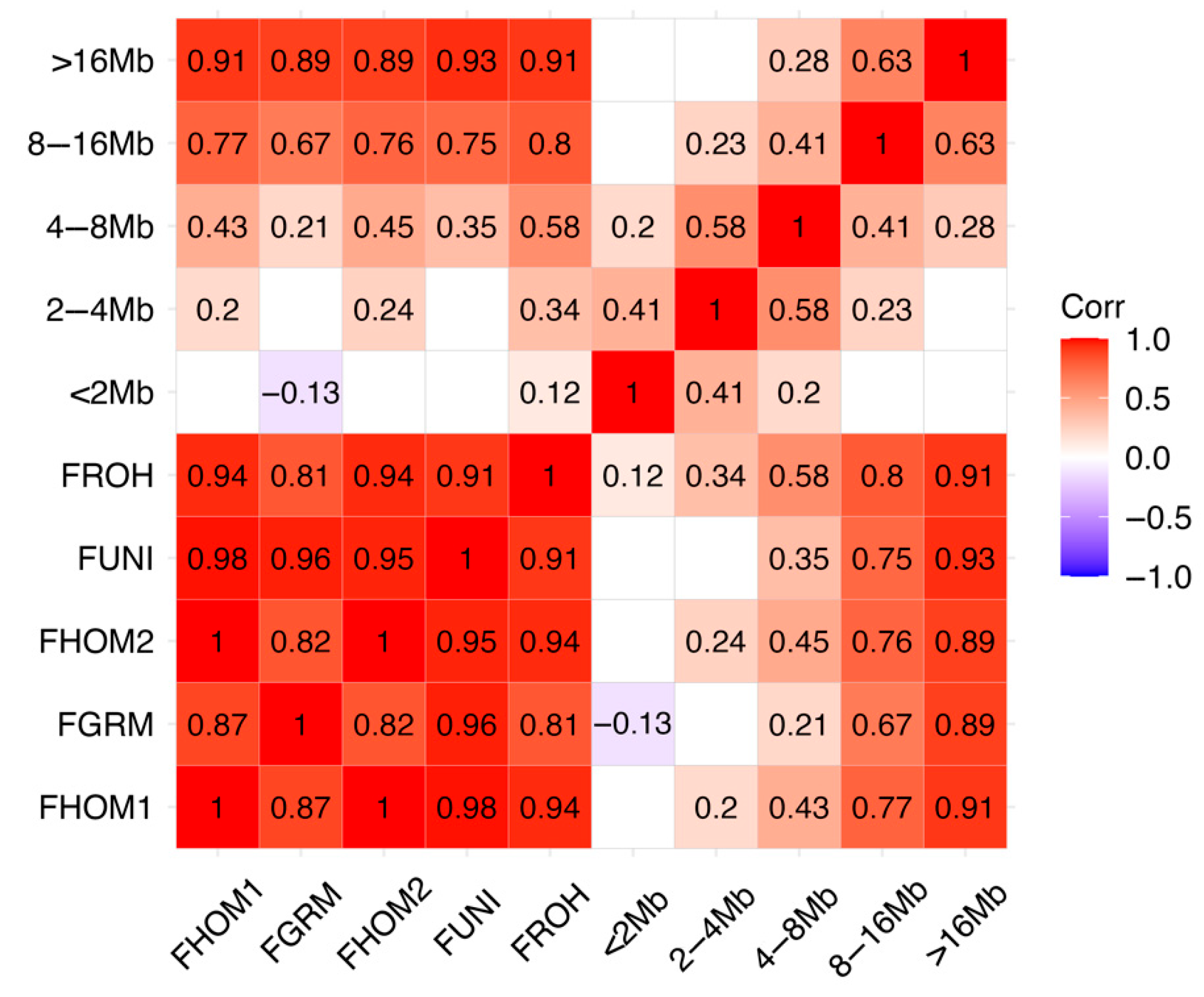
3.1. Population Structure and Runs of Homozygosity
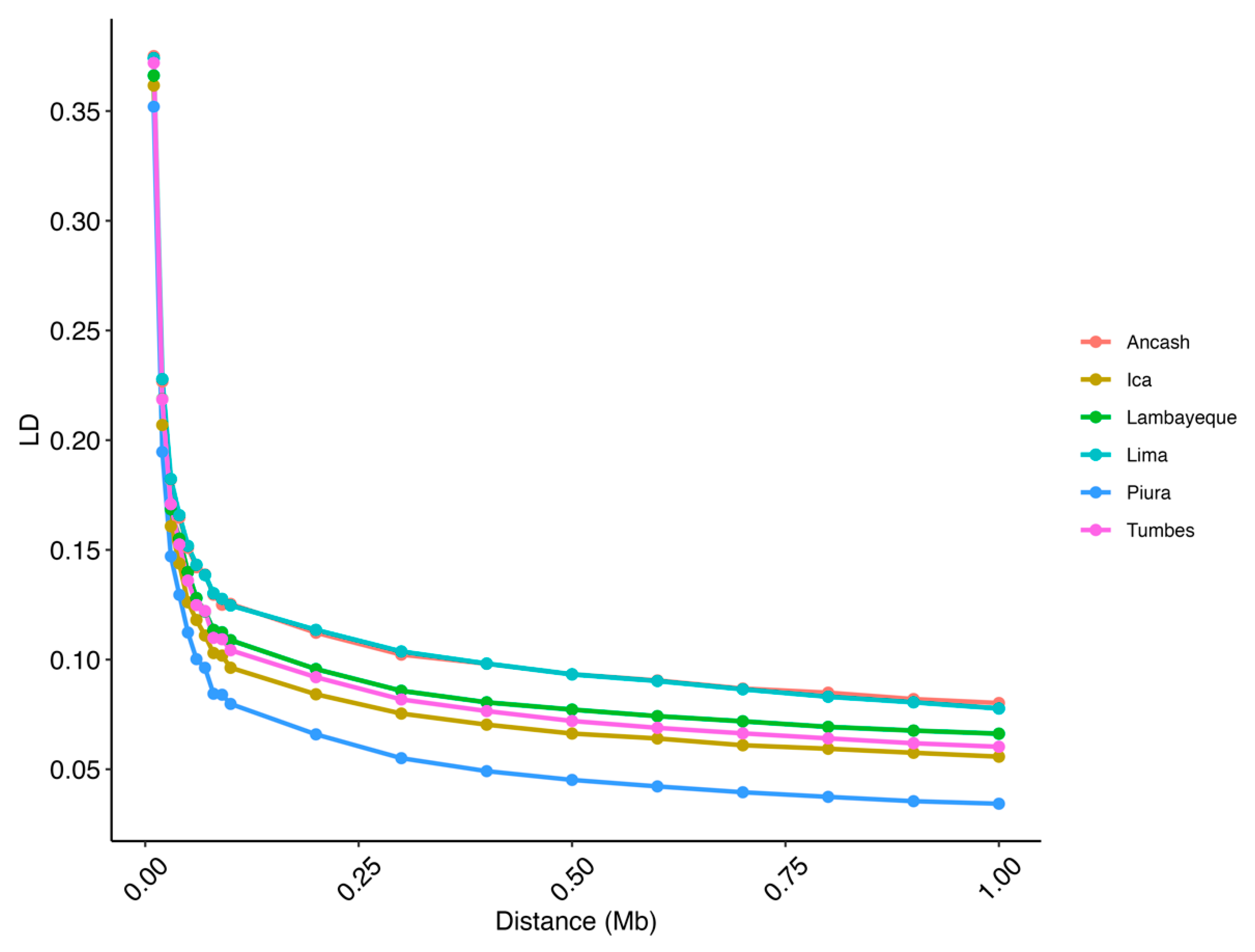
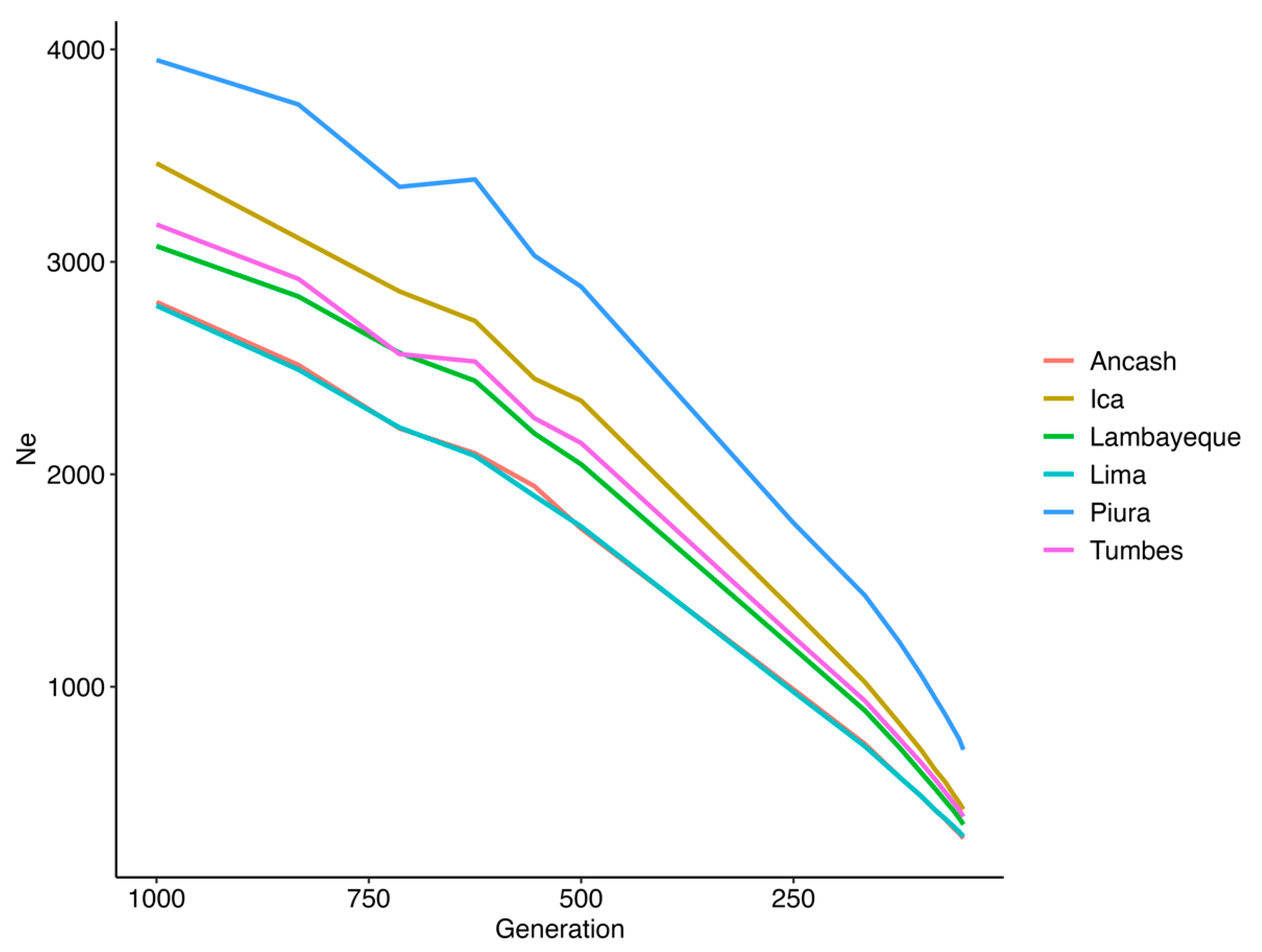
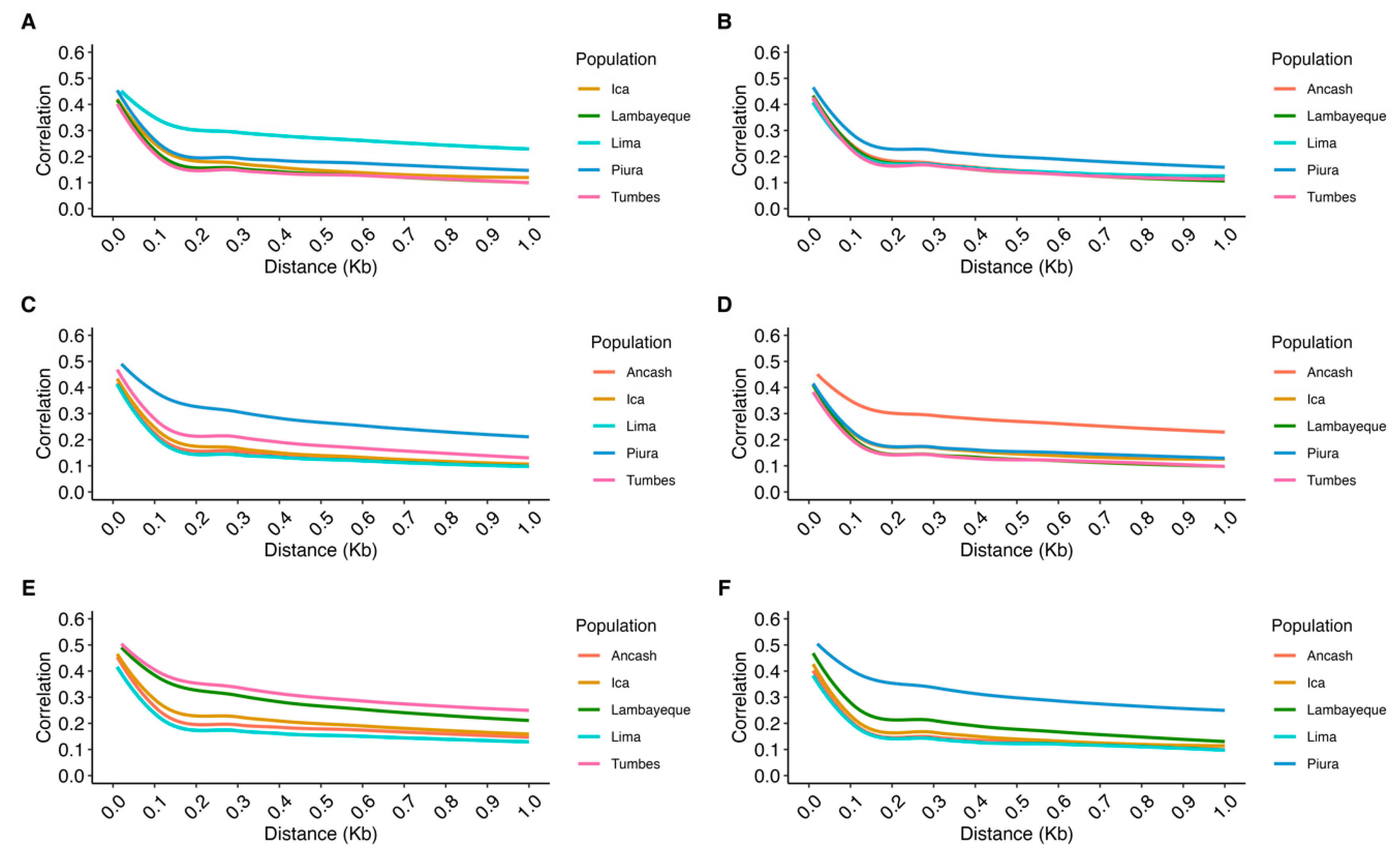
3.2. Population Metrics
4. Discussion
4.1. Population Structure and Runs of Homozygosity
4.2. Population Metrics
4.3. Conservation and Breeding Implications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCG | Peruvian Creole goats |
| SNP | Single nucleotide polymorphism |
| ROH | Runs of homozygosity |
| PCA | Principal component analysis |
| FST | Fixation index |
| MAF | Minor allele frequency |
| HWE | Hardy–Weinberg equilibrium |
| LD | Linkage disequilibrium |
| Ne | Effective population size |
| GPRIN3 | GPRIN family member 3 |
| TIGD2 | Tigger transposable element derived 2 |
| FAM13A | Family with sequence similarity 13 member A |
| HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| NAP1L5 | Nucleosome assembly protein 1 like 5 |
| PYURF | PIGY upstream open reading frame |
| HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| HERC6 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| PPM1K | Protein phosphatase, Mg2+/Mn2+ dependent 1K |
| ABCG2 | ATP binding cassette subfamily G member 2 (JR blood group) |
| PKD2 | Polycystin 2, transient receptor potential cation channel |
| SPP1 | Secreted phosphoprotein 1 |
| MEPE | Matrix extracellular phosphoglycoprotein |
| IBSP | Integrin binding sialoprotein |
| LAP3 | Leucine aminopeptidase 3 |
| MED28 | Mediator complex subunit 28 |
| FAM184B | Family with sequence similarity 184 member B |
| DCAF16 | DDB1 and CUL4 associated factor 16 |
| NCAPG | Non-SMC condensin I complex subunit G |
| LCORL | Ligand dependent nuclear receptor corepressor like |
References
- Gómez-Urviola, N.C.; Gómez-Urviola, J.W.; Celi-Mariátegui, I.D.R.; Milán-Sendra, M.J.; Jordana-Vidal, J. La Cabra Criolla Peruana, Situación Actual y Perspectivas Conservacionistas. In Biodiversidad Caprina Iberoamericana, 1st ed.; Vargas, J.E., Zaragoza, L., Delgado, J.V., Rodríguez, G., Eds.; Ediciones Universidad Cooperativa de Colombia: Bogotá, Colombia, 2016; pp. 163–168. [Google Scholar]
- Paredes Chocce, M.E. (Instituto Nacional de Innovación Agraria, Lima, Peru). Personal communication. 2023. [Google Scholar]
- Whaley, O.Q.; Orellana-Garcia, A.; Pecho-Quispe, J.O. An Annotated Checklist to Vascular Flora of the Ica Region, Peru—With Notes on Endemic Species, Habitat, Climate and Agrobiodiversity. Phytotaxa 2019, 389, 1–125. [Google Scholar] [CrossRef]
- Sessarego, E.A.; Trillo, F.C.; Godoy, D.J.; Palomino-Guerrera, W.; Cruz, J.A. Characterization and Typology of Goat Production Systems in the Southern Highlands of Peru. Vet. World 2025, 18, 220. [Google Scholar] [CrossRef]
- Arroyo, Ó. Situación Actual y Proyecciones de La Crianza de Caprinos En El Perú. Available online: https://utoronto.scholaris.ca/server/api/core/bitstreams/10b467fd-9027-4067-9ea1-606965bf1bc2/content (accessed on 10 November 2024).
- Sarria, J.A.; Ruiz, F.A.; Mena, Y.; Castel, J.M. Caracterización y Propuestas de Mejora de Los Sistemas de Producción Caprina de La Costa Central de Perú. Rev. Mex. Cienc. Pecu. 2014, 5, 487–504. [Google Scholar] [CrossRef]
- Paredes Chocce, M.E.; Ramírez-Vergara, R.; Trillo-Zárate, F.T.; Cruz Luis, J. Characterization of Dairy Goat Production Systems in Coastal Valleys of the Lima Region. Trop. Anim. Health Prod. 2024, 56, 351. [Google Scholar] [CrossRef] [PubMed]
- Perevolotsky, A. Goat Production Systems in Piura, Peru: A Multidisciplinary Analysis. Agric. Syst. 1990, 32, 55–81. [Google Scholar] [CrossRef]
- Socola, T.; Alexander, V. Sistema de Producción de Caprinos En Tres Zonas Vulnerables Al Cambio Climático de La Región Piura. Master’s Thesis, Universidad Nacional Agraria La Molina, Lima, Peru, 2019. [Google Scholar]
- Temoche, V.; Acosta, I.; Gonzales, P.; Godoy Padilla, D.; Jibája, O.; Cruz, J.; Corredor, F.-A. Characterization of Goat Production Systems in the Northern Dry Forest of Peru Using a Multivariate Analysis. Animals 2025, 15, 567. [Google Scholar] [CrossRef]
- Ramirez Antaurco, M.F. (Instituto Nacional de Innovación Agraria, Lima, Peru). Personal communication. 2020. [Google Scholar]
- Giridhar, K.; Samireddypalle, A. Impact of Climate Change on Forage Availability for Livestock. In Climate Change Impact on Livestock: Adaptation and Mitigation; Springer: New Delhi, India, 2015; pp. 97–112. [Google Scholar]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of Homozygosity: Current Knowledge and Applications in Livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and Runs of Homozygosity: A Possible Solution to an Old Problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Ceballos, F.C.; Joshi, P.K.; Clark, D.W.; Ramsay, M.; Wilson, J.F. Runs of Homozygosity: Windows into Population History and Trait Architecture. Nat. Rev. Genet. 2018, 19, 220–234. [Google Scholar] [CrossRef]
- Corredor, F.-A.; Figueroa, D.; Estrada, R.; Burgos-Paz, W.; Salazar, W.; Cruz, W.; Lobato, R.; Injante, P.; Godoy, D.; Barrantes, C. Genome-Wide Single Nucleotide Polymorphisms Reveal the Genetic Diversity and Population Structure of Creole Goats from Northern Peru. Livest. Sci. 2024, 283, 105473. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística e Informática IV Censo Nacional Agropecuario 2012. Available online: http://censos1.inei.gob.pe/Cenagro/redatam/ (accessed on 14 March 2022).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Purcell, S.; Chang, C. PLINK 1.9. Available online: https://www.cog-genomics.org/plink/1.9 (accessed on 14 March 2024).
- Howrigan, D.P.; Simonson, M.A.; Keller, M.C. Detecting Autozygosity through Runs of Homozygosity: A Comparison of Three Autozygosity Detection Algorithms. BMC Genom. 2011, 12, 460. [Google Scholar] [CrossRef]
- Lencz, T.; Lambert, C.; DeRosse, P.; Burdick, K.E.; Morgan, T.V.; Kane, J.M.; Kucherlapati, R.; Malhotra, A.K. Runs of Homozygosity Reveal Highly Penetrant Recessive Loci in Schizophrenia. Proc. Natl. Acad. Sci. USA 2007, 104, 19942–19947. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package BiomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- R core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Li, C.C.; Horvitz, D.G. Some Methods of Estimating the Inbreeding Coefficient. Am. J. Hum. Genet. 1953, 5, 107. [Google Scholar]
- Vanraden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W. Common SNPs Explain a Large Proportion of the Heritability for Human Height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.-L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Corbin, L.J.; Liu, A.Y.H.; Bishop, S.C.; Woolliams, J.A. Estimation of Historical Effective Population Size Using Linkage Disequilibria with Marker Data. J. Anim. Breed. Genet. 2012, 129, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Ginja, C.; Gama, L.T.; Martínez, A.; Sevane, N.; Martin-Burriel, I.; Lanari, M.R.; Revidatti, M.A.; Aranguren-Méndez, J.A.; Bedotti, D.O.; Ribeiro, M.N.; et al. Genetic Diversity and Patterns of Population Structure in Creole Goats from the Americas. Anim. Genet. 2017, 48, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Riofrio, L.; Maza-Tandazo, T.; Quezada-Padilla, M.; Albito-Balcazar, O.; Flores-Gonzalez, A.; Camacho-Enriquez, O.; Martinez-Martinez, A.; Consortium, B.; Delgado-Bermejo, J.V. Genetic Characterization of the “Chusca Lojana”, a Creole Goat Reared in Ecuador, and Its Relationship with Other Goat Breeds. Animals 2020, 10, 1026. [Google Scholar] [CrossRef]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of Homozygosity and Population History in Cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, F.C.; Hazelhurst, S.; Ramsay, M. Runs of Homozygosity in Sub-Saharan African Populations Provide Insights into Complex Demographic Histories. Hum. Genet. 2019, 138, 1123–1142. [Google Scholar] [CrossRef]
- Kirin, M.; McQuillan, R.; Franklin, C.S.; Campbell, H.; McKeigue, P.M.; Wilson, J.F. Genomic Runs of Homozygosity Record Population History and Consanguinity. PLoS ONE 2010, 5, e13996. [Google Scholar] [CrossRef]
- Zhao, Q.; Huang, C.; Chen, Q.; Su, Y.; Zhang, Y.; Wang, R.; Su, R.; Xu, H.; Liu, S.; Ma, Y.; et al. Genomic Inbreeding and Runs of Homozygosity Analysis of Cashmere Goat. Animals 2024, 14, 1246. [Google Scholar] [CrossRef] [PubMed]
- Sumreddee, P.; Hay, E.H.; Toghiani, S.; Roberts, A.; Aggrey, S.E.; Rekaya, R. Grid Search Approach to Discriminate Between Old and Recent Inbreeding Using Phenotypic, Pedigree and Genomic Information. BMC Genom. 2021, 22, 538. [Google Scholar] [CrossRef]
- Tao, L.; He, X.; Jiang, Y.; Liu, Y.; Ouyang, Y.; Shen, Y.; Hong, Q.; Chu, M. Genome-Wide Analyses Reveal Genetic Convergence of Prolificacy Between Goats and Sheep. Genes 2021, 12, 480. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, E.; Vera, B.; Peraza, P.; Ciappesoni, G.; Damián, J.P.; Van Lier, E. Identification of Candidate Genes and Pathways Linked to the Temperament Trait in Sheep. Genes 2024, 15, 229. [Google Scholar] [CrossRef]
- Li, X.; Ran, L.; Li, Y.; Wang, Y.; Xiong, Y.; Wang, Y.; Xing, J.; Lin, Y. Molecular Characterizations of FAM13A and Its Functional Role in Inhibiting the Differentiation of Goat Intramuscular Adipocytes Through RIG-I Receptor Signaling Pathway. Genes 2024, 15, 1143. [Google Scholar] [CrossRef]
- Kowalewska-Łuczak, I.; Kulig, H. Polymorphism of the FAM13A, ABCG2, OPN, LAP3, HCAP-G, PPARGC1A Genes and Somatic Cell Count of Jersey Cows–Preliminary Study. Res. Vet. Sci. 2013, 94, 252–255. [Google Scholar] [CrossRef]
- Al Kalaldeh, M.; Gibson, J.; Lee, S.H.; Gondro, C.; Van Der Werf, J.H.J. Detection of Genomic Regions Underlying Resistance to Gastrointestinal Parasites in Australian Sheep. Genet. Sel. Evol. 2019, 51, 37. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Bissonnette, N.; Lacasse, P.; Miglior, F.; Sargolzaei, M.; Zhao, X.; Ibeagha-Awemu, E.M. Genome-Wide Association Analysis and Pathways Enrichment for Lactation Persistency in Canadian Holstein Cattle. J. Dairy Sci. 2017, 100, 1955–1970. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, V.B.; Schenkel, F.S.; Chen, S.-Y.; Oliveira, H.R.; Casey, T.M.; Melka, M.G.; Brito, L.F. Genomewide Association Analyses of Lactation Persistency and Milk Production Traits in Holstein Cattle Based on Imputed Whole-Genome Sequence Data. Genes 2021, 12, 1830. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.; Zhang, X.; Li, M.; Luo, N.; Zhao, Y. Screening of Candidate Housekeeping Genes in Uterus Caruncle by RNA-Sequence and QPCR Analyses in Different Stages of Goat (Capra hircus). Animals 2023, 13, 1897. [Google Scholar] [CrossRef]
- Modesto, P.; Peletto, S.; Pisoni, G.; Cremonesi, P.; Castiglioni, B.; Colussi, S.; Caramelli, M.; Bronzo, V.; Moroni, P.; Acutis, P.L. Evaluation of Internal Reference Genes for Quantitative Expression Analysis by Real-Time Reverse Transcription-PCR in Somatic Cells from Goat Milk. J. Dairy Sci. 2013, 96, 7932–7944. [Google Scholar] [CrossRef]
- Takasuga, A. PLAG1 and NCAPG-LCORL in Livestock. Anim. Sci. J. 2016, 87, 159–167. [Google Scholar] [CrossRef]
- La, Y.; Zhang, X.; Li, F.; Zhang, D.; Li, C.; Mo, F.; Wang, W. Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep. Genes 2019, 10, 616. [Google Scholar] [CrossRef] [PubMed]
- Majeres, L.E.; Dilger, A.C.; Shike, D.W.; McCann, J.C.; Beever, J.E. Defining a Haplotype Encompassing the LCORL-NCAPG Locus Associated with Increased Lean Growth in Beef Cattle. Genes 2024, 15, 576. [Google Scholar] [CrossRef] [PubMed]
- Buaban, S.; Lengnudum, K.; Boonkum, W.; Phakdeedindan, P. Genome-Wide Association Study on Milk Production and Somatic Cell Score for Thai Dairy Cattle Using Weighted Single-Step Approach with Random Regression Test-Day Model. J. Dairy Sci. 2022, 105, 468–494. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Farahani, A.H.K.; Moradi, M.H.; Mastrangelo, S.; Di Gerlando, R.; Sardina, M.T.; Scatassa, M.L.; Portolano, B.; Tolone, M. Weighted Single-Step Genome-Wide Association Study Uncovers Known and Novel Candidate Genomic Regions for Milk Production Traits and Somatic Cell Score in Valle Del Belice Dairy Sheep. Animals 2022, 12, 1155. [Google Scholar] [CrossRef]
- Tarekegn, G.M.; Khayatzadeh, N.; Liu, B.; Osama, S.; Haile, A.; Rischkowsky, B.; Zhang, W.; Tesfaye, K.; Dessie, T.; Mwai, O.A.; et al. Ethiopian Indigenous Goats Offer Insights into Past and Recent Demographic Dynamics and Local Adaptation in Sub-Saharan African Goats. Evol. Appl. 2021, 14, 1716–1731. [Google Scholar] [CrossRef]
- Michailidou, S.; Tsangaris, G.T.; Tzora, A.; Skoufos, I.; Banos, G.; Argiriou, A.; Arsenos, G. Analysis of Genome-Wide DNA Arrays Reveals the Genomic Population Structure and Diversity in Autochthonous Greek Goat Breeds. PLoS ONE 2019, 14, e0226179. [Google Scholar] [CrossRef]
- Alderson, L. Breeds at Risk: Definition and Measurement of the Factors Which Determine Endangerment. Livest. Sci. 2009, 123, 23–27. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Tolone, M.; Di Gerlando, R.; Fontanesi, L.; Sardina, M.T.; Portolano, B. Genomic Inbreeding Estimation in Small Populations: Evaluation of Runs of Homozygosity in Three Local Dairy Cattle Breeds. Animal 2016, 10, 746–754. [Google Scholar] [CrossRef]
- Alemu, S.W.; Kadri, N.K.; Harland, C.; Faux, P.; Charlier, C.; Caballero, A.; Druet, T. An Evaluation of Inbreeding Measures Using a Whole-Genome Sequenced Cattle Pedigree. Heredity 2020, 126, 410–423. [Google Scholar] [CrossRef]
- Monau, P.I.; Visser, C.; Muchadeyi, F.C.; Okpeku, M.; Nsoso, S.J.; Van Marle-Köster, E. Population Structure of Indigenous Southern African Goats Based on the Illumina Goat50K SNP Panel. Trop. Anim. Health Prod. 2020, 52, 1795–1802. [Google Scholar] [CrossRef] [PubMed]
- Amills, M.; Capote, J.; Tosser-Klopp, G. Goat Domestication and Breeding: A Jigsaw of Historical, Biological and Molecular Data with Missing Pieces. Anim. Genet. 2017, 48, 631–644. [Google Scholar] [CrossRef]
- Kumar, C.; Song, S.; Dewani, P.; Kumar, M.; Parkash, O.; Ma, Y.; Malhi, K.K.; Yang, N.; Mwacharo, J.M.; He, X.; et al. Population Structure, Genetic Diversity and Selection Signatures Within Seven Indigenous Pakistani Goat Populations. Anim. Genet. 2018, 49, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, D.; Martínez, R.; Manrique, C.; Parra, L.M.; Rocha, J.F.; Gómez, Y.; Abuabara, Y.; Gallego, J. Linkage Disequilibrium Levels and Allele Frequency Distribution in Blanco Orejinegro and Romosinuano Creole Cattle Using Medium Density SNP Chip Data. Genet. Mol. Biol. 2018, 41, 426–433. [Google Scholar] [CrossRef]
- Caivio-Nasner, S.; López-Herrera, A.; González-Herrera, L.G.; Rincón, J.C. Diversity Analysis, Runs of Homozygosity and Genomic Inbreeding Reveal Recent Selection in Blanco Orejinegro Cattle. J. Anim. Breed. Genet. 2021, 138, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Bottani, G. Bolivian Creole Cattle: Population Structure, Genetic Diversity and Management Practices. Acta Univ. Agric. Sueciae 2020, 2020, 7. [Google Scholar]
- Pérez O’Brien, A.M.; Mészáros, G.; Utsunomiya, Y.T.; Sonstegard, T.S.; Garcia, J.F.; Van Tassell, C.P.; Carvalheiro, R.; da Silva, M.V.B.; Sölkner, J. Linkage Disequilibrium Levels in Bos Indicus and Bos Taurus Cattle Using Medium and High Density SNP Chip Data and Different Minor Allele Frequency Distributions. Livest. Sci. 2014, 166, 121–132. [Google Scholar] [CrossRef]
- Karimi, K.; Esmailizadeh Koshkoiyeh, A.; Gondro, C. Comparison of Linkage Disequilibrium Levels in Iranian Indigenous Cattle Using Whole Genome SNPs Data. J. Anim. Sci. Technol. 2015, 57, 47. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Bradshaw, C.J.A.; Brook, B.W. Genetics in Conservation Management: Revised Recommendations for the 50/500 Rules, Red List Criteria and Population Viability Analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Kichamu, N.; Wanjala, G.; Toma Cziszter, L.; Strausz, P.; Kusuma Astuti, P.; Bagi, Z.; Kusza, S. Assessing the Population Structure and Genetic Variability of Kenyan Native Goats Under Extensive Production System. Sci. Rep. 2024, 14, 16342. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Baneh, H.; Roy, R.; Notter, D.R. Genetic Diversity and Population Structure of Jamunapari Goat in India Using Pedigree Analysis. Trop. Anim. Health Prod. 2021, 53, 218. [Google Scholar] [CrossRef]
- Rashidi, A.; Mokhtari, M.S.; Gutiérrez, J.P. Pedigree Analysis and Inbreeding Effects on Early Growth Traits and Greasy Fleece Weight in Markhoz Goat. Small Rumin. Res. 2015, 124, 1–8. [Google Scholar] [CrossRef]
- Vermette, S.; Jafarikia, M.; Maignel, L.; Wyss, S.; Sullivan, B.; Brito, L.; Schenkel, F.; Weaver, K.; Girouard, S. Final Report Goat Herd Improvement on Productivity and Health Using Genomics. Available online: https://www.researchgate.net/publication/303311547_Goat_Herd_improvement_on_productivity_and_health_using_genomics (accessed on 10 November 2024).
- Teissier, M.; Brito, L.F.; Schenkel, F.S.; Bruni, G.; Fresi, P.; Bapst, B.; Robert-Granie, C.; Larroque, H. Genetic Characterization and Population Connectedness of North American and European Dairy Goats. Front. Genet. 2022, 13, 862838. [Google Scholar] [CrossRef]
- Brito, L.F.; Jafarikia, M.; Grossi, D.A.; Kijas, J.W.; Porto-Neto, L.R.; Ventura, R.V.; Salgorzaei, M.; Schenkel, F.S. Characterization of Linkage Disequilibrium, Consistency of Gametic Phase and Admixture in Australian and Canadian Goats. BMC Genet. 2015, 16, 67. [Google Scholar] [CrossRef]
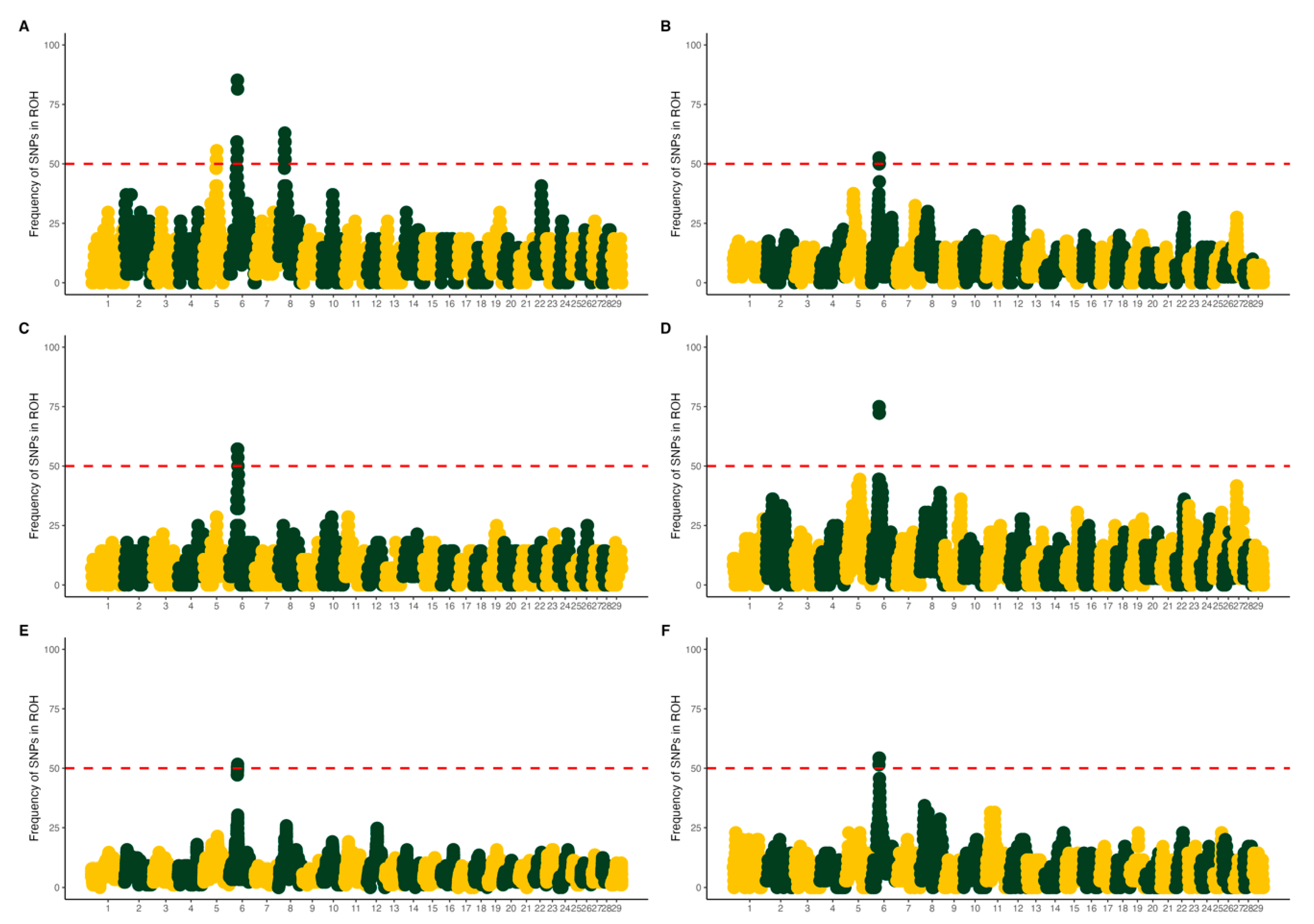
| Chr | Start (bp) | End (bp) | Length | Gene | Description |
|---|---|---|---|---|---|
| 6 | 36,070,368 | 36,072,710 | 2342 | GPRIN3 | GPRIN family member 3 |
| 6 | 36,205,904 | 36,207,478 | 1574 | TIGD2 | Tigger transposable element derived 2 |
| 6 | 36,424,597 | 36,551,359 | 126,762 | FAM13A | Family with sequence similarity 13 member A |
| 6 | 36,572,520 | 36,707,017 | 134,497 | HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| 6 | 36,593,698 | 36,594,279 | 581 | NAP1L5 | Nucleosome assembly protein 1 like 5 |
| 6 | 36,759,209 | 36,761,094 | 1885 | PYURF | PIGY upstream open reading frame |
| 6 | 36,765,648 | 36,813,335 | 47,687 | HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| 6 | 36,819,857 | 36,872,364 | 52,507 | HERC6 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| 6 | 36,967,148 | 36,981,000 | 13,852 | PPM1K | Protein phosphatase, Mg2+/Mn2+ dependent 1K |
| 6 | 36,987,612 | 37,118,958 | 131,346 | ABCG2 | ATP binding cassette subfamily G member 2 (JR blood group) |
| 6 | 37,126,098 | 37,193,068 | 66,970 | PKD2 | Polycystin 2, transient receptor potential cation channel |
| 6 | 37,213,669 | 37,221,650 | 7981 | SPP1 | Secreted phosphoprotein 1 |
| 6 | 37,364,698 | 37,368,465 | 3767 | MEPE | Matrix extracellular phosphoglycoprotein |
| 6 | 37,397,003 | 37,414,860 | 17,857 | IBSP | Integrin binding sialoprotein |
| 6 | 37,669,173 | 37,694,927 | 25,754 | LAP3 | Leucine aminopeptidase 3 |
| 6 | 37,696,130 | 37,702,512 | 6382 | MED28 | Mediator complex subunit 28 |
| 6 | 37,708,219 | 37,826,187 | 117,968 | FAM184B | Family with sequence similarity 184 member B |
| 6 | 37,846,165 | 37,848,349 | 2184 | DCAF16 | DDB1 and CUL4 associated factor 16 |
| 6 | 37,858,170 | 37,903,004 | 44,834 | NCAPG | Non-SMC condensin I complex subunit G |
| 6 | 37,905,295 | 38,068,616 | 163,321 | LCORL | Ligand dependent nuclear receptor corepressor like |
| Inbreeding Metric | Ancash | Ica | Lambayeque | Lima | Piura | Tumbes |
|---|---|---|---|---|---|---|
| FHOM1 | 0.06 | 0.05 | 0.04 | 0.05 | 0.04 | 0.05 |
| FGRM | 0.05 | 0.07 | 0.04 | 0.03 | 0.04 | 0.04 |
| FHOM2 | 0.07 | 0.04 | 0.04 | 0.05 | 0.04 | 0.05 |
| FUNI | 0.06 | 0.06 | 0.04 | 0.04 | 0.04 | 0.05 |
| FROH | 1.04× 10−4 | 7.60 × 10−5 | 6.66 × 10−5 | 9.35 × 10−5 | 6.13 × 10−5 | 7.14 × 10−5 |
| <2 Mb | 4.80 × 10−6 | 3.09 × 10−6 | 3.35 × 10−6 | 5.35 × 10−6 | 3.63 × 10−6 | 4.24 × 10−6 |
| 2–4 Mb | 2.41 × 10−5 | 1.49 × 10−5 | 1.68 × 10−5 | 3.26 × 10−5 | 1.64 × 10−5 | 1.96 × 10−5 |
| 4–8 Mb | 2.58 × 10−5 | 1.71 × 10−5 | 1.57 × 10−5 | 3.20 × 10−5 | 1.61 × 10−5 | 1.66 × 10−5 |
| 8–16 Mb | 1.86 × 10−5 | 1.50 × 10−5 | 1.47 × 10−5 | 1.60 × 10−5 | 1.08 × 10−5 | 1.40 × 10−5 |
| >16 Mb | 3.09 × 10−5 | 2.59 × 10−5 | 1.60 × 10−5 | 7.54 × 10−6 | 1.44 × 10−5 | 1.70 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corredor, F.-A.; Godoy-Padilla, D.; Sessarego, E.A.; Temoche-Socola, V.; Paredes Chocce, M.E.; Escobar Robledo, H.; Ramírez Antaurco, M.F.; Burgos-Paz, W.; Ruiz, J.; Cruz, J.; et al. Genomic Characterization of Peruvian Creole Goats: Insights into Population Structure and Runs of Homozygosity. Animals 2025, 15, 2577. https://doi.org/10.3390/ani15172577
Corredor F-A, Godoy-Padilla D, Sessarego EA, Temoche-Socola V, Paredes Chocce ME, Escobar Robledo H, Ramírez Antaurco MF, Burgos-Paz W, Ruiz J, Cruz J, et al. Genomic Characterization of Peruvian Creole Goats: Insights into Population Structure and Runs of Homozygosity. Animals. 2025; 15(17):2577. https://doi.org/10.3390/ani15172577
Chicago/Turabian StyleCorredor, Flor-Anita, David Godoy-Padilla, Emmanuel Alexander Sessarego, Víctor Temoche-Socola, Miguel Enrique Paredes Chocce, Héctor Escobar Robledo, Máximo Fabricio Ramírez Antaurco, William Burgos-Paz, José Ruiz, Juancarlos Cruz, and et al. 2025. "Genomic Characterization of Peruvian Creole Goats: Insights into Population Structure and Runs of Homozygosity" Animals 15, no. 17: 2577. https://doi.org/10.3390/ani15172577
APA StyleCorredor, F.-A., Godoy-Padilla, D., Sessarego, E. A., Temoche-Socola, V., Paredes Chocce, M. E., Escobar Robledo, H., Ramírez Antaurco, M. F., Burgos-Paz, W., Ruiz, J., Cruz, J., Mulim, H. A., & Oliveira, H. R. d. (2025). Genomic Characterization of Peruvian Creole Goats: Insights into Population Structure and Runs of Homozygosity. Animals, 15(17), 2577. https://doi.org/10.3390/ani15172577








