A Preliminary Investigation of the Gastrointestinal Bacterial Microbiomes of Barred Owls (Strix varia) Admitted to a Wildlife Hospital
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Microbiome Analysis
2.3. Antimicrobial Resistance Gene Analysis
3. Results
3.1. Treatment
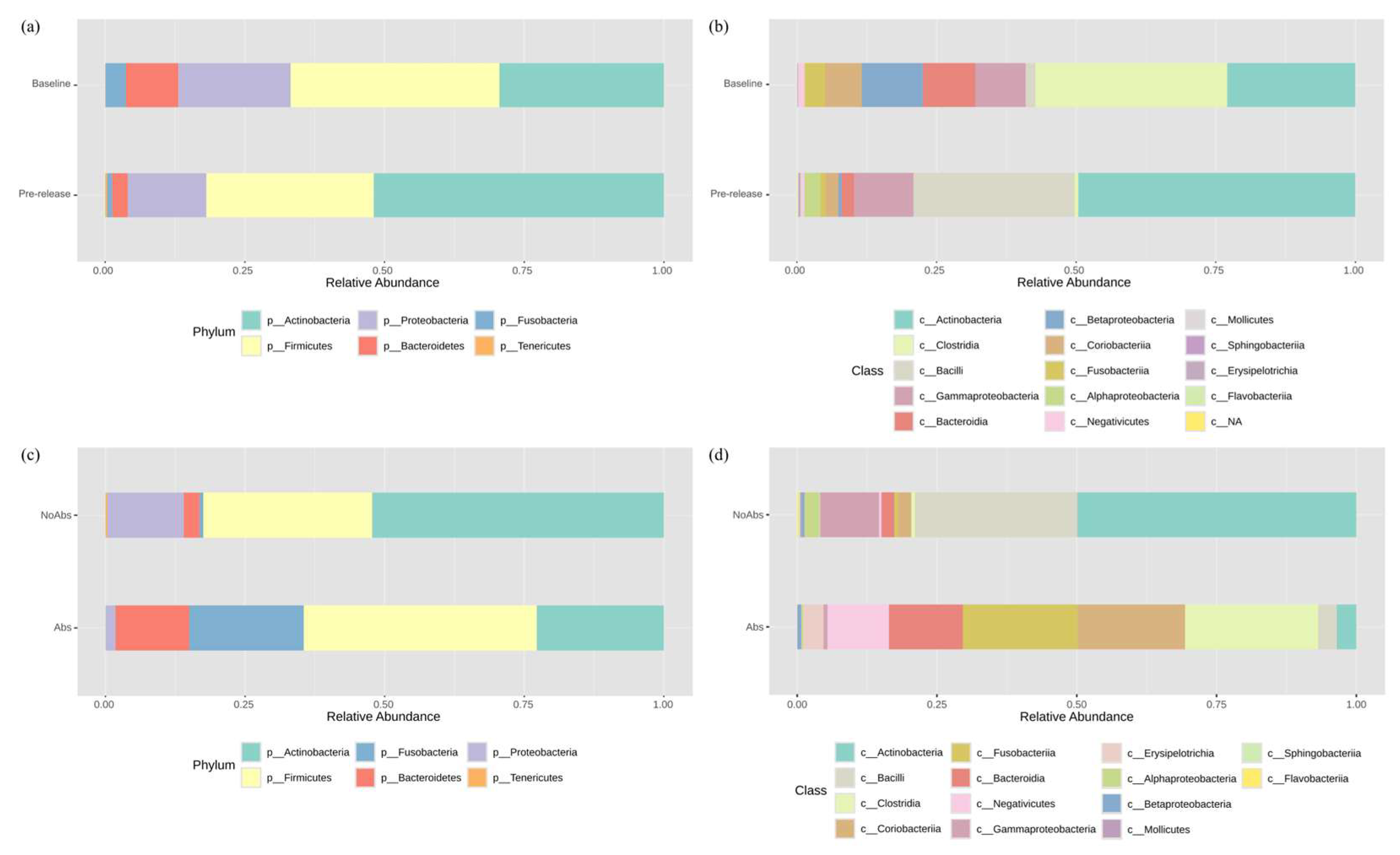
3.2. Bacterial Composition of the Gut Microbiome
3.3. Diversity Analysis
3.4. Antimicrobial Resistance Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmeller, D.S.; Courchamp, F.; Killeen, G. Biodiversity Loss, Emerging Pathogens and Human Health Risks. Biodivers. Conserv. 2020, 29, 3095–3102. [Google Scholar] [CrossRef]
- Bodawatta, K.H.; Hird, S.M.; Grond, K.; Poulsen, M.; Jønsson, K.A. Avian Gut Microbiomes Taking Flight. Trends Microbiol. 2022, 30, 268–280. [Google Scholar] [CrossRef]
- Trevelline, B.K.; Fontaine, S.S.; Hartup, B.K.; Kohl, K.D. Conservation Biology Needs a Microbial Renaissance: A Call for the Consideration of Host-Associated Microbiota in Wildlife Management Practices. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182448. [Google Scholar] [CrossRef] [PubMed]
- Garcês, A.; Pires, I.; Pacheco, F.; Fernandes, L.S.; Soeiro, V.; Lóio, S.; Prada, J.; Cortes, R.; Queiroga, F. Natural and Anthropogenic Causes of Mortality in Wild Birds in a Wildlife Rehabilitation Centre in Northern Portugal: A Ten-Year Study. Bird Study 2019, 66, 484–493. [Google Scholar] [CrossRef]
- Rhim, H.; Gahng, J.; Baek, G.; Kim, M.; Han, J.-I. Morbidity of Rescued Wild Birds by Admission Causes in the Republic of Korea. Animals 2024, 14, 2071. [Google Scholar] [CrossRef]
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2016, 6, 1543. [Google Scholar] [CrossRef]
- Elokil, A.A.; Abouelezz, K.F.M.; Ahmad, H.I.; Pan, Y.; Li, S. Investigation of the Impacts of Antibiotic Exposure on the Diversity of the Gut Microbiota in Chicks. Animals 2020, 10, 896. [Google Scholar] [CrossRef]
- Bell, B.G.; Schellevis, F.; Stobberingh, E.; Goossens, H.; Pringle, M. A Systematic Review and Meta-Analysis of the Effects of Antibiotic Consumption on Antibiotic Resistance. BMC Infect. Dis. 2014, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial Resistance in Humans, Livestock and the Wider Environment. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial Resistance: Its Surveillance, Impact, and Alternative Management Strategies in Dairy Animals. Front. Vet. Sci. 2018, 4, 237. [Google Scholar] [CrossRef]
- Charani, E.; Holmes, A. Antibiotic Stewardship—Twenty Years in the Making. Antibiotics 2019, 8, 7. [Google Scholar] [CrossRef]
- Davey, P.; Brown, E.; Charani, E.; Fenelon, L.; Gould, I.M.; Holmes, A.; Ramsay, C.R.; Wiffen, P.J.; Wilcox, M. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst. Rev. 2013, 4. [Google Scholar] [CrossRef]
- Mitchell, M.; Dahlgran, E.; Roy, A. Characterizing the Antibiotic Resistance Patterns of Microbes Isolated from Wildlife Presented to the Wildlife Hospital of Louisiana (Baton Rouge, La). Wildl. Rehabil. Bull. 2004, 22, 31–36. [Google Scholar] [CrossRef]
- Rhim, H.; Kim, H.-C.; Na, K.-J.; Han, J.-I. Multidrug Resistance of Coagulase-Negative Staphylococci Isolated from Rescued Wild Animals. Korean J. Vet. Serv. 2019, 42, 251–255. [Google Scholar] [CrossRef]
- Carpenter, J.W.; Harms, C.A. Chapter 5: Birds. In Carpenter’s Exotic Animal Formulary, 6th ed; Elsevier: St. Louis, MO, USA, 2023. [Google Scholar]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. Microbiomeanalyst 2.0: Comprehensive Statistical, Functional and Integrative Analysis of Microbiome Data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef] [PubMed]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading Players in the Maintenance of Gut Homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef]
- Simon, K.; Verwoolde, M.B.; Zhang, J.; Smidt, H.; de Vries Reilingh, G.; Kemp, B.; Lammers, A. Long-Term Effects of Early Life Microbiota Disturbance on Adaptive Immunity in Laying Hens. Poult. Sci. 2016, 95, 1543–1554. [Google Scholar] [CrossRef]
- Oliveira, B.C.M.; Murray, M.; Tseng, F.; Widmer, G. The Fecal Microbiota of Wild and Captive Raptors. Anim. Microbiome 2020, 2, 15. [Google Scholar] [CrossRef]
- Song, H.; Yi, S.; Kim, W.-H.; Guk, J.-H.; Ha, M.; Kwak, I.; Han, J.; Yeon, S.-C.; Cho, S. Environmental Perturbations During the Rehabilitation of Wild Migratory Birds Induce Gut Microbiome Alteration and Antibiotic Resistance Acquisition. Microbiol. Spectr. 2022, 10, e01163-22. [Google Scholar] [CrossRef]
- Corl, A.; Charter, M.; Rozman, G.; Toledo, S.; Turjeman, S.; Kamath, P.L.; Getz, W.M.; Nathan, R.; Bowie, R.C.K. Movement Ecology and Sex Are Linked to Barn Owl Microbial Community Composition. Mol. Ecol. 2020, 29, 1358–1371. [Google Scholar] [CrossRef]
- Bartlow, A.W.; Moser, S.K.; Ellis, J.E.; Hathcock, C.D.; Fair, J.M. Comparing Western (Megascops kennicottii) and Whiskered (M. trichopsis) Screech-Owl Microbiomes in Southern Arizona Using a Novel 16s rRNA Sequencing Method. Anim. Microbiome 2022, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Van der Waaij, D.; Berghuis-de Vries, J.M.; Lekkerkerk-Van der Wees, J.E.C. Colonization Resistance of the Digestive Tract in Conventional and Antibiotic-Treated Mice. Epidemiol. Infect. 1971, 69, 405–411. [Google Scholar] [CrossRef]
- Vital, M.; Gao, J.; Rizzo, M.; Harrison, T.; Tiedje, J.M. Diet Is a Major Factor Governing the Fecal Butyrate-Producing Community Structure across Mammalia, Aves and Reptilia. ISME J. 2015, 9, 832–843. [Google Scholar] [CrossRef]
- Roggenbuck, M.; Bærholm Schnell, I.; Blom, N.; Bælum, J.; Bertelsen, M.F.; Sicheritz-Pontén, T.; Sørensen, S.J.; Gilbert, M.T.P.; Graves, G.R.; Hansen, L.H. The Microbiome of New World Vultures. Nat. Commun. 2014, 5, 5498. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Lu, S.; Yang, J.; Jin, D.; Wang, X.; Bai, X.; Wen, Y.; Wang, Y.; Niu, L.; Ye, C.; et al. Metataxonomics Reveal Vultures as a Reservoir for Clostridium perfringens. Emerg. Microbes Infect. 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, L.; McMahon, B.H.; Berendzen, J.; Longmire, J.; Gleasner, C.; Hengartner, N.W.; Vuyisich, M.; Cohn, J.R.; Jenkins, M.; Bartlow, A.W.; et al. California Condor Microbiomes: Bacterial Variety and Functional Properties in Captive-Bred Individuals. PLoS ONE 2019, 14, e0225858. [Google Scholar] [CrossRef]
- Barka Essaid, A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel Gilles, P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2015, 80, 1–43. [Google Scholar] [CrossRef]
- Hernandez, J.; Escallón, C.; Medina, D.; Vernasco, B.J.; Walke, J.B.; Belden, L.K.; Moore, I.T. Cloacal Bacterial Communities of Tree Swallows (Tachycineta bicolor): Similarity within a Population, but Not between Pair-Bonded Social Partners. PLoS ONE 2020, 15, e0228982. [Google Scholar] [CrossRef]
- Ahmed, Z. Characterization of the External Microbiomes of Central Il Passerines and Woodpeckers Using Culture Techniques. Master's Thesis, Bradley University, Peoria, IL, USA, 2024. [Google Scholar]
- Costanzo, A.; Ambrosini, R.; Franzetti, A.; Romano, A.; Cecere, J.G.; Morganti, M.; Rubolini, D.; Gandolfi, I. The Cloacal Microbiome of a Cavity-Nesting Raptor, the Lesser Kestrel (Falco naumanni). PeerJ 2022, 10, e13927. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, H.; Gong, Y.; Ge, J.; Bao, L. The Gut Microbiota in the Common Kestrel (Falco tinnunculus): A Report from the Beijing Raptor Rescue Center. PeerJ 2020, 8, e9970. [Google Scholar] [CrossRef]
- Waite, D.W.; Taylor, M.W. Characterizing the Avian Gut Microbiota: Membership, Driving Influences, and Potential Function. Front. Microbiol. 2014, 5, 223. [Google Scholar] [CrossRef] [PubMed]
- Turjeman, S.; Pekarsky, S.; Corl, A.; Kamath, P.L.; Getz, W.M.; Bowie, R.C.K.; Markin, Y.; Nathan, R. Comparing Invasive and Noninvasive Faecal Sampling in Wildlife Microbiome Studies: A Case Study on Wild Common Cranes. Mol. Ecol. Resour. 2023, 23, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ingala, M.R.; Simmons, N.B.; Wultsch, C.; Krampis, K.; Speer, K.A.; Perkins, S.L. Comparing Microbiome Sampling Methods in a Wild Mammal: Fecal and Intestinal Samples Record Different Signals of Host Ecology, Evolution. Front. Microbiol. 2018, 9, 803. [Google Scholar] [CrossRef] [PubMed]
- Videvall, E.; Strandh, M.; Engelbrecht, A.; Cloete, S.; Cornwallis, C.K. Measuring the Gut Microbiome in Birds: Comparison of Faecal and Cloacal Sampling. Mol. Ecol. Resour. 2018, 18, 424–434. [Google Scholar] [CrossRef]
- Berlow, M.; Kohl, K.D.; Derryberry, E.P. Evaluation of Non-Lethal Gut Microbiome Sampling Methods in a Passerine Bird. Ibis 2019, 162, 911–923. [Google Scholar] [CrossRef]
- Tilocca, B.; Burbach, K.; Heyer, C.M.E.; Hoelzle, L.E.; Mosenthin, R.; Stefanski, V.; Camarinha-Silva, A.; Seifert, J. Dietary Changes in Nutritional Studies Shape the Structural and Functional Composition of the Pigs’ Fecal Microbiome—From Days to Weeks. Microbiome 2017, 5, 144. [Google Scholar] [CrossRef]
- Kim, J.; An, J.-U.; Kim, W.; Lee, S.; Cho, S. Differences in the Gut Microbiota of Dogs (Canis lupus familiaris) Fed a Natural Diet or a Commercial Feed Revealed by the Illumina Miseq Platform. Gut Pathog. 2017, 9, 68. [Google Scholar] [CrossRef]
- Ganz Holly, H.; Doroud, L.; Firl Alana, J.; Hird Sarah, M.; Eisen Jonathan, A.; Boyce Walter, M. Community-Level Differences in the Microbiome of Healthy Wild Mallards and Those Infected by Influenza a Viruses. mSystems 2017, 2, e00188-16. [Google Scholar] [CrossRef]
- Florkowski, M.R.; Hamer, S.A.; Yorzinski, J.L. Brief Exposure to Captivity in a Songbird Is Associated with Reduced Diversity and Altered Composition of the Gut Microbiome. FEMS Microbiol. Ecol. 2023, 99, fiad096. [Google Scholar] [CrossRef]
- van Veelen, H.P.J.; Falcão Salles, J.; Matson, K.D.; van der Velde, M.; Tieleman, B.I. Microbial Environment Shapes Immune Function and Cloacal Microbiota Dynamics in Zebra Finches Taeniopygia guttata. Anim. Microbiome 2020, 2, 21. [Google Scholar] [CrossRef]
- Murray, M.H.; Lankau, E.W.; Kidd, A.D.; Welch, C.N.; Ellison, T.; Adams, H.C.; Lipp, E.K.; Hernandez, S.M. Gut Microbiome Shifts with Urbanization and Potentially Facilitates a Zoonotic Pathogen in a Wading Bird. PLoS ONE 2020, 15, e0220926. [Google Scholar] [CrossRef] [PubMed]
- Berlow, M.; Phillips, J.N.; Derryberry, E.P. Effects of Urbanization and Landscape on Gut Microbiomes in White-Crowned Sparrows. Microb. Ecol. 2021, 81, 253–266. [Google Scholar] [CrossRef]
- Bodawatta, K.H.; Koane, B.; Maiah, G.; Sam, K.; Poulsen, M.; Jønsson, K.A. Species-Specific but Not Phylosymbiotic Gut Microbiomes of New Guinean Passerine Birds Are Shaped by Diet and Flight-Associated Gut Modifications. Proc. R. Soc. B Biol. Sci. 2021, 288. [Google Scholar] [CrossRef]
- Bohnhoff, M.; Miller, C.P. Enhanced Susceptibility to Salmonella Infection in Streptomycin-Treated Mice. J. Infect. Dis. 1962, 111, 117–127. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Medvecky, M.; Cejkova, D.; Polansky, O.; Karasova, D.; Kubasova, T.; Cizek, A.; Rychlik, I. Whole Genome Sequencing and Function Prediction of 133 Gut Anaerobes Isolated from Chicken Caecum in Pure Cultures. BMC Genom. 2018, 19, 561. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Jones, R.B.; Fodor, A.A. Inference-Based Accuracy of Metagenome Prediction Tools Varies across Sample Types and Functional Categories. Microbiome 2020, 8, 46. [Google Scholar] [CrossRef]
- Maurice, C.F.; Knowles, S.C.L.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked Seasonal Variation in the Wild Mouse Gut Microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef]
- Bobbie, C.B.; Mykytczuk, N.C.S.; Schulte-Hostedde, A.I. Temporal Variation of the Microbiome Is Dependent on Body Region in a Wild Mammal (Tamiasciurus hudsonicus). FEMS Microbiol. Ecol. 2017, 93, fix081. [Google Scholar] [CrossRef]
- Zoelzer, F.; Burger, A.L.; Dierkes, P.W. Unraveling Differences in Fecal Microbiota Stability in Mammals: From High Variable Carnivores and Consistently Stable Herbivores. Anim. Microbiome 2021, 3, 77. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16s rRNA Sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Relman, D.A. Incomplete Recovery and Individualized Responses of the Human Distal Gut Microbiota to Repeated Antibiotic Perturbation. Proc. Natl. Acad. Sci. USA 2011, 108, 4554–4561. [Google Scholar] [CrossRef]
- de La Cochetière, M.-F.; Durand, T.; Lepage, P.; Bourreille, A.; Galmiche, J.P.; Dore, J. Resilience of the Dominant Human Fecal Microbiota Upon Short-Course Antibiotic Challenge. J. Clin. Microbiol. 2005, 43, 5588–5592. [Google Scholar] [CrossRef]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-Term Ecological Impacts of Antibiotic Administration on the Human Intestinal Microbiota. ISME J. 2007, 1, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Eigeland, K.A.; Lanyon, J.M.; Trott, D.J.; Ouwerkerk, D.; Blanshard, W.; Milinovich, G.J.; Gulino, L.-M.; Martinez, E.; Merson, S.; Klieve, A.V. Bacterial Community Structure in the Hindgut of Wild and Captive Dugongs (Dugong dugon). Aquat. Mamm. 2012, 38, 402–411. [Google Scholar] [CrossRef]
- Chong, R.; Grueber, C.E.; Fox, S.; Wise, P.; Barrs, V.R.; Hogg, C.J.; Belov, K. Looking Like the Locals-Gut Microbiome Changes Post-Release in an Endangered Species. Anim. Microbiome 2019, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Hu, Y.; Liu, F.; Wang, Y.; Bi, Y.; Lv, N.; Li, J.; Zhu, B.; Gao, G.F. Metagenomic Analysis Reveals the Microbiome and Resistome in Migratory Birds. Microbiome 2020, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Assess Drug-Resistance Phenotypes, Not Just Genotypes. Nat. Microbiol. 2016, 1, 16120. [Google Scholar] [CrossRef][Green Version]
- Miller, E.A.; Amato, R.; Ponder, J.B.; Bueno, I. Survey of Antimicrobial and Probiotic Use Practices in Wildlife Rehabilitation in the United States. PLoS ONE 2024, 19, e0308261. [Google Scholar] [CrossRef]

| Drug | Antibiotic Treatment | Duration of Treatment |
|---|---|---|
| Meloxicam | Yes | 11 (9–13, 9–14) |
| No | 5 (4–9, 0–19) | |
| Hydromorphone | Yes | 2 (0–4, 0–5) |
| No | 0 (0–6, 0–8) |
 |
| Against Antibiotics | AMR Gene | Resistance Mechanism (Enzyme) | Baseline | PR-NoAbs | PR-Abs |
|---|---|---|---|---|---|
| Number of owls | 5 | 8 | 4 | ||
| Aminoglycoside | AAC(3)-IIb | Aminoglycoside acetyltransferase | 1 | 1 | - |
| aadA | Aminoglycoside nucleotidyltransferase | - | 1 | 1 | |
| APH(3′)-IIIa | Aminoglycoside phosphotransferase | 1 | - | - | |
| APH(3″)-Ib * | Aminoglycoside phosphotransferase | 3 | 1 | 1 | |
| APH(6)-Id | Aminoglycoside phosphotransferase | 2 | 1 | 1 | |
| ANT(4′)-Ib | Kanamycin nucleotidyltransferase | 1 | - | 1 | |
| Beta-lactams | blaZ | Class A beta-lactamase | - | 3 | 2 |
| mecA | Penicillin-binding protein 2a | 1 | 1 | - | |
| Fluoroquinolone | gyrA | DNA gyrase, subunit A (mutated) | - | 1 | 1 |
| MLS: macrolide, lincosamide, streptogramin | ermB | Ribosomal methylase | 2 | - | 1 |
| ermC | 23S rRNA methyltransferase | 1 | 1 | - | |
| ermX | Ribosomal RNA methyltransferase | 1 | 2 | - | |
| lnuA * | Lincosamide nucleotidyltransferase | 1 | 3 | 1 | |
| mphC | Macrolide phosphotransferase | 1 | - | - | |
| mphD | Macrolide phosphotransferase | 1 | 1 | - | |
| msrA | ABC-F ribosomal protection protein | - | 1 | - | |
| msrD | ABC-F ribosomal protection protein | 1 | - | - | |
| Phenicol | cat | Chloramphenicol acetyltransferase | - | 1 | - |
| cmx | Chloramphenicol exporter | 1 | 1 | 1 | |
| Sulfonamide | sul1 | Dihydropteroate synthase | 1 | 1 | 1 |
| sul2 | Dihydropteroate synthase | 1 | 2 | 1 | |
| Tetracycline | tetC | Tetracycline efflux pump | 1 | - | - |
| tetK | Tetracycline efflux pump | 1 | - | - | |
| tetL | Tetracycline efflux pump | 1 | - | - | |
| tetWNW | Ribosomal protection protein | 2 | - | 2 | |
| Total | 25 | 22 | 15 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rhim, H.; Aguilar, M.G.; Boykin, K.L.; Zapanta, K.; Krumbeck, J.A.; Mitchell, M.A. A Preliminary Investigation of the Gastrointestinal Bacterial Microbiomes of Barred Owls (Strix varia) Admitted to a Wildlife Hospital. Animals 2025, 15, 1643. https://doi.org/10.3390/ani15111643
Rhim H, Aguilar MG, Boykin KL, Zapanta K, Krumbeck JA, Mitchell MA. A Preliminary Investigation of the Gastrointestinal Bacterial Microbiomes of Barred Owls (Strix varia) Admitted to a Wildlife Hospital. Animals. 2025; 15(11):1643. https://doi.org/10.3390/ani15111643
Chicago/Turabian StyleRhim, Haerin, Maria G. Aguilar, Kimberly L. Boykin, Kaylie Zapanta, Janina A. Krumbeck, and Mark A. Mitchell. 2025. "A Preliminary Investigation of the Gastrointestinal Bacterial Microbiomes of Barred Owls (Strix varia) Admitted to a Wildlife Hospital" Animals 15, no. 11: 1643. https://doi.org/10.3390/ani15111643
APA StyleRhim, H., Aguilar, M. G., Boykin, K. L., Zapanta, K., Krumbeck, J. A., & Mitchell, M. A. (2025). A Preliminary Investigation of the Gastrointestinal Bacterial Microbiomes of Barred Owls (Strix varia) Admitted to a Wildlife Hospital. Animals, 15(11), 1643. https://doi.org/10.3390/ani15111643





