Simple Summary
Heat shock proteins (HSPs) are a family of highly conserved proteins identified in diverse living organisms that protect cells from stress. HSPs also participate in the activation of defense mechanisms in the body, promoting the activation of the immune system. Previous studies have shown that the administration of DnaK (bacterial homolog of HSP70) may be a strategy to potentiate the immune response and survival of aquatic organisms against bacterial infections. Thus, this study aimed to evaluate the effect of cells overexpressing DnaK on the mortality and immune-related gene expression in gnotobiotic sea bass larvae challenged with Vibrio anguillarum. The results showed lower mortality in the sea bass larvae treated with DnaK and challenged with V. anguillarum. This response would be promoted by an increase in the expression of genes associated with antimicrobial, pro-inflammatory, and chemotaxis responses. These findings suggest that DnaK administration induces a potent innate immune response, enhancing the survival of sea bass larvae challenged with V. anguillarum. The use of DnaK could be used as a pathogen resistance agent, allowing the prevention of diseases in aquatic organisms used for aquaculture.
Abstract
Heat shock proteins (HSPs), particularly HSP70, play a vital role in fish immune defense against pathogens. The administration of DnaK (bacterial homolog of HSP70) may be a strategy to potentiate the immune response and survival of aquatic organisms. This study evaluates the effect of cells overexpressing DnaK on mortality and immune-related gene expression in gnotobiotic sea bass larvae challenged with Vibrio anguillarum. Larvae were subjected to different treatments: NB (no bacteria), YS0 (E. coli with no plasmid), YS1 (E. coli expressing truncated DnaK), and YS2 (E. coli expressing DnaK), and then infected with V. anguillarum at 7 days post-hatching (dph). Mortality was monitored, and RT-qPCR was used to evaluate immune gene expression at 0, 18, 24, 36, and 120 hpc. While no significant variations were recorded in the non-challenged larvae, constant and sustained mortality was observed in challenged larvae from 60 to 120 hpc. However, lower mortality was observed in the larvae treated with DnaK. DnaK treatment promoted the expression of antimicrobial (hepcidin, transferrin) and chemotaxis genes (ccl4), which was further enhanced after a challenge with V. anguillarum, in conjunction with the modulation of il1β and il-8 at 120 hpc. These findings suggest that DnaK induces a potent innate immune response, improving survival against V. anguillarum and supporting its potential use as a disease-preventive strategy in aquaculture.
1. Introduction
Stress is a state in which intrinsic and/or extrinsic stressors disrupt or threaten the organism’s homeostasis, which may harm its integrity [1,2]. Once the stressor is perceived, the hypothalamic-pituitary-interrenal (HPI) axis is activated, releasing cortisol, considered the main glucocorticoid [3]. The physiological stress response mediates the release of stress hormones and is accompanied by a rapid increase in the synthesis of stress-specific proteins to regulate the overall reaction to this allostatic load. Heat shock proteins (HSPs) synthesis is also promoted at the cellular level [4,5,6]. HSPs are a family of highly conserved molecular chaperones identified in diverse living organisms, from bacteria to humans [7]. They are essential in various biological processes, such as the transporting, folding, and assembly of degraded or misfolded proteins. Moreover, several reports have shown that HSPs participate in host defense by potentiating inflammatory responses and promoting cytokine production [8,9,10,11,12]. Due to these immunomodulatory properties, HSPs have been explored as potential antigens for infection control [13].
The members of this superfamily are classified according to their relative molecular mass, structural homology, and function [9]. Three HSP families are described: HSP90 (85–90 kDa), HSP70 (68–73 kDa), and low molecular weight HSP (16–47 kDa) [14]. The HSP70 family is the most abundant and best studied [10]. HSP70 is involved in numerous functions in various cellular processes, including protecting proteins against stress, and has been identified in several fish species [7,9]. Traditionally, this protein is considered a biomarker of environmental stress in teleost fish. However, its expression has been documented to increase with dietary additives in aquaculture [15,16]. According to previous studies, the use of dietary probiotics and prebiotics leads to increased expression of HSP70 in the intestine of rainbow trout [17] and Asian seabass (Lates calcarifer) [18], respectively. This upregulation has improved fish health and enhanced resistance to pathogen infections. In fact, HSP70 has gained increasing attention in aquaculture fish species for its involvement in immune response against pathogens [19], enhancing resistance against Vibrio campbellii and V. proteolyticus [1].
The prokaryotic HSP70, also known as DnaK, can induce innate immune responses and survival in several aquatic organisms, including fish, shrimp, and mollusks, against different bacterial infections [20,21]. DnaK homologs are present in most bacterial species and play protective and regulatory roles in the immune response [22]. Such is the case of DnaK from Mycobacterium tuberculosis, which acts as an immunomodulator during infection by polarizing macrophages to an M2 phenotype [22]. These microbial proteins have been widely used as relevant antigens in defense against infectious diseases by potent immune system activators [23]. The existing literature on the in vivo effect of DnaK in fish has been mainly focused on administering the recombinant or encapsulated protein through microparticles [20,24]. However, it is essential to note that microparticle production and application entail significant costs and involve more complex logistics compared to other administration techniques.
Fish represent excellent models for studying HSP expression, as various stressors are encountered in their environment and/or can be exposed to them in an experimental setting [25]. In response to stress, teleosts were the first vertebrates to develop a response involving a complex network of signals from the central regulatory system: neuronal, endocrine, and immune, mainly through the HPI and Brain-Sympathetic-Chromaffin (BSC) axes and their interaction with networks associated with the defense response [2,26]. Thus, the perception of a stressor promotes the activation of multiple mechanisms in the central nervous system that trigger the secretion of specific neuropeptides and hormones responsible for the immune effector action [27]. Additionally, several studies have demonstrated that pro-inflammatory cytokines (IL-6, IL-1, TNFα) play an active role in regulating certain stress hormones, demonstrating the interconnectedness of these regulatory systems and their role in fish physiology [2,28].
During the fish life cycle, the larval stage undergoes differentiation and activation of many organs. This process requires early neuro-immune-endocrine integration using signaling molecules that may be different from those of adult fish. The immune-endocrine system has been studied in several fish species, including the sea bass (Dicentrarchus labrax) [20]. Sea bass is a species of great commercial interest; being one of the three main species bred in Europe, it is also among the most studied models of teleost fish [14]. To achieve high production rates, these fish may be subjected to stressful conditions that trigger the appearance of pathogenic diseases such as vibriosis, mainly caused by Vibrio anguillarum in these fish [29].
Infectious diseases are one of the most critical issues for the aquaculture industry, resulting in significant economic losses due to high mortality. V. anguillarum (the etiological agent of vibriosis) is a gram-negative marine bacterium considered pathogenic for several fish species relevant to the aquaculture industry, including sea bass [30]. To date, few treatments are available to improve the immunity and health of fish infected with V. anguillarum. For this reason, this study aimed to evaluate the potential protective action of cells overexpressing DnaK and its effect on the cumulative mortality and immune-related gene expression pattern in gnotobiotic full-sibling sea bass larvae challenged with V. anguillarum. The study was conducted under gnotobiotic conditions because this approach enhanced the experimental reproducibility [20]. Experimentally, sea bass larvae were incubated on day 3 post-hatching with a unique pulse of one of the following treatments: YS0 (E. coli without plasmid), YS1 (E. coli expressing truncated DnaK), and YS2 (E. coli expressing DnaK). Then, on day 7 post-hatching, sea bass larvae were bath-incubated with V. anguillarum strain HI610. The cumulative survival and the expression of a set of immune-related genes were evaluated at different time points (0, 18, 24, 24, 36, and 120 h post-challenge). This study sheds light on the mechanisms modulated by DnaK in sea bass larvae exposed to a pathogen relevant to the aquaculture industry.
2. Materials and Methods
2.1. Bacterial Strain and Culture Conditions
Vibrio anguillarum strain HI 610 serovar O2a was used to evaluate the effect of DnaK as an immune protective molecule. The bacteria were initially isolated by O. Bergh (Institute of Marine Research, Bergen, Norway) from cod with vibriosis symptoms at the Parisvatnet research facility in Norway. The strain was made rifampicin-resistant by natural selection, allowing its viability in the axenic system. The bacteria were grown in 10% marine broth (Difco Laboratories, Detroit, MI, USA) with the addition of NaCl to obtain the same salinity as the water in the fish larvae experiment (36 g L−1) and kept on a horizontal shaker at 150 rpm at 16 °C. The density of the bacterial suspension was determined with a spectrophotometer (Genesys 20, Thermospectronic, Thermo Electron, Waltham, MA, USA) at 550 nm according to the McFarland standard (BioMérieux, Marcy L’Etoile, France).
2.2. Induction and Synthesis of DnaK
The plasmid constructs for synthesizing DnaK have been developed and described elsewhere, as has their protective effect against bacterial infection [31]. Briefly, the DnaK cDNA sequence from E. coli str K-12 substr. MG1655 (GenBank: AIZ92815.1) was amplified with specific primers, cloned into the TOPO cloning vector, and transformed into E. coli One Shot TOP10 cells and grown on LB agar containing 100 μg·mL−1 ampicillin at 37 °C. A bacterial-positive clone containing DnaK cDNA (henceforth YS2) was isolated from the LB plates. A construct containing a short DNA fragment with two in-frame stop codons that make protein production unviable was used as a negative control for DnaK expression (henceforth YS1). In addition, E. coli with no plasmid (henceforth YS0) was also included in the study. All bacterial cell cultures were grown in LB broth at 37 °C. Overnight cultures were diluted (1:50) in 5 mL fresh LB broth and incubated at 37 °C (220 rpm) until the cultures reached an OD600 value of 0.5–0.7. Then, DnaK synthesis was induced with 0.5 mg·mL−1 ʟ-arabinose for 4 h. The presence of DnaK in the culture was verified by analyzing the protein extract by SDS-PAGE and Western blot. After induction, bacteria were harvested by centrifugation at 2200× g for 15 min at room temperature. Pellets were washed once with seawater (previously autoclaved and filtered) and then suspended in 30 mL of autoclaved seawater. The bacterial cell culture was quantified spectrophotometrically at 550 nm and used to immediately feed the sea bass larvae at 4 days post-hatching (dph) using a concentration of 107 cells/mL, as described in previous studies [31,32].
2.3. Protein Extraction, SDS-PAGE, and Western Blot to Verify DnaK Synthesis
The bacterial suspension was homogenized with 0.1 mm diameter glass beads in cold buffer K (150 mm sorbitol, 70 mm potassium gluconate, 5 mm MgCl2, 5 mm NaH2PO4, 40 mm HEPES, pH 7.4) containing protease inhibitor cocktail (Sigma-Aldrich, Diegem, Belgium) at the highest recommended level. Samples were centrifuged at 2200× g for 1 min at 4 °C. The supernatant containing the crude protein extract was then collected to determine its concentration using the Quick Start Bradford Protein Assay (Bio-Rad, Hercules, CA, USA). DnaK was also purified as described elsewhere [20] and used as a positive control. For SDS-PAGE, 50 µg of total protein extract was combined with 2 × SDS polyacrylamide gel buffer (ratio 1:1), incubated at 95 °C for 5 min, and loaded in 10% SDS polyacrylamide gels (Bio-Rad) and then stained with Coomassie Biosafe (BioRad). The proteins were transferred to polyvinylidene fluoride membranes (PVDF, Bio-Rad) to detect DnaK synthesis by Western blot. Membranes were incubated for 60 min with blocking buffer (PBS 1×, 0.2% v/v Tween-20, 5% w/v BSA). DnaK detection was performed with the monoclonal antibody 8E2/2 mouse anti-DnaK at a dilution of 1:1000 (Stressgen Bioreagents, Victoria, BC, Canada), and the horseradish peroxidase-conjugated donkey anti-mouse IgG was used as a secondary antibody at a dilution of 1:4000 (Affinity Bioreagents, Golden, CO, USA). Detection was performed with a chemiluminescent western blot detection kit (Bio-Rad). Images were captured with the ChemiDoc MP system (Bio-Rad).
2.4. Gnotobiotic Full-Sibling Sea Bass Larvae
Sea bass eggs of 2 days post-fertilization were obtained from Ecloserie Marine in Gravelines, France. One batch of full-sibling eggs was acquired for this experiment. Upon arrival, the eggs were acclimatized in UV-sterilized seawater for 4 h in a cylindro-conical tank at 16 ± 1 °C and a salinity of 36 g L−1. The disinfection of full-sibling sea bass eggs, hatching, and testing for axenity were performed according to Dierckens et al. (2009) [33]. Handling and procedures, including hatching, stocking, and treatments, were carried out under a laminar flow hood and in a temperature-regulated room (16 ± 1 °C) with dim light (100 lux), as described elsewhere [20].
2.5. Full-Sibling Sea Bass Larvae Incubated with E. coli Overexpressing DnaK
On day 3 post-hatching (3 dph), n = 12 full-sib sea bass larvae were stocked individually in each 10 mL sterile screw cap vial (n = 4 replicate vials per treatment) with 10 mg·L−1 rifampicin. On day 4 post-hatching, sea bass larvae were incubated one single time with one of the following different treatments: YS0 (E. coli with no plasmid), YS1 (E. coli expressing truncated DnaK), and YS2 (E. coli expressing DnaK). For this procedure, 107 cells/mL suspended in autoclaved seawater (obtained in Section 2.2) were added to each vial. The procedure was conducted in a laminar flow hood to keep the culture under gnotobiotic conditions. As an experimental control of the E. coli administration, sea bass larvae incubated with no bacteria (NB group) were also included in the analysis.
2.6. Full-Sibling Sea Bass Larvae Incubated with E. coli Overexpressing DnaK and Challenged with Vibrio anguillarum
Sea bass larvae were bath-incubated with V. anguillarum strain HI610 (1 × 107 CFU/mL suspended in autoclaved seawater, as described in Section 2.1) on day three of incubation with E. coli overexpressing DnaK (equivalent to 7 dph). The experimental design included the incubation with V. anguillarum-free seawater (mock-incubated (MI)) for all the treatments analyzed as an experimental control of the bacterial challenge, including sea bass larvae incubated with no bacteria (NB-MI), E. coli with no plasmid (YS0-MI), E. coli expressing truncated DnaK (YS1-MI), and E. coli expressing DnaK (YS2-MI). For sampling, live larvae were collected at 0, 18, 24, 36, and 120 h post-challenge (hpc). To do this, n = 12 larvae were randomly taken from the four vials assigned to each treatment and immediately stored at −80 °C. The chosen sampling times were determined based on our previous experience analyzing the gene expression profile of sea bass larvae [34]. Survival was monitored in all vials at all sampling points. The experimental set-up overview is shown in Figure 1.
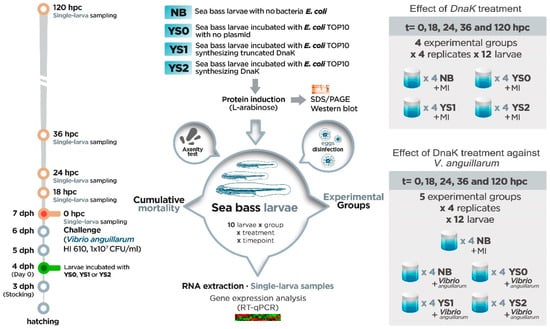
Figure 1.
Experimental set-up overview. The timeline shows that on day 3 post-hatching (3 dph), full-sib sea bass larvae were stocked individually in each of the 10 mL sterile screw cap vials (n = 4 replicate vials per treatment; n = 12 larvae each vial). Then, sea bass larvae were incubated at 4 dph one single time with one of the following different treatments: YS0 (E. coli with no plasmid), YS1 (E. coli expressing truncated DnaK), YS2 (E. coli expressing DnaK), and NB (sea bass larvae with no E. coli). Three days after incubation (equivalent to 7 dph) with NB, YS0, YS1, or YS2, the effect of DnaK treatment against V. anguillarum was evaluated. For this purpose, sea bass larvae were infected with V. anguillarum strain HI610 (1 × 107 CFU/mL). Also, the individual effect of treatment was evaluated: for that, all the groups were mock-infected (MI). Samples (12 single larvae × treatment) were collected at 0, 18, 24, 36, and 120 h post-challenge (hpc; orange bold circles in timeline).
2.7. Ethics Statement
The experiment was evaluated and approved by the Ethical Committee of the Faculty of Veterinary Medicine and the Faculty of Bioscience Engineering, Ghent University (no. EC2015_02). The protocol was carried out according to the recommendations of the European Union Ethical Guidelines for experimental animal care and other scientific purposes (2010/63/EU).
2.8. RNA Extraction and cDNA Synthesis of Single Full-Sibling Sea Bass Larvae
Total RNA was extracted from single whole larvae with TriReagent following the manufacturer’s instructions. Total RNA concentration was determined by NanoDrop-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the integrity was measured by Experion RNA StdSens (Bio-Rad). Samples with RNA quality indicator (RQI) values greater than 8 were chosen for gene expression analysis. Five hundred ng of total RNA from each single larva was used to synthesize cDNA with iScript™ cDNA Synthesis Kit (BioRad). The cDNA was used as a template for gene expression analysis.
2.9. Gene Expression Analysis by Real-Time PCR
Real-time PCR (Bio-Rad) was performed to analyze the expression pattern of immune-related genes associated with innate immunity (pentraxin, lysozyme, hepcidin, transferrin, hsp70, il-1β, il-8, il-10, and ccl4). The reference candidate genes (ribosomal protein l13 (rpl13), elongation factor 1α (ef1α), and 40S ribosomal protein SA (rpsa)) were tested using the BestKeeper software [35]. According to previous antecedents on sea bass infected with V. anguillarum [34,36], rpl13 was chosen in our study as a reference gene because of its lower variation tested upon all the samples included in our study. The specific primers used for gene expression analysis are detailed in previous work [34]. The reaction mix (10 μL final volume) consisted of 5 μL of iQ SYBR Green supermix (Bio-Rad), 0.5 μL of forward and reverse primer (500 nM final concentration), 1.5 μL of H2O, and 2.5 μL of 1:10 cDNA sample dilution. The running conditions were one step at 95 °C for 3 min, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C, following a melting curve dissociation analysis from 65 °C to 95 °C with increments of 0.5 °C. Ten single larvae per time point and treatment were analyzed. Values for each treatment were expressed as normalized relative expression (NRE), calculated in relation to all treatments’ values at time zero, normalized against rpl13 expression following the Pfaffl method, and corrected for the efficiency of each primer set [37]. The results are expressed as the mean value ± standard deviation (SD) for each treatment and time point evaluated.
2.10. Statistical Analysis
Cumulative survival curves were compared using the log-rank test corrected for multiple comparisons (p < 0.05). Given the rapidity with which the fish of the NB group were dying compared to the rest of the treatments, a Hazard ratio (Mantel-Haenszel test) was carried out. Differences at the gene expression level were evaluated by two-way ANOVA considering the two main effects: treatment and time. Also, the interaction between two factors was considered for the model since we expected differences between groups at different times. Thus, a Tukey’s post-test was performed to test the significant differences between all the groups at each time point included in the study. All the statistical analyses considered a p < 0.05 as significant.
3. Results
3.1. DnaK Induction and Synthesis
In order to check the DnaK synthesis through the induction by ʟ-arabinose, all the constructs included in this study were incubated with ʟ-arabinose to observe the DnaK synthesis by SDS-PAGE and Western blot (Figure 2). No band associated with DnaK synthesis was observed for YS0 (non-induced; ʟ-arabinose-induced) (Figure 2A, lanes 2 and 3, respectively) and YS1 (Figure 2A, lanes 4 and 5, respectively). In the case of YS2, no band associated with the over-synthesis of DnaK was registered in the non-induced condition (Figure 2A lane 6); by contrast, a positive band representative of the DnaK over-synthesis was observed in the YS2 cell culture induced with ʟ-arabinose (Figure 2A lane 7). The presence and specificity of DnaK on the SDS-PAGE were confirmed by Western blot on the YS2 cell culture incubated with ʟ-arabinose (Figure 2B lane 7). A slight positive band was also noted on the YS2 non-induced cell culture (Figure 2B lane 6). Altogether, these results indicate that the ʟ-arabinose-induced YS2 cell culture over-synthesizes DnaK.
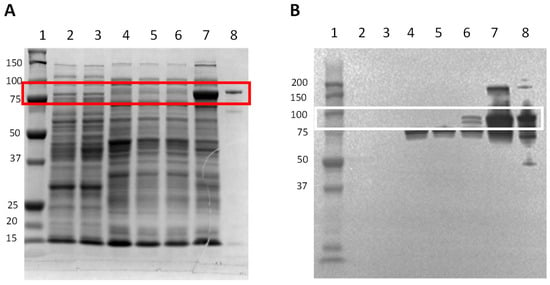
Figure 2.
DnaK synthesis induction in cells incubated with ʟ-arabinose. The bacterial cell cultures with an OD600 value of 0.5–0.7 were induced with 0.5 mg mL−1 of ʟ-arabinose for 4 h. Total bacterial crude protein extract was obtained and was resolved in (A) SDS-PAGE stained with Coomassie Biosafe, or (B) by Western blot. Fifty micrograms of protein were loaded in each lane. Lane 1: Precision plus protein standard; lane 2: YS0; lane 3: YS0+ ʟ-arabinose; lane 4: YS1; lane 5: YS1+ ʟ-arabinose; lane 6: YS2; lane 7: YS2+ ʟ-arabinose; lane 8: DnaK purified (positive control). Protein molecular weight (kDa) is indicated on the left. Red (SDS-PAGE) and white (WB) boxes show the presence of recombinant DnaK. YS0: E. coli TOP10 with no plasmid. YS1: E. coli TOP10 expressing truncated DnaK. YS2: E. coli TOP10 overexpressing DnaK.
3.2. Cumulative Survival in Gnotobiotic Full-Sibling European Sea Bass Larvae Incubated with DnaK
In order to evaluate whether DnaK has a protective effect on the survival of full-sibling sea bass larvae challenged with V. anguillarum, the cumulative survival of sea bass larvae was recorded after the pathogen exposure (Figure 3). On the non-challenged condition, only slight variations were registered after the mock incubation (Figure 3A). No significant differences were determined between the cumulative survival curves from all the treatments tested during the experiment despite the higher survival trend in fish incubated with DnaK (YS2-MI group). Regarding how rapidly fish are dying, the hazard ratio of YS2-MI compared to the NB-MI group was 1.67 (95% confidence interval [CI], 0.39 to 6.87). These results suggest that DnaK may have a protective effect on sea bass larvae growth under gnotobiotic conditions.
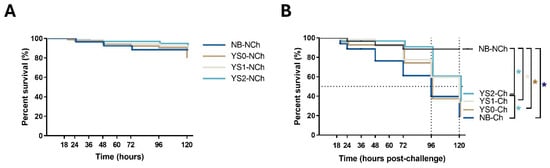
Figure 3.
Kaplan-Meier cumulative survival curves of full-sibling European sea bass larvae during 120 h after the pathogen exposure. Larvae were incubated at day 4 post-hatching (YS0, YS1, YS2) and then exposed to V. anguillarum on day 7 post-hatching. The graphs represent the cumulative mortality in the first 120 h post-challenge. (A) Cumulative survival for non-challenged sea bass larvae. (B) Cumulative survival for sea bass larvae challenged with V. anguillarum. The time scale indicated in (A) represents the same time shown in (B). The dotted black line refers to median survival. Asterisk (*) represents the significant difference (p < 0.05) in survival (log-rank test corrected for multiple comparisons) compared to control (NB). The line-type details for each group are indicated on the right of each graph, as follows: NB: Sea bass larvae mock-incubated ((A): solid line and (B): solid black line). YS0: Sea bass larvae incubated with E. coli TOP10 with no plasmid (solid brown line). YS1: Sea bass larvae incubated with E. coli TOP10 expressing truncated DnaK (solid grey line). YS2: Sea bass larvae incubated with E. coli TOP10 overexpressing DnaK (solid light blue line).
When the sea bass larvae incubated with NB, YS0, YS1, or YS2 were challenged with V. anguillarum, a different cumulative survival curve pattern was obtained (Figure 3B). Based on these data, the median survival was determined at 96 hpc for the NB-Ch and YS0-Ch groups and 120 hpc for the YS1-Ch and YS2-Ch groups (Figure 3B, dotted black line). Notably, the cumulative survival curves of YS2-Ch but not YS0-Ch and YS1-Ch were significant compared to the NB-Ch group. This statistical difference was also noted in the rapidity of dying fish of the NB-Ch group compared to YS2-Ch, with a hazard ratio of 2.26 (95% confidence interval [CI], 1.23 to 4.17); meanwhile, a lower ratio was determined for YS1-Ch (2.0; 95% confidence interval [CI], 1.06 to 3.74). These results indicated that DnaK confers protection on gnotobiotic sea bass larvae challenged with V. anguillarum.
3.3. Innate Immune-Related Gene Expression Profile in Gnotobiotic Full-Sibling European Sea Bass Larvae Incubated with DnaK
The expression profile of several genes related to innate immunity was conducted in non-challenged fish to determine whether the administration of DnaK had a differential modulatory effect on sea bass larvae. For this purpose, DnaK was administered to sea bass larvae at day 4 post-hatching. Then, the expression profile was assessed three days after the DnaK administration (equivalent to 7 days post-hatching and indicated as zero hours post-challenge, 0 hpc). In the case of non-challenged fish (MI), no variations were registered for the expression of pentraxin (Figure 4A), il-1β (Figure 4E), il-8 (Figure 4F), il-10 (Figure 4H), and hsp70 (Figure 4I) at any of the time points assessed. For the other innate-related genes, only punctual variations were registered. In this way, the treatment with DnaK promoted the upregulation of lysozyme at 0 hpc in the gnotobiotic sea bass larvae treated with YS2-MI compared to the NB-MI group (Figure 4B), to then return its value to the basal level. On the other hand, the expression of transferrin (involved in removing iron from the bloodstream) showed the downregulation of YS1-MI at 18 hpc and 120 hpc compared to YS0-MI (Figure 4C). In the same way, the expression of transferrin for YS1-MI was also downregulated at 36 hpc. Importantly, the upregulation of transferrin was identified for YS2-MI at 120 hpc compared to the NB-MI group (Figure 4C). A similar effect was also observed for YS0-MI (Figure 4C).
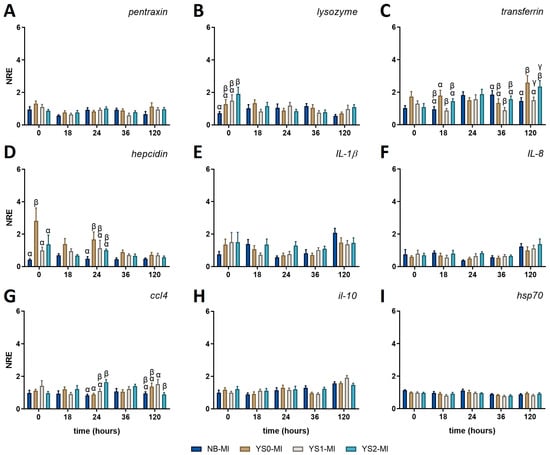
Figure 4.
Normalized relative expression (NRE) of innate immune-related genes in full-sibling European sea bass larvae treated with DnaK. The graphs represent the normalized relative expression (NRE; mean value ± SD) for (A) pentraxin; (B) lysozyme; (C) transferrin; (D) hepcidin; (E) interleukin (il)-1β; (F) il-8; (G) CC motif chemokine 4 (ccl4); (H) il-10; and (I) heat shock protein 70 (hsp70). The obtained values were statistically analyzed by two-way ANOVA and Tukey’s post-test for significant differences (p-value < 0.05). The statistical difference between the control group (NB-MI) and YS0-MI, YS1-MI, or YS2-MI at each time point analyzed are indicated by different Greek letters. NB-MI: sea bass larvae incubated with no bacteria and Mock Infected (NB-MI group; blue bar). YS0-MI: Sea bass larvae incubated with E. coli TOP10 with no plasmid and Mock Infected (brown bar). YS1-MI: Sea bass larvae incubated with E. coli TOP10 expressing truncated DnaK and Mock Infected (fawn bar). YS2-MI: Sea bass larvae incubated with E. coli TOP10 expressing DnaK and Mock Infected (light blue bar).
Hepcidin modulation (the principal circulating regulator of iron absorption and distribution across tissues) was also noted, registering an increase at 0 hpc in the YS0-MI group compared with all the rest of the conditions (Figure 4D). In the same line, the hepcidin expression for YS0-MI at 24 hpc was also upregulated compared with the NB-MI group.
The expression of the inflammatory chemokine ccl4 (also known as macrophage inflammatory protein-1β [MIP-1β]) was upregulated at 24 h in YS2-MI compared to NB-MI and YS0-MI groups. Then, the expression for YS2-MI was downregulated compared to YS1-MI, but no difference was observed compared to the NB-MI group (Figure 4G). The NRE (mean ± SD) for all genes evaluated and the p-values obtained for each comparison can be found in Table A1 and Table A2, respectively.
Collectively, our results suggest that DnaK does not activate a robust immune response in gnotobiotic sea bass larvae but seems to promote bacteriolytic, iron regulation, and chemotaxis.
3.4. Innate Immune-Related Gene Expression Profile in Gnotobiotic Full-Sibling European Sea Bass Larvae Incubated with DnaK and Challenged with V. anguillarum
In order to evaluate whether the administration of DnaK modulated the response of sea bass larvae challenged with V. anguillarum, we analyzed the expression pattern of a set of genes related to innate immunity. No variations were observed in pentraxin at any of the time points evaluated (Figure 5A). All the other genes evaluated showed at least one significant upregulation at 120 hpc in the challenged groups (NB-Ch; YS0-Ch; YS1-Ch; YS2-Ch) compared with the mock-incubated group (NB-MI) (Figure 5). Importantly, YS2-Ch was the only experimental group that showed a significant upregulation in all the genes assessed, suggesting the role of DnaK in promoting defensive mechanisms in sea bass against V. anguillarum (Figure 5). In comparing all time points evaluated, an upregulation of lysozyme expression was observed at 120 h post-challenge (hpc) in all the challenged groups except YS1-Ch compared to the NB-MI group (Figure 5B).
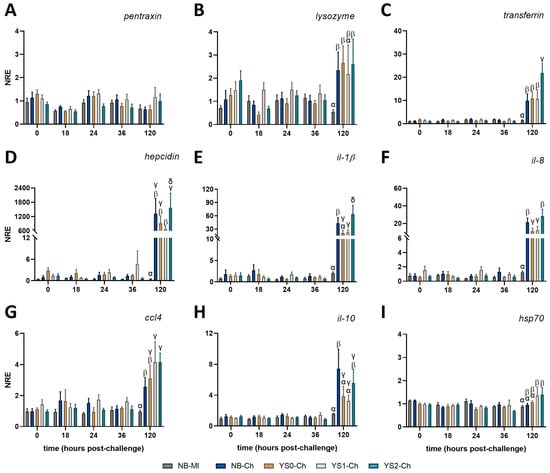
Figure 5.
Normalized relative expression of innate immune-related genes in full-sibling European sea bass larvae treated with DnaK and challenged with V. anguillarum. Larvae were incubated at day 4 post-hatching and then exposed to V. anguillarum at day 7 post-hatching. The graphs represent the normalized relative expression (NRE; mean value ± SD) for (A) pentraxin; (B) lysozyme; (C) transferrin; (D) hepcidin; (E) interleukin (il)-1β; (F) il-8; (G) CC motif chemokine 4 (ccl4); (H) il-10; and (I) heat shock protein 70 (hsp70). The obtained values were evaluated by two-way ANOVA and Tukey’s post-test for significant differences (p < 0.05). Different Greek letters indicate statistical differences between the control group (NB-MI) and NB-Ch, YS0-Ch, YS1-Ch, or YS2-Ch at each time point analyzed. NB: Sea bass larvae incubated with no bacteria (NB group; mock-incubated; grey and blue bar). YS0: Sea bass larvae incubated with E. coli TOP10 with no plasmid (brown bar). YS1: Sea bass larvae incubated with E. coli TOP10 expressing truncated DnaK (fawn bar). YS2: Sea bass larvae incubated with E. coli TOP10 expressing DnaK (light blue bar).
The upregulation in the expression of transferrin was observed at 120 h post-challenge (hpc) in larvae incubated with YS2-Ch (Figure 5C) compared with all other treatments (NB-MI; NB-Ch; YS0-Ch; YS1-Ch). A notorious marked increase in hepcidin expression compared with the NB-MI group was observed at 120 hpc in all the treatments challenged with V. anguillarum (Figure 5D). However, only a significant upregulation of hepcidin was identified in sea bass larvae incubated with YS2-Ch compared with the YS1-Ch group (Figure 5D).
A similar expression pattern was observed at the pro-inflammatory level. Thus, the expression of il-1β (Figure 5E) and il-8 (Figure 5F) at 120 hpc showed their upregulation in fish incubated with YS2-Ch compared with YS0-Ch, YS1-Ch, and NB-Ch groups. An increase in the expression of il-8 was observed in all treatments challenged with V. anguillarum compared with the NB-MI group at 120 hpc (Figure 5F). Importantly, the expression of both cytokines was downregulated in the YS0-Ch and YS1-Ch treatments compared with the NB-Ch group. The expression of ccl4 was upregulated in all the challenged groups, even showing a significant increase in their expression for YS1-Ch and YS2 compared to the NB-Ch group. Despite these differences, no significant differences were registered between YS2-Ch and YS1-Ch or YS0-Ch groups. However, an increase in ccl4 expression was observed in all treatments compared to the NB-MI group at 120 hpc (Figure 5G).
The anti-inflammatory cytokine il-10 showed a higher expression in NB-Ch at 120 hpc compared to YS0-Ch and YS1-Ch. On the other hand, the YS2-Ch group showed an augment in its expression profile compared with the NB-MI group, but no differences compared with the other challenged treatments (Figure 5H). None of the treatments in the study modified the expression of hsp70, although an increase was noted for the YS1-Ch and YS2-Ch compared to the NB-MI group (Figure 5I). The gene expression data (mean ± SD) for all evaluated genes and the p-values obtained for each comparison in full-sibling European sea bass larvae treated with DnaK and challenged with V. anguillarum can be found in Table A3 and Table A4, respectively.
Taken together, these results showed that sea bass larvae incubated with DnaK had an increased modulatory effect on genes related to iron regulation and pro-inflammatory cytokines at 120 hpc, which is related to the observed protection of DnaK on larval survival at 120 hpc.
4. Discussion
Several stress factors affect fish in the aquatic environment. It has been widely described that stress and immune response are closely connected, in which a stressor can induce alterations in innate immune responses. These stress factors can make fish more sensitive to pathogen attacks, such as Vibrio anguillarum [14,38]. This bacterium generates major problems in aquaculture due to high mortality, causing significant economic losses [34]. Therefore, studying the interaction between the nervous, endocrine, and immune systems is essential to allow the survival of these fish [14].
Bacterial endocytic shock proteins, such as DnaK, stand out for their ability as potent immunomodulators and present considerable potential as therapeutic agents in infection control in aquaculture, as documented in previous studies [21,39]. This effect is partly due to their presence both in the cytoplasm and on the surface of bacteria, facilitating their delivery to larvae by making them more accessible [40]. Thus, feeding with DnaK-enriched bacteria to aquatic organisms emerges as a novel strategy in disease management in aquaculture [1,41]. The advantages of this strategy are that using bacteria that can autonomously produce and secrete DnaK makes it more sustainable, efficient, and economical in the long term compared to direct protein production and administration. Thus, the novelty of this work lies in a different approach consisting of providing DnaK to gnotobiotic sea bass larvae. However, one limitation of the study is the absence of a direct comparison between the E. coli-based delivery system and the conventional encapsulated microparticle approach previously used in sea bass larvae [20]. The analysis of this comparison in future studies will provide valuable insight into the relative efficacy of both methods. Prospectively, the scalability and applicability of this novel strategy in aquaculture farms should be explored and analyzed in future studies.
In this study, we observed that sea bass larvae presented high mortality against V. anguillarum infection. This effect is probably attributable to fish larvae’s defense relying specifically on their innate immune system, as their adaptive immune system is not yet sufficiently developed [34,42]. Consequently, larvae are more vulnerable to pathogens present in the aquatic environment than adults, leading to high mortality rates [42]. Also, during the first two weeks after hatching, sea bass larvae in gnotobiotic conditions do not present mucus-producing cells (goblet cells), so they do not present this physical barrier in the gut, remaining exposed [43]. In fact, previous works have found high numbers of V. anguillarum in the gastrointestinal tract of gnotobiotic sea bass larvae when exposed to the bacterium [43]. In our study, we observed that gnotobiotic sea bass larvae incubated with DnaK showed a higher survival against V. anguillarum infection than larvae not incubated with this protein. These results agree with previous studies that demonstrated that V. anguillarum induces a gradual decrease in larval survival of sea bass under gnotobiotic conditions and that DnaK has a protective role in the survival of aquatic organisms [31,34]. Importantly, significant differences in the expression of genes related to pro-inflammatory (lysozyme, transferrin, hepcidin) and antimicrobial (il-1β, il-8, ccl4, il-10) response in larvae incubated with DnaK also occurred at 120 hpc. These antecedents are consistent with the significant difference in cumulative survival mentioned above. Taken together, we can infer that DnaK has a protective effect on the growth of sea bass larvae under gnotobiotic conditions against pathogen infection by activating the expression of different genes related to the innate immune response.
The heat shock protein 70 is involved in many cellular processes, including synthesis, translocation, proper folding, and degradation of proteins. In addition, it participates in antigen presentation and activation of immune cells such as lymphocytes and dendritic cells. In fish, HSP70 is induced by stress factors such as high temperatures, microbial infections, and heavy metals [14]. It has been described in teleost fish larvae that HSP70 protein expression can be used as a marker of larval stress [25]. Regarding cell stress response, this protein plays a vital role in cortisol binding to the glucocorticoid receptor (GR), considered the primary receptor for glucocorticoid activity in teleost fish. In this regard, a correlation between the positive regulation of glucocorticoid receptors (gr1 and gr2) and hsp70 gene expression has been described [34]. An unexpected finding was that hsp70 expression was not upregulated at any time point analyzed after infection with V. anguillarum. This differs from the results obtained by Dang et al. (2010) [7], who reported that infection with V. anguillarum increased hsp70 expression in rainbow trout (Oncorhynchus mykiss). However, basal hsp70 expression has been reported in gnotobiotic sea bass larvae even after exposure to this bacterium [34]. These results are consistent with other research indicating that increased expression of HSP70 is only evident in 40-day-old stressed sea bass larvae [14]. Furthermore, several studies have specified that cortisol may not be associated with increased gr or hsp70 expression [44,45]. This background would indicate that cortisol does not modulate hsp70 expression in gnotobiotic sea bass larvae challenged with V. anguillarum at these early stages of larval development [34].
It has been reported that fish rely mainly on innate immune defense in their first days after hatching (64–68 days). Such a response is crucial in the early fight against infections by pathogenic microorganisms. Also, previous studies in fish have described that, at hatching and subsequent weeks, the expression of components with antimicrobial activity associated with the humoral arm of innate immunity (pentraxin, lysozyme, transferrin, hepcidin) is induced [34,46]. This background suggests that these molecules play a role in preparing the organism against potential environmental pathogens that may be encountered during the early stage of larval development. Consequently, the modulation of genes associated with innate immunity in the response of sea bass larvae against V. anguillarum is of great importance [34].
Like in mammals, fish can produce acute phase proteins (APPs) during infection or because of the presence of danger or stress signals, being one of the most common pentraxins. These APPs activate crucial signaling cascades to induce a pro-inflammatory response [34,47]. Pentraxins are phylogenetically conserved soluble pattern recognition receptors (PRRs) that are associated with the acute phase response (APR) and mediate agglutination, complement system activation, and opsonization [34,47]. Our study found no variation in pentraxin gene expression in both bacterially challenged and unchallenged larvae. These results are in line with previous studies, which described that sea bass larvae under gnotobiotic conditions do not positively regulate pentraxin expression after infection with V. anguillarum [34], unlike other teleost fish such as Oncorhynchus mykiss [48], Plecoglossus altivelis [49] and Gadus morhua [50]. Thus, the non-modulatory response of pentraxin in sea bass larvae in response to the incubation with V. anguillarum suggests that other receptors and molecules of innate immunity might be involved in the recognition and response against this pathogenic bacterium [34]. Furthermore, these results suggest that administration with DnaK does not modulate pentraxin gene expression pathways at times analyzed. Consequently, gene expression analysis should be conducted at more time points to support this hypothesis.
Lysozyme is a vital immune system bacteriolytic enzyme with a broad bactericidal spectrum. It cleaves the bacterial wall by hydrolyzing the β-[1,4]-glucosidic linkage of peptidoglycans [29,34]. This study found that uninfected fish incubated with YS2 presented a significant increase in lysozyme expression at 0 h (equivalent to 7 dph). Meanwhile, larvae incubated with YS0 and YS1 maintained a constant basal expression of this enzyme compared to other conditions. These findings are in agreement with those described by Cecchini et al. (2000) [51], who indicated that the expression of lysozyme in sea bass (Dicentrarchus labrax) larvae is observed up to 24 h after hatching, experiencing a significant decrease in its expression after that time. The results also showed an increase in lysozyme expression at 120 hpc, with no effect on the expression of this enzyme in larvae incubated with DnaK. This suggests that sea bass larvae would not recognize this bacterium early during the first 36 hpc [34].
Transferrin and hepcidin are another group of acute-phase proteins involved in the innate immune response and the regulation of iron homeostasis. Transferrin is a plasma iron-binding glycoprotein that controls free iron levels in biological fluids. This glycoprotein has bacteriostatic activity by chelating available iron to make it inaccessible to invading pathogenic bacteria [30,34]. Hepcidin is an antimicrobial peptide (AMP) with a wide range of actions. In mammals, it has been described to be involved in the body’s defense against pathogens, being considered a significant player in the innate immune system. Hepcidin has been identified in several teleost fish species, including European sea bass (Dicentrarchus labrax), Atlantic salmon (Salmo salar), and rainbow trout (Oncorhynchus mykiss), among others. Its structure is conserved in these species, suggesting its participation in defense responses in fish against pathogenic infections [52].
An interesting finding of our study is that gnotobiotic sea bass larvae incubated with DnaK presented at 120 hpc a significant overexpression of transferrin and hepcidin against V. anguillarum infection. Such a response would promote antimicrobial activity (driven by hepcidin) and the regulation and restriction of iron in the bloodstream by transferrin, thus reducing its bioavailability for V. anguillarum growth. These results are related to previous studies in sea bass, which indicate that synthetic hepcidin has an inhibitory effect on the growth of this bacterium and presents elevated expression levels when stimulated with DnaK [20,52]. Iron is crucial for bacteria to grow and achieve infection, so the ability to compete for iron availability in the bloodstream is crucial in establishing the ability of the fish to overcome pathogen infection. Taken together, these data suggest that sea bass larvae incubated with DnaK have enhanced survival by favoring an antimicrobial environment against V. anguillarum infection.
The strategy of delivering immune molecules in expression vectors has been used previously. Various strategies have been used, including the administration of proinflammatory cytokines (IL-β, IL-6, TNFα, IL-8, IL-22, IFN-1, IL-17C) [53,54,55,56,57], activators of innate immunity (C-type lectin, PGRP, NCCRP) [58,59], transcription factors (IRF3) [60] in different fish species. Thus, the administration of immune-related molecules improves the health status of fish and protects them against bacterial pathogens [60] by stimulating the expression of proinflammatory molecules. Precisely, a key mechanism in initiating antibacterial responses is the co-expression of pro-inflammatory cytokines, such as IL-1β, IL-8 and CCL4 [34]. Importantly, in our study, we observed a positive upregulation at 120 hpc of the previously mentioned innate effectors, which are directly associated with the positive upregulation of il-1β, il-8, and ccl4. IL-1β is a cytokine released in the early stage of response against infection and initiates pro-inflammatory responses, allowing the synthesis of other cytokines and the activation of different immune cells. In teleost fish, il-1β expression in stressed fish has been reported to increase up to 8-fold [3]. Chistiakov et al. (2010) [61] demonstrated that il-1β is involved in the resistance process of European sea bass against V. anguillarum.
Moreover, in the context of DnaK administration, it has been described that this protein is recognized as an immunodominant antigen that induces a strong humoral and cellular immune response in fish. This response appears to activate Toll-like receptors (TLR2, TLR4) that induce pro-inflammatory signals and promote resistance against disease [22,31]. Positive regulation of il-1β is also accompanied by the expression of il-8 and increased expression of ccl4. IL-8 and CCL4 are chemokines produced by macrophages that have chemoattractant activity for neutrophils and T cells at sites of infection [34]. Therefore, the increased survival of sea bass larvae against this bacterium observed up to 120 hpc suggests that DnaK promotes elevated expression of these pro-inflammatory cytokines and chemoattractants that favor the activation of an antimicrobial response.
The pro-inflammatory response is controlled by anti-inflammatory cytokines that regulate its expression and promote tissue repair. IL-10 is an anti-inflammatory cytokine that contributes to the resolution of pathogen-mediated inflammation and the reduction of tissue damage caused by inflammation [34]. This study observed a higher value of normalized relative expression at 120 hpc in the NB-Ch and incubated groups with DnaK (YS2-Ch). On the other hand, the groups incubated only with plasmid (YS0-Ch) and YS1-Ch (truncated DnaK) showed underexpression compared to the NB-Ch group. The higher expression of il-10 in the NB-Ch group would suggest that V. anguillarum infection promotes the control of the pro-inflammatory response in sea bass larvae. This relates to previous studies indicating that this bacterium can evade the fish’s immune system by inhibiting the respiratory burst of M1 macrophages [20]. Indeed, this result agrees with previous studies in untreated sea bass larvae in which an increase in il-10 expression at 120 hpc was identified in sea bass larvae challenged with V. anguillarum [34]. Interestingly, such an increase would be linked to the physiological stress response mounted by the fish and the increase in glucocorticoids at 120 hpc, and the induction of a systemic anti-inflammatory environment [34]. On the other hand, the increased value of il-10 (although not significant) in the YS2-Ch group would suggest that the activation of mechanisms controlling the pro-inflammatory response could take place in sea bass larvae challenged with V. anguillarum. This non-significant but increased effect on il-10 expression could be due to a temporal effect and the times chosen for the analysis, as suggested for the modulation of immune response-associated genes reported in other studies [62]. Apparently, promoting control of the pro-inflammatory response could also be linked to increased survival in sea bass larvae challenged with V. anguillarum by regulating inflammation and activating eventual tissue damage mechanisms [3].
We hypothesize that the delay in activating the immune response promoted by the pro-inflammatory and antimicrobial genes involved in the response against V. anguillarum may have contributed to the high mortality observed up to 120 hpc. This delay in the response could have caused a delay in the processes necessary to sequester available iron from the bloodstream, thereby inhibiting this bacterium’s growth. This may be due to the fact that V. anguillarum uses iron acquisition mechanisms for its growth even if transferrin is present in fish [20]. Likewise, it has been described that this pathogenic bacterium has the ability to evade the immune system of sea bass by interfering negatively with leukocytes, thus explaining the delayed response to this pathogen even with DnaK pretreatment [63,64]. Despite these antecedents, larvae incubated with DnaK before being challenged with V. anguillarum showed increased expression at 120 hpc of a set of genes involved in the organism’s defense. This immune response activation suggests that DnaK administration induces a potent innate immune response that enhances the survival of sea bass larvae challenged with V. anguillarum. We highlight that this response takes place in an organism even before the full maturation of the immunological system in fish larvae, thus reinforcing the immunoprotective role of DnaK against pathogenic infections in sea bass larvae. A graphical summary of our main results describing the modulatory effect of DnaK on sea bass larvae is shown in Figure 6.
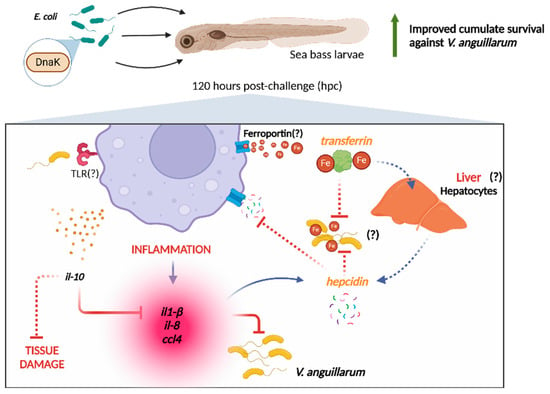
Figure 6.
Summary of the effects promoted by the administration of DnaK to gnotobiotic sea bass larvae challenged with Vibrio anguillarum. The administration of DnaK modulates the expression of pro-inflammatory (il-1β, IL-8, CCL4) and anti-inflammatory (IL-10) cytokines in gnotobiotic sea larvae after 120 hpc with V. anguillarum. We hypothesize that TLR triggers these mechanisms in response to recognizing V. anguillarum pathogen-associated molecular patterns (PAMPs). These pro-inflammatory cytokines stimulate hepcidin production. Hepcidin inhibits ferroportin, the principal iron transporter, limiting the iron availability in the environment, while transferrin restricts the remaining iron. The action of both proteins prevents iron uptake by V. anguillarum, influencing its proliferation. The question marks and dashed lines represent topics we propose based on our results, but need validation in future studies. Figure created with Biorender (https://biorender.com).
5. Conclusions
This study describes the effect of DnaK administration in modulating the immune response in gnotobiotic sea bass larvae challenged with V. anguillarum, demonstrating its potential as a pathogen resistance agent in fish by improving fish survival. The results showed a significant upregulation of antimicrobial (transferrin, hepcidin) and proinflammatory (il-1β, il-8, ccl4) genes after DnaK administration at 120 hpc, despite the underdeveloped immune system of the larvae. This strategy would enable improved disease management in aquaculture farms; however, future studies are needed to evaluate its efficacy on other pathogens and in different fish species.
Author Contributions
Conceptualization, P.B., K.D., L.T. and F.E.R.-L.; methodology, E.V.-V., C.F.-C., A.R.K., K.D. and F.E.R.-L.; formal analysis, E.V.-V., S.R.-C. and F.E.R.-L.; investigation, E.V.-V., C.F.-C., M.J.S.-A., M.G., J.C.B., A.R.K., K.D., P.B., L.T. and F.E.R.-L.; data curation, E.V.-V., C.F.-C., A.R.K. and F.E.R.-L.; writing—original draft preparation, E.V.-V., M.J.S.-A. and F.E.R.-L.; writing—review and editing, E.V.-V., C.F.-C., M.J.S.-A., M.G., S.R.-C., J.C.B., A.R.K., K.D., P.B., L.T. and F.E.R.-L.; visualization, E.V.-V., S.R.-C., J.C.B. and F.E.R.-L.; supervision, P.B., L.T. and F.E.R.-L.; project administration, E.V.-V., K.D., P.B., L.T. and F.E.R.-L.; funding acquisition, E.V.-V., K.D., P.B., L.T. and F.E.R.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Aquaexcel EU project [0133/09/14/31] (FERL), MINECO-Spain [AGL2016-76069-C2-2-R] (LT), Fondecyt regular 1252231 (ANID; Chile) (FERL), Fondecyt iniciacion 11221308 (ANID; Chile) (EVV), and DICYT-USACH [082344RL_Postdoc; 082541RL_Ayudante] (FERL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
The animal study protocol was approved by the Ethical Committee of the Faculty of Veterinary Medicine and the Faculty of Bioscience Engineering, Ghent University (no. EC2015_02).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Appendix A. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare that they do not have any conflicts of interest when submitting this article.
Appendix A

Table A1.
Normalized relative expression (NRE) values of innate immune-related genes in full-sibling European sea bass larvae treated with DnaK at different time points (0, 18, 24, 36, and 120 h).
Table A1.
Normalized relative expression (NRE) values of innate immune-related genes in full-sibling European sea bass larvae treated with DnaK at different time points (0, 18, 24, 36, and 120 h).
| Genes | Time (hpc) | NB-MI | YS0-MI | YS1-MI | YS2-MI | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| pentraxin | 0 | 0.94 | 0.48 | 1.31 | 0.53 | 1.10 | 0.51 | 0.87 | 0.25 |
| 18 | 0.57 | 0.16 | 0.76 | 0.35 | 0.64 | 0.24 | 0.77 | 0.35 | |
| 24 | 0.93 | 0.42 | 0.82 | 0.24 | 0.93 | 0.34 | 1.01 | 0.33 | |
| 36 | 0.93 | 0.30 | 0.89 | 0.31 | 0.59 | 0.43 | 0.78 | 0.31 | |
| 120 | 0.67 | 0.50 | 1.13 | 0.68 | 0.94 | 0.36 | 0.97 | 0.34 | |
| lysozyme | 0 | 0.71 | 0.26 | 1.27 | 0.90 | 1.48 | 1.17 | 1.90 | 1.24 |
| 18 | 1.02 | 0.74 | 1.34 | 0.63 | 0.82 | 0.46 | 1.16 | 0.67 | |
| 24 | 1.05 | 0.66 | 0.89 | 0.41 | 1.18 | 0.64 | 0.84 | 0.35 | |
| 36 | 1.16 | 0.46 | 1.06 | 0.59 | 0.75 | 0.40 | 0.77 | 0.45 | |
| 120 | 0.55 | 0.29 | 0.71 | 0.26 | 0.98 | 0.64 | 1.10 | 0.46 | |
| trasnsferrin | 0 | 1.03 | 0.41 | 1.74 | 0.97 | 1.30 | 0.54 | 1.10 | 0.65 |
| 18 | 0.96 | 0.50 | 1.79 | 1.00 | 0.88 | 0.38 | 1.46 | 0.47 | |
| 24 | 1.83 | 0.64 | 1.51 | 0.50 | 1.57 | 0.89 | 1.89 | 0.93 | |
| 36 | 1.85 | 0.75 | 1.34 | 0.79 | 0.89 | 0.54 | 1.57 | 0.63 | |
| 120 | 1.47 | 0.63 | 2.60 | 1.26 | 1.51 | 0.55 | 2.36 | 1.15 | |
| hepcidin | 0 | 0.43 | 0.14 | 2.82 | 2.52 | 0.99 | 0.61 | 1.37 | 1.73 |
| 18 | 0.69 | 0.34 | 1.39 | 1.02 | 0.95 | 0.48 | 0.68 | 0.19 | |
| 24 | 0.48 | 0.40 | 1.68 | 1.45 | 1.13 | 1.54 | 1.01 | 0.21 | |
| 36 | 0.45 | 0.26 | 0.89 | 0.42 | 0.71 | 0.25 | 0.67 | 0.35 | |
| 120 | 0.48 | 0.17 | 0.72 | 0.45 | 0.69 | 0.36 | 0.57 | 0.24 | |
| il-1β | 0 | 0.75 | 0.51 | 1.36 | 1.05 | 1.50 | 1.69 | 1.51 | 1.80 |
| 18 | 1.38 | 0.86 | 1.07 | 0.68 | 0.73 | 0.35 | 1.35 | 1.03 | |
| 24 | 0.57 | 0.24 | 0.70 | 0.41 | 0.77 | 0.54 | 1.30 | 0.75 | |
| 36 | 0.81 | 0.69 | 0.71 | 0.37 | 1.02 | 0.47 | 1.10 | 0.47 | |
| 120 | 2.08 | 0.89 | 1.48 | 0.88 | 1.37 | 0.68 | 1.46 | 0.96 | |
| il-8 | 0 | 0.76 | 0.77 | 0.61 | 0.20 | 0.79 | 0.57 | 0.69 | 0.38 |
| 18 | 0.85 | 0.33 | 0.67 | 0.46 | 0.56 | 0.42 | 0.79 | 0.62 | |
| 24 | 0.38 | 0.08 | 0.49 | 0.25 | 0.64 | 0.50 | 0.82 | 0.29 | |
| 36 | 0.57 | 0.30 | 0.54 | 0.31 | 0.65 | 0.29 | 0.64 | 0.22 | |
| 120 | 1.23 | 0.58 | 0.99 | 0.68 | 1.12 | 0.53 | 1.39 | 0.98 | |
| ccl4 | 0 | 0.98 | 0.49 | 1.11 | 0.31 | 1.42 | 1.00 | 0.97 | 0.32 |
| 18 | 0.94 | 0.51 | 1.20 | 0.45 | 0.90 | 0.35 | 1.23 | 0.64 | |
| 24 | 0.83 | 0.25 | 0.89 | 0.24 | 1.11 | 0.38 | 1.65 | 0.48 | |
| 36 | 1.07 | 0.57 | 1.06 | 0.40 | 1.22 | 0.51 | 1.40 | 0.39 | |
| 120 | 0.95 | 0.32 | 1.38 | 0.64 | 1.53 | 0.83 | 0.89 | 0.37 | |
| il-10 | 0 | 1.00 | 0.42 | 1.17 | 0.40 | 1.00 | 0.25 | 1.22 | 0.46 |
| 18 | 0.89 | 0.30 | 0.92 | 0.40 | 1.11 | 0.34 | 1.12 | 0.41 | |
| 24 | 1.15 | 0.45 | 1.29 | 0.54 | 1.17 | 0.37 | 1.21 | 0.57 | |
| 36 | 1.29 | 0.38 | 0.96 | 0.20 | 0.92 | 0.25 | 1.24 | 0.31 | |
| 120 | 1.57 | 0.31 | 1.59 | 0.24 | 1.92 | 0.38 | 1.48 | 0.33 | |
| hsp70 | 0 | 1.14 | 0.08 | 0.99 | 0.14 | 0.99 | 0.15 | 0.97 | 0.13 |
| 18 | 0.96 | 0.30 | 0.92 | 0.26 | 0.81 | 0.21 | 0.93 | 0.22 | |
| 24 | 1.12 | 0.27 | 1.02 | 0.38 | 0.97 | 0.18 | 0.96 | 0.08 | |
| 36 | 0.89 | 0.11 | 0.85 | 0.11 | 0.77 | 0.19 | 0.80 | 0.16 | |
| 120 | 0.87 | 0.18 | 0.95 | 0.20 | 0.81 | 0.20 | 0.93 | 0.18 | |

Table A2.
Summary of statistical differences (p values) from Tuckey’s multiple comparison test for the relative expression of different genes evaluated in sea bass larvae treated with DnaK at different time points (0, 18, 24, 36, and 120 h). Significant differences (p < 0.05) are highlighted in bold.
Table A2.
Summary of statistical differences (p values) from Tuckey’s multiple comparison test for the relative expression of different genes evaluated in sea bass larvae treated with DnaK at different time points (0, 18, 24, 36, and 120 h). Significant differences (p < 0.05) are highlighted in bold.
| Genes | Group Comparison | Hours Post Administration of DnaK | ||||
|---|---|---|---|---|---|---|
| 0 | 18 | 24 | 36 | 120 | ||
| pentraxin | NB-MI vs. YS0-MI | 0.2129 | 0.7074 | 0.9298 | 0.996 | 0.0506 |
| NB-MI vs. YS1-MI | 0.8373 | 0.9732 | >0.9999 | 0.2203 | 0.4512 | |
| NB-MI vs. YS2-MI | 0.9816 | 0.6739 | 0.9636 | 0.8262 | 0.3233 | |
| YS0-MI vs. YS1-MI | 0.6127 | 0.9144 | 0.9286 | 0.3008 | 0.7478 | |
| YS0-MI vs. YS2-MI | 0.0633 | >0.9999 | 0.6987 | 0.9146 | 0.7897 | |
| YS1-MI vs. YS2-MI | 0.5575 | 0.894 | 0.9644 | 0.6908 | 0.9992 | |
| lysozyme | NB-MI vs. YS0-MI | 0.29 | 0.6633 | 0.9493 | 0.9864 | 0.9529 |
| NB-MI vs. YS1-MI | 0.0751 | 0.903 | 0.9741 | 0.5091 | 0.4884 | |
| NB-MI vs. YS2-MI | 0.0018 | 0.959 | 0.9051 | 0.561 | 0.2287 | |
| YS0-MI vs. YS1-MI | 0.8867 | 0.2641 | 0.736 | 0.705 | 0.81 | |
| YS0-MI vs. YS2-MI | 0.1457 | 0.9278 | 0.9988 | 0.7548 | 0.5441 | |
| YS1-MI vs. YS2-MI | 0.4821 | 0.6493 | 0.646 | 0.9998 | 0.9815 | |
| transferrin | NB-MI vs. YS0-MI | 0.2237 | 0.0818 | 0.7765 | 0.4656 | 0.0074 |
| NB-MI vs. YS1-MI | 0.8907 | 0.9952 | 0.8722 | 0.0327 | 0.9996 | |
| NB-MI vs. YS2-MI | 0.9977 | 0.4819 | 0.9978 | 0.8574 | 0.0458 | |
| YS0-MI vs. YS1-MI | 0.5808 | 0.0459 | 0.9975 | 0.5429 | 0.0174 | |
| YS0-MI vs. YS2-MI | 0.2523 | 0.7831 | 0.668 | 0.9177 | 0.8982 | |
| YS1-MI vs. YS2-MI | 0.9423 | 0.3465 | 0.7815 | 0.2148 | 0.086 | |
| hepcidin | NB-MI vs. YS0-MI | <0.0001 | 0.358 | 0.0289 | 0.7354 | 0.943 |
| NB-MI vs. YS1-MI | 0.6525 | 0.9251 | 0.4246 | 0.9286 | 0.963 | |
| NB-MI vs. YS2-MI | 0.1929 | >0.9999 | 0.5981 | 0.9567 | 0.9958 | |
| YS0-MI vs. YS1-MI | 0.0003 | 0.7279 | 0.5578 | 0.9741 | >0.9999 | |
| YS0-MI vs. YS2-MI | 0.0047 | 0.3662 | 0.3834 | 0.952 | 0.9864 | |
| YS1-MI vs. YS2-MI | 0.8373 | 0.9215 | 0.9918 | 0.9996 | 0.9934 | |
| il-1β | NB-MI vs. YS0-MI | 0.4449 | 0.8649 | 0.9901 | 0.9953 | 0.4291 |
| NB-MI vs. YS1-MI | 0.2986 | 0.3479 | 0.9613 | 0.9532 | 0.2998 | |
| NB-MI vs. YS2-MI | 0.2632 | 0.9999 | 0.2538 | 0.8965 | 0.3791 | |
| YS0-MI vs. YS1-MI | 0.9841 | 0.8235 | 0.9981 | 0.8582 | 0.9926 | |
| YS0-MI vs. YS2-MI | 0.979 | 0.8928 | 0.4181 | 0.7685 | >0.9999 | |
| YS1-MI vs. YS2-MI | >0.9999 | 0.3856 | 0.5025 | 0.9971 | 0.9951 | |
| il-8 | NB-MI vs. YS0-MI | 0.9271 | 0.8564 | 0.9605 | 0.9995 | 0.6896 |
| NB-MI vs. YS1-MI | 0.9988 | 0.5566 | 0.6531 | 0.9833 | 0.9597 | |
| NB-MI vs. YS2-MI | 0.9925 | 0.9936 | 0.1957 | 0.9866 | 0.8876 | |
| YS0-MI vs. YS1-MI | 0.8659 | 0.9599 | 0.9029 | 0.9618 | 0.9448 | |
| YS0-MI vs. YS2-MI | 0.9862 | 0.9509 | 0.417 | 0.9675 | 0.2748 | |
| YS1-MI vs. YS2-MI | 0.9728 | 0.7245 | 0.8285 | >0.9999 | 0.6411 | |
| ccl4 | NB-MI vs. YS0-MI | 0.9366 | 0.6931 | 0.9942 | >0.9999 | 0.2572 |
| NB-MI vs. YS1-MI | 0.2473 | 0.9985 | 0.6058 | 0.9154 | 0.0809 | |
| NB-MI vs. YS2-MI | >0.9999 | 0.6155 | 0.0019 | 0.4821 | 0.9932 | |
| YS0-MI vs. YS1-MI | 0.5279 | 0.5916 | 0.7607 | 0.9016 | 0.9317 | |
| YS0-MI vs. YS2-MI | 0.9252 | 0.9993 | 0.0047 | 0.446 | 0.1559 | |
| YS1-MI vs. YS2-MI | 0.2144 | 0.5129 | 0.0776 | 0.8535 | 0.0429 | |
| il-10 | NB-MI vs. YS0-MI | 0.7923 | 0.9984 | 0.8448 | 0.2293 | 0.9995 |
| NB-MI vs. YS1-MI | >0.9999 | 0.5506 | 0.9998 | 0.1309 | 0.199 | |
| NB-MI vs. YS2-MI | 0.6378 | 0.5435 | 0.9834 | 0.9901 | 0.9423 | |
| YS0-MI vs. YS1-MI | 0.7655 | 0.677 | 0.8742 | 0.9916 | 0.2638 | |
| YS0-MI vs. YS2-MI | 0.9924 | 0.6665 | 0.9658 | 0.3563 | 0.9111 | |
| YS1-MI vs. YS2-MI | 0.5956 | >0.9999 | 0.9917 | 0.2178 | 0.0613 | |
| hsp70 | NB-MI vs. YS0-MI | 0.4408 | 0.9584 | 0.6403 | 0.9663 | 0.8582 |
| NB-MI vs. YS1-MI | 0.4316 | 0.3345 | 0.3182 | 0.5489 | 0.9171 | |
| NB-MI vs. YS2-MI | 0.3211 | 0.9805 | 0.2748 | 0.8029 | 0.9326 | |
| YS0-MI vs. YS1-MI | >0.9999 | 0.6666 | 0.95 | 0.8145 | 0.5146 | |
| YS0-MI vs. YS2-MI | 0.9936 | 0.9995 | 0.9241 | 0.9697 | 0.9967 | |
| YS1-MI vs. YS2-MI | 0.9947 | 0.5926 | 0.9998 | 0.9729 | 0.622 | |

Table A3.
Normalized relative expression values of innate immune-related genes in full-sibling European sea bass larvae treated with DnaK and challenged with V. anguillarum at different time points (0, 18, 24, 36, and 120 h).
Table A3.
Normalized relative expression values of innate immune-related genes in full-sibling European sea bass larvae treated with DnaK and challenged with V. anguillarum at different time points (0, 18, 24, 36, and 120 h).
| Genes | Time (hpc) | NB-Ch | YS0-Ch | YS1-Ch | YS2-Ch | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| pentraxin | 0 | 1.13 | 0.69 | 1.31 | 0.53 | 1.10 | 0.51 | 0.87 | 0.25 |
| 18 | 0.74 | 0.16 | 0.55 | 0.10 | 0.66 | 0.32 | 0.55 | 0.21 | |
| 24 | 1.22 | 0.64 | 1.21 | 0.52 | 1.32 | 0.55 | 0.78 | 0.28 | |
| 36 | 1.06 | 0.55 | 0.78 | 0.35 | 1.07 | 0.69 | 0.72 | 0.44 | |
| 120 | 0.63 | 0.32 | 0.63 | 0.41 | 1.17 | 1.09 | 1.00 | 0.89 | |
| lysozyme | 0 | 1.08 | 1.08 | 1.27 | 0.90 | 1.48 | 1.17 | 1.90 | 1.24 |
| 18 | 0.85 | 0.47 | 0.43 | 0.34 | 1.51 | 0.90 | 0.69 | 0.29 | |
| 24 | 1.12 | 0.75 | 0.91 | 0.59 | 1.51 | 0.99 | 1.26 | 0.62 | |
| 36 | 1.03 | 0.62 | 0.92 | 0.39 | 1.35 | 1.10 | 1.06 | 0.74 | |
| 120 | 2.35 | 2.33 | 2.67 | 1.77 | 2.17 | 3.59 | 2.61 | 3.26 | |
| trasnferrin | 0 | 1.14 | 0.50 | 1.74 | 0.97 | 1.46 | 0.73 | 1.10 | 0.65 |
| 18 | 1.72 | 0.69 | 1.04 | 0.66 | 1.50 | 0.61 | 0.99 | 0.37 | |
| 24 | 1.89 | 0.98 | 1.23 | 0.73 | 1.76 | 1.14 | 1.40 | 0.47 | |
| 36 | 1.56 | 0.70 | 1.03 | 0.36 | 1.86 | 1.16 | 1.20 | 0.49 | |
| 120 | 9.89 | 8.92 | 10.81 | 7.48 | 10.76 | 11.12 | 21.92 | 10.50 | |
| hepcidin | 0 | 0.92 | 1.40 | 2.82 | 2.52 | 1.40 | 1.21 | 1.37 | 1.73 |
| 18 | 1.02 | 1.12 | 2.12 | 2.89 | 0.82 | 0.31 | 0.55 | 0.23 | |
| 24 | 1.50 | 2.59 | 1.70 | 1.71 | 2.35 | 2.40 | 0.96 | 0.43 | |
| 36 | 1.31 | 1.22 | 1.43 | 0.88 | 4.67 | 11.64 | 0.67 | 0.54 | |
| 120 | 1320.33 | 1673.60 | 917.26 | 699.12 | 659.76 | 355.95 | 1564.23 | 1757.90 | |
| il-1β | 0 | 1.77 | 3.08 | 1.36 | 1.05 | 1.50 | 1.69 | 1.51 | 1.80 |
| 18 | 2.65 | 4.09 | 1.84 | 2.88 | 1.14 | 0.83 | 0.73 | 0.49 | |
| 24 | 1.12 | 0.81 | 0.65 | 0.41 | 1.77 | 1.67 | 1.03 | 0.29 | |
| 36 | 1.40 | 1.49 | 1.02 | 0.48 | 1.22 | 0.98 | 0.73 | 0.49 | |
| 120 | 43.22 | 39.27 | 20.67 | 16.47 | 22.23 | 17.56 | 64.18 | 52.47 | |
| il-8 | 0 | 0.76 | 0.77 | 0.61 | 0.20 | 1.55 | 1.67 | 0.69 | 0.38 |
| 18 | 1.04 | 1.04 | 0.96 | 1.13 | 0.67 | 0.36 | 0.45 | 0.17 | |
| 24 | 0.73 | 0.63 | 0.69 | 0.65 | 1.58 | 1.48 | 0.81 | 0.54 | |
| 36 | 1.31 | 1.54 | 0.58 | 0.25 | 0.99 | 0.68 | 0.64 | 0.56 | |
| 120 | 21.33 | 14.37 | 11.18 | 8.94 | 12.34 | 12.47 | 28.56 | 21.35 | |
| ccl4 | 0 | 0.98 | 0.49 | 1.11 | 0.31 | 1.42 | 1.00 | 0.97 | 0.32 |
| 18 | 1.67 | 1.71 | 1.64 | 2.11 | 1.25 | 0.71 | 1.21 | 0.66 | |
| 24 | 1.52 | 0.89 | 0.95 | 0.66 | 1.73 | 1.01 | 1.08 | 0.33 | |
| 36 | 1.15 | 0.53 | 1.19 | 0.28 | 1.63 | 0.79 | 1.11 | 0.70 | |
| 120 | 2.56 | 1.83 | 3.10 | 2.17 | 4.17 | 3.69 | 4.17 | 1.66 | |
| il-10 | 0 | 1.22 | 0.74 | 1.17 | 0.40 | 1.00 | 0.25 | 1.22 | 0.46 |
| 18 | 1.21 | 0.36 | 1.24 | 0.44 | 1.09 | 0.58 | 1.02 | 0.22 | |
| 24 | 1.44 | 0.45 | 1.26 | 0.76 | 1.04 | 0.26 | 1.13 | 0.25 | |
| 36 | 1.10 | 0.46 | 1.06 | 0.43 | 1.46 | 0.90 | 0.89 | 0.29 | |
| 120 | 7.42 | 7.60 | 3.85 | 3.13 | 3.26 | 2.10 | 5.58 | 5.39 | |
| hsp70 | 0 | 1.14 | 0.08 | 0.99 | 0.14 | 0.99 | 0.15 | 0.97 | 0.13 |
| 18 | 0.85 | 0.12 | 0.89 | 0.23 | 0.93 | 0.16 | 0.97 | 0.17 | |
| 24 | 1.02 | 0.34 | 0.78 | 0.15 | 0.90 | 0.12 | 0.84 | 0.10 | |
| 36 | 0.97 | 0.25 | 0.87 | 0.09 | 0.94 | 0.70 | 0.69 | 0.09 | |
| 120 | 0.95 | 0.20 | 1.07 | 0.13 | 1.36 | 1.06 | 1.40 | 0.94 | |

Table A4.
Summary of statistical differences (p values) from Tuckey’s multiple comparison test for the relative expression of different genes evaluated in sea bass larvae treated with DnaK and challenged with V. anguillarum at different time points (0, 18, 24, 36, and 120 h). Significant differences (p < 0.05) are highlighted in bold.
Table A4.
Summary of statistical differences (p values) from Tuckey’s multiple comparison test for the relative expression of different genes evaluated in sea bass larvae treated with DnaK and challenged with V. anguillarum at different time points (0, 18, 24, 36, and 120 h). Significant differences (p < 0.05) are highlighted in bold.
| Genes | Group Comparison | Hours Post Challenge | ||||
|---|---|---|---|---|---|---|
| 0 | 18 | 24 | 36 | 120 | ||
| pentraxin | NB-MI vs. NB-Ch | 0.9483 | 0.95 | 0.7373 | 0.984 | 0.9998 |
| NB-MI vs. YS0-Ch | 0.5548 | >0.9999 | 0.7564 | 0.968 | >0.9999 | |
| NB-MI vs. YS1-Ch | 0.9665 | 0.9958 | 0.401 | 0.9748 | 0.2522 | |
| NB-MI vs. YS2-Ch | 0.9984 | >0.9999 | 0.9666 | 0.8925 | 0.5979 | |
| NB-Ch vs. YS0-Ch | 0.9384 | 0.9382 | >0.9999 | 0.771 | >0.9999 | |
| NB-Ch vs. YS1-Ch | >0.9999 | 0.997 | 0.9926 | >0.9999 | 0.2062 | |
| NB-Ch vs. YS2-Ch | 0.8134 | 0.93 | 0.3555 | 0.6113 | 0.5171 | |
| YS0-Ch vs. YS1-Ch | 0.8761 | 0.9923 | 0.9902 | 0.7058 | 0.3046 | |
| YS0-Ch vs. YS2-Ch | 0.2969 | >0.9999 | 0.3744 | 0.9987 | 0.6318 | |
| YS1-Ch vs. YS2-Ch | 0.8463 | 0.991 | 0.1174 | 0.529 | 0.9621 | |
| lysozyme | NB-MI vs. NB-Ch | 0.9816 | 0.9988 | >0.9999 | 0.9996 | 0.0249 |
| NB-MI vs. YS0-Ch | 0.9057 | 0.8752 | 0.9996 | 0.9942 | 0.0157 | |
| NB-MI vs. YS1-Ch | 0.7512 | 0.9204 | 0.9469 | 0.9979 | 0.0695 | |
| NB-MI vs. YS2-Ch | 0.3662 | 0.9833 | 0.997 | 0.9998 | 0.0063 | |
| NB-Ch vs. YS0-Ch | 0.998 | 0.9614 | 0.9977 | 0.9997 | 0.9895 | |
| NB-Ch vs. YS1-Ch | 0.9674 | 0.8196 | 0.9714 | 0.9866 | 0.9987 | |
| NB-Ch vs. YS2-Ch | 0.693 | 0.999 | 0.9994 | >0.9999 | 0.9932 | |
| YS0-Ch vs. YS1-Ch | 0.9965 | 0.4253 | 0.8727 | 0.947 | 0.9537 | |
| YS0-Ch vs. YS2-Ch | 0.8291 | 0.9933 | 0.9803 | 0.9993 | >0.9999 | |
| YS1-Ch vs. YS2-Ch | 0.9549 | 0.6697 | 0.9933 | 0.9874 | 0.9589 | |
| transferrin | NB-MI vs. NB-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 |
| NB-MI vs. YS0-Ch | >0.99 | >0.99 | >0.99 | 0.99 | <0.001 | |
| NB-MI vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| NB-MI vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| NB-Ch vs. YS0-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.99 | |
| NB-Ch vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.99 | |
| NB-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| YS0-Ch vs. YS1-Ch | >0.99 | >0.99 | >0.99 | 0.99 | >0.99 | |
| YS0-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| YS1-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| hepcidin | NB-MI vs. NB-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | <0.0001 |
| NB-MI vs. YS0-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.0039 | |
| NB-MI vs. YS1-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.0368 | |
| NB-MI vs. YS2-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | <0.0001 | |
| NB-Ch vs. YS0-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.5799 | |
| NB-Ch vs. YS1-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.0659 | |
| NB-Ch vs. YS2-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.8506 | |
| YS0-Ch vs. YS1-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.8799 | |
| YS0-Ch vs. YS2-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.1113 | |
| YS1-Ch vs. YS2-Ch | >0.9999 | >0.9999 | >0.9999 | >0.9999 | 0.0022 | |
| il-1β | NB-MI vs. NB-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 |
| NB-MI vs. YS0-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.05 | |
| NB-MI vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.02 | |
| NB-MI vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| NB-Ch vs. YS0-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.01 | |
| NB-Ch vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.02 | |
| NB-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.02 | |
| YS0-Ch vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
| YS0-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| YS1-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| il-8 | NB-MI vs. NB-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 |
| NB-MI vs. YS0-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.01 | |
| NB-MI vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| NB-MI vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| NB-Ch vs. YS0-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.01 | |
| NB-Ch vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.02 | |
| NB-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | 0.08 | |
| YS0-Ch vs. YS1-Ch | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
| YS0-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| YS1-Ch vs. YS2-Ch | >0.99 | >0.99 | >0.99 | >0.99 | <0.001 | |
| ccl4 | NB-MI vs. NB-Ch | >0.9999 | 0.6909 | 0.7139 | 0.9999 | 0.031 |
| NB-MI vs. YS0-Ch | 0.9992 | 0.7504 | 0.9997 | 0.9994 | 0.0054 | |
| NB-MI vs. YS1-Ch | 0.9346 | 0.9821 | 0.4441 | 0.8471 | <0.0001 | |
| NB-MI vs. YS2-Ch | >0.9999 | 0.989 | 0.9917 | >0.9999 | <0.0001 | |
| NB-Ch vs. YS0-Ch | 0.9992 | >0.9999 | 0.8898 | >0.9999 | 0.9099 | |
| NB-Ch vs. YS1-Ch | 0.9346 | 0.9434 | 0.9956 | 0.9198 | 0.0481 | |
| NB-Ch vs. YS2-Ch | >0.9999 | 0.9243 | 0.9315 | >0.9999 | 0.0369 | |
| YS0-Ch vs. YS1-Ch | 0.979 | 0.9625 | 0.7075 | 0.9258 | 0.4689 | |
| YS0-Ch vs. YS2-Ch | 0.999 | 0.9481 | 0.9996 | 0.9999 | 0.437 | |
| YS1-Ch vs. YS2-Ch | 0.9248 | >0.9999 | 0.7538 | 0.8713 | >0.9999 | |
| il-10 | NB-MI vs. NB-Ch | 0.9995 | 0.9969 | 0.9982 | 0.9997 | <0.0001 |
| NB-MI vs. YS0-Ch | 0.9998 | 0.9964 | >0.9999 | 0.9991 | 0.1938 | |
| NB-MI vs. YS1-Ch | >0.9999 | 0.9995 | >0.9999 | 0.9998 | 0.4035 | |
| NB-MI vs. YS2-Ch | 0.9995 | >0.9999 | >0.9999 | 0.993 | 0.0003 | |
| NB-Ch vs. YS0-Ch | >0.9999 | >0.9999 | 0.9998 | >0.9999 | 0.009 | |
| NB-Ch vs. YS1-Ch | 0.9994 | >0.9999 | 0.9929 | 0.996 | 0.0004 | |
| NB-Ch vs. YS2-Ch | >0.9999 | 0.9996 | 0.9974 | 0.9995 | 0.3103 | |
| YS0-Ch vs. YS1-Ch | 0.9998 | 0.9999 | 0.9995 | 0.9923 | 0.9832 | |
| YS0-Ch vs. YS2-Ch | >0.9999 | 0.9995 | >0.9999 | 0.9997 | 0.4897 | |
| YS1-Ch vs. YS2-Ch | 0.9994 | >0.9999 | >0.9999 | 0.9715 | 0.1339 | |
| hsp70 | NB-MI vs. NB-Ch | >0.9999 | 0.9556 | 0.9703 | 0.9901 | 0.9902 |
| NB-MI vs. YS0-Ch | 0.9112 | 0.9921 | 0.2385 | >0.9999 | 0.814 | |
| NB-MI vs. YS1-Ch | 0.9081 | 0.9996 | 0.6239 | 0.9972 | 0.0352 | |
| NB-MI vs. YS2-Ch | 0.8633 | >0.9999 | 0.3933 | 0.7379 | 0.0124 | |
| NB-Ch vs. YS0-Ch | 0.9112 | 0.9993 | 0.651 | 0.9745 | 0.9652 | |
| NB-Ch vs. YS1-Ch | 0.9081 | 0.9889 | 0.9568 | 0.9999 | 0.1275 | |
| NB-Ch vs. YS2-Ch | 0.8633 | 0.9543 | 0.8359 | 0.4585 | 0.0582 | |
| YS0-Ch vs. YS1-Ch | >0.9999 | 0.9993 | 0.9467 | 0.9894 | 0.5684 | |
| YS0-Ch vs. YS2-Ch | 0.9999 | 0.9912 | 0.9947 | 0.7928 | 0.4038 | |
| YS1-Ch vs. YS2-Ch | >0.9999 | 0.9994 | 0.9962 | 0.4974 | 0.9992 | |
References
- Sung, Y.Y.; Pineda, C.; MacRae, T.H.; Sorgeloos, P.; Bossier, P. Exposure of Gnotobiotic Artemia franciscana Larvae to Abiotic Stress Promotes Heat Shock Protein 70 Synthesis and Enhances Resistance to Pathogenic Vibrio campbellii. Cell Stress. Chaperones 2008, 13, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Tort, L. Stress and Immune Modulation in Fish. Dev. Comp. Immunol. 2011, 35, 1366–1375. [Google Scholar] [CrossRef]
- Khansari, A.R.; Balasch, J.C.; Vallejos-Vidal, E.; Teles, M.; Fierro-Castro, C.; Tort, L.; Reyes-López, F.E. Comparative Study of Stress and Immune-Related Transcript Outcomes Triggered by Vibrio anguillarum Bacterin and Air Exposure Stress in Liver and Spleen of Gilthead Seabream (Sparus aurata), Zebrafish (Danio rerio) and Rainbow Trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2019, 86, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, S. The Heat-Shock Response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Steinert, S.A.; Pickwell, G.V. Induction of HSP70 Proteins in Mussels by Ingestion of Tributyltin. Mar. Environ. Res. 1993, 35, 89–93. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Loeschcke, V. Larval Crowding in Drosophila Melanogaster Induces Hsp70 Expression, and Leads to Increased Adult Longevity and Adult Thermal Stress Resistance. J. Insect Physiol. 2001, 47, 1301–1307. [Google Scholar] [CrossRef]
- Dang, W.; Hu, Y.H.; Zhang, M.; Sun, L. Identification and Molecular Analysis of a Stress-Inducible Hsp70 from Sciaenops Ocellatus. Fish Shellfish Immunol. 2010, 29, 600–607. [Google Scholar] [CrossRef]
- Athman, R.; Philpott, D. Innate Immunity via Toll-like Receptors and Nod Proteins. Curr. Opin. Microbiol. 2004, 7, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, S.; Wang, Y.; Lu, W.; Zhang, Q.; Cheng, J. Genomic and Transcriptomic Landscape and Evolutionary Dynamics of Heat Shock Proteins in Spotted Sea Bass (Lateolabrax maculatus) under Salinity Change and Alkalinity Stress. Biology 2022, 11, 353. [Google Scholar] [CrossRef]
- Li, Z.H.; Zhong, L.Q.; Wu, Y.H.; Mu, W.N. Alteration of Cytochrome P450 1 Regulation and HSP 70 Level in Brain of Juvenile Common Carp (Cyprinus carpio) after Chronic Exposure to Tributyltin. Fish. Physiol. Biochem. 2016, 42, 287–294. [Google Scholar] [CrossRef]
- Wieten, L.; Broere, F.; van der Zee, R.; Koerkamp, E.K.; Wagenaar, J.; van Eden, W. Cell Stress Induced HSP Are Targets of Regulatory T Cells: A Role for HSP Inducing Compounds as Anti-Inflammatory Immuno-Modulators? FEBS Lett. 2007, 581, 3716–3722. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, V.; Soza, C.; Martínez, R.; Rosemblatt, M.; Burzio, L.O.; Valenzuela, P.D.T. Production and Immune Response of Recombinant Hsp60 and Hsp70 from the Salmon Pathogen Piscirickettsia Salmonis. Biol. Res. 2005, 38, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Josepriya, T.A.; Chien, K.H.; Lin, H.Y.; Huang, H.N.; Wu, C.J.; Song, Y.L. Immobilization Antigen Vaccine Adjuvanted by Parasitic Heat Shock Protein 70C Confers High Protection in Fish against Cryptocaryonosis. Fish Shellfish Immunol. 2015, 45, 517–527. [Google Scholar] [CrossRef]
- Pederzoli, A.; Mola, L. The Early Stress Responses in Fish Larvae. Acta Histochem. 2016, 118, 443–449. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, L.; Liu, B.S.; Guo, H.Y.; Zhu, K.C.; Zhang, N.; Jiang, S.G.; Zhang, D.C. Effects of Stocking Density on the Growth Performance, Serum Biochemistry, Muscle Composition and HSP70 Gene Expression of Juvenile Golden Pompano Trachinotus Ovatus (Linnaeus, 1758). Aquaculture 2020, 518, 734841. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, J.H.; Jo, A.H.; Choi, Y.J.; Choi, C.Y.; Kang, J.C.; Kim, J.H. Microplastic Polyamide Toxicity: Neurotoxicity, Stress Indicators and Immune Responses in Crucian Carp, Carassius carassius. Ecotoxicol. Environ. Saf. 2023, 265, 115469. [Google Scholar] [CrossRef]
- Ahani, S.; Ahani, S.; Taheri, A.; Morteza, S.; Pagheh, E.; Arghideh, M.; Yousefi, M. Probiotic, Fructooligosaccharide and Yeast Extract Mixture Improves Gut Health in Rainbow Trout, Oncorhynchus mykiss. J. Anim. Physiol. Anim. Nutr. 2025, 109, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.C.; Lee, J.W.; Hu, Y.F.; Ballantyne, R.; Liu, C.H. Effects of Aspergillus-Meal Prebiotic Diet on the Growth Performance, Health Status and Gut Microbiota of Asian Seabass, Lates calcarifer. Fish Shellfish Immunol. 2023, 136, 108696. [Google Scholar] [CrossRef]
- Reyes-López, F.E.; Romeo, J.S.; Vallejos-Vidal, E.; Reyes-Cerpa, S.; Sandino, A.M.; Tort, L.; Mackenzie, S.; Imarai, M. Differential Immune Gene Expression Profiles in Susceptible and Resistant Full-Sibling Families of Atlantic Salmon (Salmo salar) Challenged with Infectious Pancreatic Necrosis Virus (IPNV). Dev. Comp. Immunol. 2015, 53, 210–221. [Google Scholar] [CrossRef]
- Yaacob, E.N.; Norouzitallab, P.; De Geest, B.G.; Bajek, A.; Dierckens, K.; Bossier, P.; Vanrompay, D. Recombinant DnaK Orally Administered Protects Axenic European Sea Bass Against Vibriosis. Front. Immunol. 2020, 10, 492419. [Google Scholar] [CrossRef]
- Sinnasamy, S.; Mat Noordin, N.; Macrae, T.H.; Ikhwanuddin bin Abdullah, M.; Bossier, P.; bin Abdul Wahid, M.E.; Noriaki, A.; Sung, Y.Y. Ingestion of Food Pellets Containing Escherichia Coli Overexpressing the Heat-Shock Protein DnaK Protects Penaeus vannamei (Boone) against Vibrio harveyi (Baumann) Infection. J. Fish Dis. 2016, 39, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, W.; Hou, S.; Fu, W.; Cai, J.; Xia, L.; Lu, Y. Comparison of Protective Efficacy between Two DNA Vaccines Encoding DnaK and GroEL against Fish Nocardiosis. Fish Shellfish Immunol. 2019, 95, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, P.K.; Bansal, A.; Sagi, S.S.K.; Sairam, M. Intraperitoneal Immunization of Recombinant HSP70 (DnaK) of Salmonella Typhi Induces a Predominant Th2 Response and Protective Immunity in Mice against Lethal Salmonella infection. Vaccine 2011, 29, 6532–6539. [Google Scholar] [CrossRef]
- Hu, B.; Phuoc, L.H.; Sorgeloos, P.; Bossier, P. Bacterial HSP70 (DnaK) Is an Efficient Immune Stimulator in Litopenaeus vannamei. Aquaculture 2014, 418–419, 87–93. [Google Scholar] [CrossRef]
- Deane, E.E.; Woo, N.Y.S. Advances and Perspectives on the Regulation and Expression of Piscine Heat Shock Proteins. Rev. Fish Biol. Fish. 2010, 21, 153–185. [Google Scholar] [CrossRef]
- Engelsma, M.Y.; Huising, M.O.; Van Muiswinkel, W.B.; Flik, G.; Kwang, J.; Savelkoul, H.F.J.; Verburg-Van Kemenade, B.M.L. Neuroendocrine-Immune Interactions in Fish: A Role for Interleukin-1. Vet. Immunol. Immunopathol. 2002, 87, 467–479. [Google Scholar] [CrossRef]
- Nardocci, G.; Navarro, C.; Cortés, P.P.; Imarai, M.; Montoya, M.; Valenzuela, B.; Jara, P.; Acuña-Castillo, C.; Fernández, R. Neuroendocrine Mechanisms for Immune System Regulation during Stress in Fish. Fish Shellfish Immunol. 2014, 40, 531–538. [Google Scholar] [CrossRef]
- Engelsma, M.Y.; Stet, R.J.M.; Saeij, J.P.; Verburg-Van Kemenade, B.M.L. Differential Expression and Haplotypic Variation of Two Interleukin-1β Genes in the Common Carp (Cyprinus carpio L.). Cytokine 2003, 22, 21–32. [Google Scholar] [CrossRef]
- Monzón-Atienza, L.; Bravo, J.; Fernández-Montero, Á.; Charlie-Silva, I.; Montero, D.; Ramos-Vivas, J.; Galindo-Villegas, J.; Acosta, F. Dietary Supplementation of Bacillus Velezensis Improves Vibrio anguillarum Clearance in European Sea Bass by Activating Essential Innate Immune Mechanisms. Fish Shellfish Immunol. 2022, 124, 244–253. [Google Scholar] [CrossRef]
- Hickey, M.E.; Lee, J.L. A Comprehensive Review of Vibrio (Listonella) Anguillarum: Ecology, Pathology and Prevention. Rev. Aquac. 2018, 10, 585–610. [Google Scholar] [CrossRef]
- Sung, Y.; Ashame, M.; Chen, S.; MacRae, T.; Sorgeloos, P.; Bossier, P. Feeding Artemia franciscana (Kellogg) Larvae with Bacterial Heat Shock Protein, Protects from Vibrio campbellii Infection. J. Fish Dis. 2009, 32, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Baruah, K.; Ranjan, J.; Sorgeloos, P.; MacRae, T.H.; Bossier, P. Priming the Prophenoloxidase System of Artemia franciscana by Heat Shock Proteins Protects against Vibrio campbellii Challenge. Fish Shellfish Immunol. 2011, 31, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Dierckens, K.; Rekecki, A.; Laureau, S.; Sorgeloos, P.; Boon, N.; Van den Broeck, W.; Bossier, P. Development of a Bacterial Challenge Test for Gnotobiotic Sea Bass (Dicentrarchus labrax) Larvae. Environ. Microbiol. 2009, 11, 526–533. [Google Scholar] [CrossRef]
- Reyes-López, F.E.; Aerts, J.; Vallejos-Vidal, E.; Ampe, B.; Dierckens, K.; Tort, L.; Bossier, P. Modulation of Innate Immune-Related Genes and Glucocorticoid Synthesis in Gnotobiotic Full-Sibling European Sea Bass (Dicentrarchus labrax) Larvae Challenged with Vibrio anguillarum. Front. Immunol. 2018, 9, 914. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Mitter, K.; Kotoulas, G.; Magoulas, A.; Mulero, V.; Sepulcre, P.; Figueras, A.; Novoa, B.; Sarropoulou, E. Evaluation of Candidate Reference Genes for QPCR during Ontogenesis and of Immune-Relevant Tissues of European Seabass (Dicentrarchus labrax). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 153, 340–347. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Ma, F.; Luo, L.T.; Wang, Q. Hsp60/10 and SHsp Families of Heat Shock Protein Genes in Rainbow Trout (Oncorhynchus mykiss) and Their Expression under Heat Stress. Aquac. Int. 2022, 30, 1–18. [Google Scholar] [CrossRef]
- Lange, M.D.; Beck, B.H.; Brown, J.D.; Farmer, B.D.; Barnett, L.M.; Webster, C.D. Missing the Target: DNAk Is a Dominant Epitope in the Humoral Immune Response of Channel Catfish (Ictalurus punctatus) to Flavobacterium Columnare. Fish Shellfish Immunol. 2016, 51, 170–179. [Google Scholar] [CrossRef]
- Camberg, J.L.; Doyle, S.M.; Jhonston, D.M.; Wickner, S. Molecular Chaperones. In Brenner’s Encyclopedia of Genetics; Elsevier: Amsterdam, The Netherlands, 2013; pp. 456–460. [Google Scholar]
- Sung, Y.; MacRae, T. Heat Shock Proteins and Disease Control in Aquatic Organisms. J. Aquac. Res. Dev. 2011, S2, 006. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a Fish Pathogen: Virulence Factors, Diagnosis and Prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Rekecki, A.; Gunasekara, R.A.Y.S.A.; Dierckens, K.; Laureau, S.; Boon, N.; Favoreel, H.; Cornelissen, M.; Sorgeloos, P.; Ducatelle, R.; Bossier, P.; et al. Bacterial Host Interaction of GFP-Labelled Vibrio anguillarum HI-610 with Gnotobiotic Sea Bass, Dicentrarchus labrax (L.), Larvae. J. Fish Dis. 2012, 35, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Nakano, T.; Grau, E.G.; Iwama, G.K. The Effects of Cortisol on Heat Shock Protein 70 Levels in Two Fish Species. Gen. Comp. Endocrinol. 2001, 124, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Stolte, E.H.; Chadzinska, M.; Przybylska, D.; Flik, G.; Savelkoul, H.; Verburg-van Kemenade, B.L. The Immune Response Differentially Regulates Hsp70 and Glucocorticoid Receptor Expression in Vitro and in Vivo in Common Carp (Cyprinus carpio L.). Fish Shellfish Immunol. 2009, 27, 9–16. [Google Scholar] [CrossRef]
- Valero, Y.; Arizcun, M.; Cortés, J.; Ramírez-Cepeda, F.; Guzmán, F.; Mercado, L.; Esteban, M.Á.; Chaves-Pozo, E.; Cuesta, A. NK-Lysin, Dicentracin and Hepcidin Antimicrobial Peptides in European Sea Bass. Ontogenetic Development and Modulation in Juveniles by Nodavirus. Dev. Comp. Immunol. 2020, 103, 103516. [Google Scholar] [CrossRef]
- Buchmann, K.; Secombes, C.J. Principles of Fish Immunology, 1st ed.; Buchmann, K., Secombes, C.J., Eds.; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Talbot, A.T.; Pottinger, T.G.; Smith, T.J.; Cairns, M.T. Acute Phase Gene Expression in Rainbow Trout (Oncorhynchus mykiss) after Exposure to a Confinement Stressor: A Comparison of Pooled and Individual Data. Fish Shellfish Immunol. 2009, 27, 309–317. [Google Scholar] [CrossRef]
- Shi, Y.H.; Chen, K.; Ma, W.J.; Chen, J. Ayu C-Reactive Protein/Serum Amyloid P Agglutinates Bacteria and Inhibits Complement-Mediated Opsonophagocytosis by Monocytes/Macrophages. Fish Shellfish Immunol. 2018, 76, 58–67. [Google Scholar] [CrossRef]
- Magnadottir, B.; Gudmundsdottir, B.K.; Groman, D. Immuno-Histochemical Determination of Humoral Immune Markers within Bacterial Induced Granuloma Formation in Atlantic Cod (Gadus morhua L.). Fish Shellfish Immunol. 2013, 34, 1372–1375. [Google Scholar] [CrossRef]
- Cecchini, S.; Terova, G.; Caricato, G.; Saroglia, M. Lysozyme Activity in Embryos and Larvae of Sea Bass (Dicentrarchus labrax L.), Spawned by Broodstock Fed with Vitamin C Enriched Diets. Bull.-Eur. Assoc. Fish Pathol. 2000, 20, 120–124. [Google Scholar]
- Álvarez, C.A.; Acosta, F.; Montero, D.; Guzmán, F.; Torres, E.; Vega, B.; Mercado, L. Synthetic Hepcidin from Fish: Uptake and Protection against Vibrio anguillarum in Sea Bass (Dicentrarchus labrax). Fish Shellfish Immunol. 2016, 55, 662–670. [Google Scholar] [CrossRef]
- Wang, X.; Li, L.; Yuan, G.; Zhu, L.; Pei, C.; Hou, L.; Li, C.; Jiang, X.; Kong, X. Interleukin (IL)-22 in Common Carp (Cyprinus carpio L.): Immune Modulation, Antibacterial Defense, and Activation of the JAK-STAT Signaling Pathway. Fish Shellfish Immunol. 2022, 131, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, D.S.; Omeka, W.K.M.; Yang, H.; Lim, C.; Choi, C.Y.; Lee, J. Molecular Characterization of Fish Cytokine IL-17C from Amphiprion Clarkii and Its Immunomodulatory Effects on the Responses to Pathogen-Associated Molecular Patterns and Bacterial Challenges. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2022, 257, 110669. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Lin, H.T.; Foung, Y.F.; Han-You Lin, J. The Bioactivity of Teleost IL-6: IL-6 Protein in Orange-Spotted Grouper (Epinephelus coioides) Induces Th2 Cell Differentiation Pathway and Antibody Production. Dev. Comp. Immunol. 2012, 38, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Villegas, J.; Mulero, I.; García-Alcazar, A.; Muñoz, I.; Peñalver-Mellado, M.; Streitenberger, S.; Scapigliati, G.; Meseguer, J.; Mulero, V. Recombinant TNFα as Oral Vaccine Adjuvant Protects European Sea Bass against Vibriosis: Insights into the Role of the CCL25/CCR9 Axis. Fish Shellfish Immunol. 2013, 35, 1260–1271. [Google Scholar] [CrossRef]
- Guo, M.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. The Effects of IL-1β, IL-8, G-CSF and TNF-α as Molecular Adjuvant on the Immune Response to an E. Tarda Subunit Vaccine in Flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2018, 77, 374–384. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Z.; Zhang, J.; Amoah, K.; Asiedu, B.; Cai, J.; Wang, B.; Jian, J. Novel C-Type Lectin Mediated Non-Specific Cytotoxic Cells Killing Activity through NCCRP-1 in Nile Tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2024, 149, 109594. [Google Scholar] [CrossRef]
- Li, J.H.; Yu, Z.L.; Xue, N.N.; Zou, P.F.; Hu, J.Y.; Nie, P.; Chang, M.X. Molecular Cloning and Functional Characterization of Peptidoglycan Recognition Protein 6 in Grass Carp Ctenopharyngodon Idella. Dev. Comp. Immunol. 2014, 42, 244–255. [Google Scholar] [CrossRef]
- Sun, Y.; Cao, Z.; Zhang, P.; Wei, C.; Li, J.; Wu, Y.; Zhou, Y. IFN Regulatory Factor 3 of Golden Pompano and Its NLS Domain Are Involved in Antibacterial Innate Immunity and Regulate the Expression of Type I Interferon (IFNa3). Front. Immunol. 2023, 14, 1128196. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Kabanov, F.V.; Troepolskaya, O.D.; Tischenko, M.M. A Variant of the Interleukin-1β Gene in European Sea Bass, Dicentrarchus labrax L., Is Associated with Increased Resistance against Vibrio anguillarum. J. Fish Dis. 2010, 33, 759–767. [Google Scholar] [CrossRef]
- Reyes-Cerpa, S.; Vallejos-Vidal, E.; Gonzalez-Bown, M.J.; Morales-Reyes, J.; Pérez-Stuardo, D.; Vargas, D.; Imarai, M.; Cifuentes, V.; Spencer, E.; Sandino, A.M.; et al. Effect of Yeast (Xanthophyllomyces dendrorhous) and Plant (Saint John’s Wort, Lemon Balm, and Rosemary) Extract Based Functional Diets on Antioxidant and Immune Status of Atlantic Salmon (Salmo salar) Subjected to Crowding Stress. Fish Shellfish Immunol. 2018, 74, 250–259. [Google Scholar] [CrossRef]
- Sepulcre, M.P.; Sarropoulou, E.; Kotoulas, G.; Meseguer, J.; Mulero, V. Vibrio anguillarum Evades the Immune Response of the Bony Fish Sea Bass (Dicentrarchus labrax L.) through the Inhibition of Leukocyte Respiratory Burst and down-Regulation of Apoptotic Caspases. Mol. Immunol. 2007, 44, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Meloni, M.; Candusso, S.; Galeotti, M.; Volpatti, D. Preliminary Study on Expression of Antimicrobial Peptides in European Sea Bass (Dicentrarchus labrax) Following in Vivo Infection with Vibrio anguillarum. A Time Course Experiment. Fish Shellfish Immunol. 2015, 43, 82–90. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).