Simple Summary
Confirmation studies for SNPs associated with milk traits, identified through genome-wide or classic association approaches, are very rare in dairy animals and, to our knowledge, have not been carried out in river buffaloes. In this species, the candidate gene approach remains the most commonly applied method for identifying markers for selective breeding. In this study, we validated and confirmed the association of three SNPs in key genes (CSN1S1, CSN3 and LPL) with milk yield, protein and fat. Our data represent a very important indication for the preselection of young bulls destined for breeding programs with a view to achieving more sustainable dairy production.
Abstract
The search for DNA polymorphisms useful for the genetic improvement of dairy farm animals has spanned more than 40 years, yielding relevant findings in cattle for milk traits, where the best combination of alleles for dairy processing has been found in casein genes and in DGAT1. Nowadays, similar results have not yet been reached in river buffaloes, despite the availability of advanced genomic technologies and accurate phenotype records. The aim of the present study was to investigate and validate the effect of four single nucleotide polymorphisms (SNP) in the CSN1S1, CSN3, SCD and LPL genes on seven milk traits in a larger buffalo population. These SNPs have previously been reported to be associated with, or affect, dairy traits in smaller populations often belonging to one farm. A total of 800 buffaloes were genotyped. The following traits were individually recorded, monthly, throughout each whole lactation period from 2010 to 2021: daily milk yield (dMY, kg), protein yield (dPY, kg) and fat yield (dFY, kg), fat and protein contents (dFP, % and dPP, %), somatic cell count (SCC, 103 cell/mL) and urea (mg/dL). A total of 15,742 individual milk test day records (2496 lactations) were available for 680 buffalo cows, with 3.6 ± 1.7 parities (from 1 to 13) and an average of 6.1 ± 1.2 test day records per lactation. Three out four SNPs in the CSN1S1, CSN3 and LPL genes were associated with at least one of analyzed traits. In particular, the CSN1S1 (AJ005430:c.578C>T) gave favorable associations with all yield traits (dMY, p = 0.022; dPY, p = 0.014; dFY, p = 0.029) and somatic cell score (SCS, p = 0.032). The CSN3 (HQ677596: c.536C>T) was positively associated with SCS (p = 0.005) and milk urea (p = 0.04). Favorable effects on daily milk yield (dMY, p = 0.028), fat (dFP, p = 0.027) and protein (dPP, p = 0.050) percentages were observed for the LPL. Conversely, the SCD did not show any association with milk traits. This is the first example of a confirmation study carried out in the Mediterranean river buffalo for genes of economic interest in the dairy field, and it represents a very important indication for the preselection of young bulls destined for breeding programs aimed at more sustainable dairy production.
1. Introduction
The domestic water buffalo (Bubalus bubalis) is a tropical animal known for its remarkable ability to adapt to the environment and its efficient use of feed in conditions of forage shortage. The species originated in Southeast Asia, where 97% of the world’s buffalo population is still reared [1], and then spread westward, arriving in Syria, Egypt and then Western Europe [2]. Therefore, these animals are of major economic and cultural importance for many populations globally, providing milk, meat and draft power. Two buffalo sub-types exist: the swamp type (2n = 48), found exclusively in its native Asian continent, and the river type (2n = 50), which is more widely distributed across other continents. These buffalo sub-types differentiate for karyological, morphological and behavioral characteristics [3,4,5].
Italy is the European country with the greatest number of buffaloes raised. In recent years, the Italian buffalo population has increased from about 12,500 head in the 1950s to over 400,000 in 2019 [1], which represent about 85% of the entire European population. Such remarkable growth has been driven by the exploitation of buffalo milk and the national and international increase in the “Mozzarella di Bufala Campana PDO” demand. Recent data show a significant growth of the whole supply chain, with a turnover estimated at EUR 500 million, involving more than 20,000 operators and observing a 5% annual increase in exports (www.ismea.it, accessed on 6 May 2024). Despite this, domestic buffalo have received less attention and economic investment compared to other ruminants, suggesting significant potential for further improvement in this species.
The achievement of high production levels and efficiency in buffalo farming requires the optimization of numerous factors and processes, including genetic improvement. In this respect, although new knowledge has been acquired [6,7], a high contiguity assembly of the reference genome has been published [8] and the first SNP array specifically designed for buffaloes has become available [9], the use of genomic data remains limited. Therefore, nowadays, the estimation of genomic breeding values and the application of genomic selection in domestic buffalo are significantly delayed, as recently highlighted by Cesarani et al. [10]. In addition, genome-wide association studies (GWAS) in buffalo using the medium density 90K SNP array often identify candidate variants in intergenic regions near many potential genes of interest [11,12,13]. However, these findings frequently lack subsequent confirmation studies. For this reason, the candidate-gene approach is still today a valid method for the identification of genetic associations with milk production traits. At the same time, this approach provides valuable information for breeders’ associations, which, in the last decade, have promoted the selection of buffalo sires with favorable genotypes for milk traits (https://www.risbufala.it/?page_id=58841, accessed on 6 May 2024).
Milk yield [14,15,16,17,18], total protein and caseins [19,20,21,22,23], fat content, fat percentage and fatty acid composition [18,20,24,25,26,27,28,29,30], milking time [14], etc., are among the most studied traits and are of great interest to breeders’ associations due to their direct link to cheese yield and economic profitability. Genetic variability and associations with dairy traits have been found for many genes of economic interest (CSN1S1, CSN1S2, CSN3, SCD, LPL, OXT, OXTR, etc.). However, many of these association studies are limited to a single buffalo farm, with a limited number of samples, or carried out using single gene variants, for instance, the association between the protein percentage and SNP AJ005430: c.578C>T in CSN1S1 (αs-1 casein) [21], or the milk yield and SNP FM876222: g.133A>C in SCD (Stearoyl-CoA Desaturase) [15]. Therefore, the aim of this study was to extend the genotyping of the four most promising SNPs in four genes of interest for selection goals (CSN1S1, CSN3, SCD, LPL) in a larger population and to validate the genetic relationships with milk traits for breeding purposes.
2. Materials and Methods
2.1. Sampling and DNA Isolation
Individual blood collection was performed in compliance with Italian national laws and regulations by official veterinarians of ASL (Local Sanitary Unit of the Ministry of Health) during routine farm prophylaxis procedures.
Sample collection was carried out on a total of 800 Italian Mediterranean river buffaloes belonging to 8 dairy farms mainly located in Campania region (Southern Italy).
Genomic DNA was isolated using the procedure described by Goossens and Kan [31]. Concentrations and OD260/280 ratios were measured with the Nanodrop ND-2000 Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA).
2.2. Genotyping
Genotyping was accomplished using PCR-based methods. In particular, a duplex Artificially Created Restriction Site (ACRS)-PCR described by Pauciullo et al. [22] was performed for the AJ005430: c.578C>T at the CSN1S1 (αs1-CN) and HQ677596: c.536C>T at the CSN3 (k-CN). Additionally, Restriction Fragment Length Polymorphism (RFLP)-PCR, as described by Gu et al. [28] and Gu et al. [27], was used for genotyping the FM876222: g.133A>C at SCD (Stearoyl-CoA desaturase) and the AWWX01438720.1: g14229A>G at LPL (Lipoprotein lipase), respectively. PCR amplification was carried out using BioRad T100 thermocycler (BioRad, Hercules, CA, USA). The digestion products were analyzed directly via electrophoresis on a 2.5% agarose gel in 0.5× TBE buffer and stained with SYBR green nucleic acid stain (Lonza Rockland Inc., Rockland, ME, USA) (Figure S1).
2.3. Phenotypes Collection and Dataset Editing
The phenotypic data for milk yield and composition were obtained from the official recording program of the Italian Association of Breeders (AIA) and they were used for this study under a cooperation agreement. Data included daily milk yield (dMY, kg), protein yield (dPY, kg) and fat yield (dFY, kg), fat and protein content (dFP, % and dPP, %), somatic cell count (SCC, 103 cell/mL) and urea (mg/dL), and they were individually recorded each month throughout the whole lactation from 2010 to 2021 (n = 16,457 records). Only animals with both complete genotypes for the 4 SNPs (MAF > 0.05, n = 762) and lactation records were retained as valid records in successive analysis. A total of 15,742 individual milk test day records (2496 lactations) were available for 680 buffalo cows with 3.6 ± 1.7 parity (from 1 to 13) and 6.1 ± 1.2 test day records per lactation on average (Table 1).

Table 1.
Genotype frequency and database structure.
Further data editing was performed prior to studying the association between SNP genotypes and milk phenotypes. The steps included the following: (i) Removal of outliers. Data that were unsound (greater than ±3.5 standard deviations) or had null values for DIM > 10 days were excluded; (ii) Transformation of SCC to SCS. Somatic cell counts (SCC) were transformed into somatic cell scores according to Ali and Shook [32] to standardize observations into a more analytical useful metric; (iii) Inclusion criteria for lactation records. Only buffalo cows with lactations that included at least 5 records were retained. Following these rigorous data preprocessing steps, the final dataset included 645 buffaloes.
2.4. Statistical Analyses
Descriptive statistics were performed on both SNP and phenotypic data. Minor allele frequencies and Hardy–Weinberg equilibrium tests were computed for all 4 genes. Pairwise Pearson correlations among milk traits were also calculated.
The effects of SNP genotypes on milk traits were assessed through mixed-model analysis, implementing 2 different genetic models using PROC MIXED from SAS® (2016 Cary, NC, USA): allelic and genotypic models. In both genetic models, the fixed effects of the contemporary group were included, along with other systematic sources of variation, as specified thereafter. The type 3 sum of squares of PROC MIXED was computed, and SNP effects were considered significant for p-values < 0.05.
2.5. Allelic Model
In the first genetic model for each of the 4 investigated polymorphisms, the phenotypic values for milk traits were regressed onto the number of B allele (0, 1 or 2) for A/A, A/B and B/B genotypes, respectively, (Table 2) according to an additive model. Moreover, the effect of dominance was assessed with a different parametrization for dominant (1) and recessive genotypes (0). The general model used both for additive and dominance parametrization was:
where y is the test-day phenotypic values for each analysed milk trait, μ is the mean of phenotypic values, SNP is the covariate of allelic count and the average substitution effect for the additive model (AM), or dominance effect for the dominance model (DM). Moreover, the fixed effect of YEAR of birth (11 levels), days in milk (DIM: 15 classes of 20 d each), parity (NL, six classes, from 1 to 6+) and season of birth (SEA, 2 classes: autumn–winter and spring–summer) were fitted. Random effects for combination of herd-test day (htd, 669 levels), buffalo cows (bcow, 645 levels) and residual were also included. Random effects were assumed independently and identically distributed.

Table 2.
Allele Frequency (minor alleles are in boldface).
2.6. Genotypic Model
The genotypic model was similar to model (1), with the main difference that the genotypes at the four loci were treated as cross-classified fixed effects instead of as covariates (3 genotypic classes for A/A, A/B, B/B) according to:
where DIM(SNP) represents the nested effect of days in milk within SNP genotype and SNP(NL) represents the genotypes nested within the parity effect. The other terms are the same as those in the previous model. In this model, a type 3 sum of square F-test for fixed effects was performed, and the marginal means of different genotypes were separated at p-values < 0.05 in post-hoc comparisons, adjusting the p-values according to Tukey HSD (adjust = Tukey of PROC MIXED).
Finally, to estimate the proportion of variance explained by the genotypes, a simplified model was used:
where SNP genotypes, htd and cow are treated as random effects and the proportions of variance explained by the SNP genotype (), herd-test day () and buffalo cows () were computed, respectively, as the ratio of the variance components for each polymorphism to the total variance.
3. Results and Discussion
In the present study, four SNPs (AJ005430:c.578C>T, HQ677596:c.536C>T, FM876222:g.133A>C and AWWX01438720.1:g14229A>G), each located in a gene of interest for selection goals (CSN1S1, CSN3, SCD and LPL, respectively) were genotyped in a population of 800 Mediterranean river buffaloes across eight farms (Figure S1). The selection of these specific SNPs was driven by the need to confirm their impact on milk traits, as identified in previous studies carried out on relatively small buffalo populations [15,21,23,27]. In addition, two of these SNPs (AJ005430:c.578C>T in the CSN1S1 and HQ677596:c.536C>T in the CSN3) were recently included in the genotyping program for buffalo sire selection by one of the two Italian Mediterranean buffalo breeders’ associations (Research Innovation and Selection for the buffalo).
The four investigated SNPs largely segregate in the buffalo population under study (MAF > 0.21, Table 2), with a variability range of 0.16–0.55 across genes or herds. With few exceptions, the four polymorphisms were in HW equilibrium both within and across herds (Figure S2, Table S1). Overall, deviation from the HW equilibrium was partially expected for SCD (χ2 = 6.19), which had previously been investigated in two different populations with similar findings (χ2 = 6.92, [15]; χ2 = 7.96, [28]). SCD FM876222: g.133A>C was associated with milk yield, and the allele substitution effect was assessed in about −1 kg/d, with 12% of the total phenotypic variance explained by polymorphism [15]. This effect is larger than that evidenced for DGAT1 on milk yield in dairy cattle [33]. Despite this, so far, no marker-assisted selection has been voluntarily applied in favour of allele A to increase buffalo milk production. Therefore, the HW deviation for SCD, with the frequency of allele A nearly reaching 80%, can be considered as the result of farmers’ directional selection for more productive animals.
Conversely, the deviation from the HW principle for CSN1S1 (χ2 = 5.06) was unexpected, given the findings of previous studies [21,22]. However, since 2021, the Italian buffalo population has been under selective pressure for the SNP AJ005430:c.578C>T (https://www.risbufala.it/?page_id=58841, accessed on 6 May 2024). Therefore, the observed HW deviation could potentially be considered the result of an artificial selection sweep.
For six milk traits, the number buffaloes with valid records were 645, with an average DIM of 153 ± 93 days. However, 29 animals were excluded from urea analysis due to missing phenotype data. The number of test days and lactation records varied slightly for the milk traits (from 20 to 22 records per animal on average). The average daily milk yield and composition, along with their pairwise phenotypic correlations (Table 3), are consistent with previous reports [10,14,15,19,34,35] and with the official average milk yield (8.70 ± 2.58 kg/d) reported for standard lactations (until 270 DIM) in 2022 [36]. Milk urea, which is important for its role in nitrogen metabolism, shows a weak correlation (<0.10) with all traits. Indeed, milk urea correlated positively with protein yield and negatively with fat content (Table 3).

Table 3.
Descriptive statistics and pairwise Pearson correlations for milk traits after data editing procedure.
This result is among the first indications of a correlation between milk urea and other milk parameters in buffaloes, as few studies are available in this species. Instead, more information is available in dairy cows, where more conflicting data have been reported. In general, a low negative genetic correlation has been found between milk urea and milk yield [37,38], but in New Zealand dairy cattle, the correlation between these two traits was reported as moderately positive [39,40]. Differences between diet formulations are considered as important factors that may cause genetic × environmental interactions that could explain such differences [37]. This could be also the case for the buffalo, whose genetic background, energy requirement and diet differ from those of dairy cattle.
With few exceptions (dFP and SCS in respect of birth season), all the fixed effects were highly significant (Table S2). Additive and dominance effects are reported in Table 4. In the allelic models, LPL showed a significant negative substitution effect on dMY when increasing the number of G alleles (p < 0.05) and a positive effect on fat and protein content of milk (dFP and dPP p < 0.05).

Table 4.
Allele effects: additive (α) and dominance (d) components of genes for the 7 analyzed phenotypic traits. Asterisks indicate significant results for p values < 0.05 (*) and values < 0.01 (**).
Considering that lipoprotein lipase (LPL) facilitates the hydrolysis of triglycerides transported via chylomicrons and very low-density lipoproteins, serving as a pivotal stage in the transportation of free fatty acids to mammary gland and adipose tissues, through its regulation of fatty-acid delivery to the mammary gland, LPL could influence the fat content of milk.
Our results are also consistent with recent findings in the Italian buffalo population. In fact, allele G showed a significant over-expression in homozygosity (~2.5-fold higher) compared with other genotypes and was associated with milk PUFA content [27]. Conversely, allele A showed higher values for milk yield in homozygosity, although the estimated difference from the other two genotypes only approached the level of significance (p = 0.07) [27]. Associations of LPL with milk fat traits and dMY have also been found in other species [41,42,43,44]. So far, no associations between LPL and milk proteins have been reported for buffaloes; however, a significant association was recently found in Czech dairy goats for this trait with the SNP LPL g.185G>T [42].
The investigated polymorphism at CSN1S1 exhibited positive additive effects on dMY, dFY, dPY and SCS at increasing dose of T alleles (Table 4), whereas no significant effects of CSN3 polymorphism was exerted on proteins (dPY and dPP) and other milk traits (dMY, dFY and urea), except for a higher SCS observed with an increasing number of T alleles (Table 4). Overall, this result confirms and reinforces the importance of the αs1-CN encoding gene in determining buffalo milk characteristics, with some important differences compared to the former study by Cosenza et al. [21]. The first difference is the higher number of dairy traits associated with the same SNP in the present study, although the protein percentage showed only a tendency in the genotypic model (p < 0.09), compared to being associated (p < 0.04) by Cosenza et al. [21]. However, the present dataset is more robust (2500 lactations, 8 farms, nearly 650 buffaloes) compared to the former study, which was numerically much smaller (500 lactations, 1 farm, 175 buffaloes). This difference also had other implications. Most notably, the allele substitution effect (cytosine into thymine) changed from the −0.014 observed by Cosenza et al. [21] to 0.011 in the present study. Differences of substitution effects across populations are possible and are influenced by several factors, such as the extent of variances (additive, dominance and additive by additive), the genetic distance of the populations and their heterozygosity [45]. The contribution of AJ005430:c.578C>T to the total phenotypic variance found by Cosenza et al. [21] was quite low (r2αs1 = 0.003) compared to the present study (r2αs1 = 0.100). Considering that Cosenza et al. [21] also found a large dominance effect (–0.028 ± 0.019), then altogether these data may explain, at least partially, the different results between the two studies.
The approached association (p < 0.06) of CSN3 (κ-CN) in the genotypic model represents a further confirmation of the importance of this locus for milk traits. The HQ677596:c.536C>T alleles X1 (p.Ile135) and X2 (Thr135) are known to play a fundamental role in buffalo-milk processing, especially in combination with the variants AJ005430:c.578C>T, alleles B (p.Ser178) and A (Leu178) at CSN1S1 [19,23]. In this context, the combined genotypes AA-X1X2 have shown better curd performance, with shorter rennet coagulation times, faster curd-firming time and greater curd firmness [19]. Conversely, the combination of alleles CSN1S1*B and CSN3*X1 resulted in higher curd yield [23]. Surprisingly, an association was also observed between both casein genes (CSN1S1 and CSN3) and SCS. The allelic and genotypic models converged in identifying the polymorphism at CSN1S1 gene for both additive (p < 0.05) and dominance (p < 0.05) effects on SCS. The average values for C/T and T/T buffalo genotypes did not differ significantly in the log-transformed somatic cell count at p < 0.05 (3.28 and 3.25), whereas the average for C/C genotypes was significantly lower than the former, thus configuring a degree of dominance of T over the C allele. Similarly, for CSN3, whose additive effect was significantly associated with SCS, a degree of dominance has also been observed for milk urea, where the heterozygous had significantly higher average values when compared to the opposite homozygous (Table 5).

Table 5.
Least square means of genotypic class for CSN1S1, CSN3, SCD and LPL genes on milk traits and proportion of variance explained by SNP, buffaloes and herd-test day effects.
Milk somatic cells consist of milk-secreting epithelial cells and immune cells. Regarding CSN3, it is known to have originated from the fibrinogen through a gene duplication event [46], and fibrinogen is one of the main mediators of the acute phase of inflammation [47]. Therefore, it is possible that the κ-casein has retained some of the functions of its ancestral gene and plays an active role as indicator of SCS and mastitis. Further support for this statement derives from the role that the κ-casein glycomacropeptide (GMP) plays in modulating the immune response, and its antibacterial and anti-inflammatory properties [48,49,50]. In addition, SNP rs43703017, located in CSN3, has recently been associated with an increase in SCS in domestic cattle [51]. Regarding CSN1S1, the association with SCS confirmed in buffalo underscores the significant impact of this gene as a promising candidate for selection to improve resistance against mastitis, as already suggested in dairy cows [52,53].
Regarding milk urea, no genes showed a significant substitution effect on this trait. The polymorphism in SCD does not appear to affect any of the investigated milk phenotypes for AM. Positive dominance effects are suggested (p < 0.05) for SCS (CSN1S1) and milk urea (CSN3).
The use of the genotypic model substantially confirmed the results of allelic model, with few differences in the significance levels for LPL (dMY, dPP), αs1-CN (dPY) κ-CN (dPP) that only approached the significant threshold (p < 0.10). However, with a good approximation, these results can still be considered suggestive of an SNP-phenotype association, as also confirmed by the proportion of variance explained by SNP effects for those trait-gene associations (from 0.2% to 0.4%) (Table 5). Indeed, the LPL polymorphism accounted for 0.3% and 0.2% of the total variability in dMY and dPP, respectively. The polymorphism at CSN1S1 explained the 0.4% of total variance for dMY and dPY and SCS. Although the percentage of variance in absolute values was relatively small (0.1% to 0.7% cumulatively across traits), this is not unusual when the genetic association of single genes is analyzed.
It is worth noting that the random effects of buffalo cows and htd explained a large part of the variance. In general, it appears that variance accounted for buffaloes is larger for SCS and urea (25–57%) and smaller for milk yield and composition (8–14%). With an opposite trend, htd largely explains the intra-herd-test-day variability (26–37%) for dMY, dPY and dFY, but less so for milk contents, SCS and urea (7.5–14.5%). In this context, the different environmental and management conditions among the eight farms might not have allowed for better control of some sources of non-genetic variation. Therefore, the high level of variability observed in the present study may be attributed to the relevant impact of environmental factors.
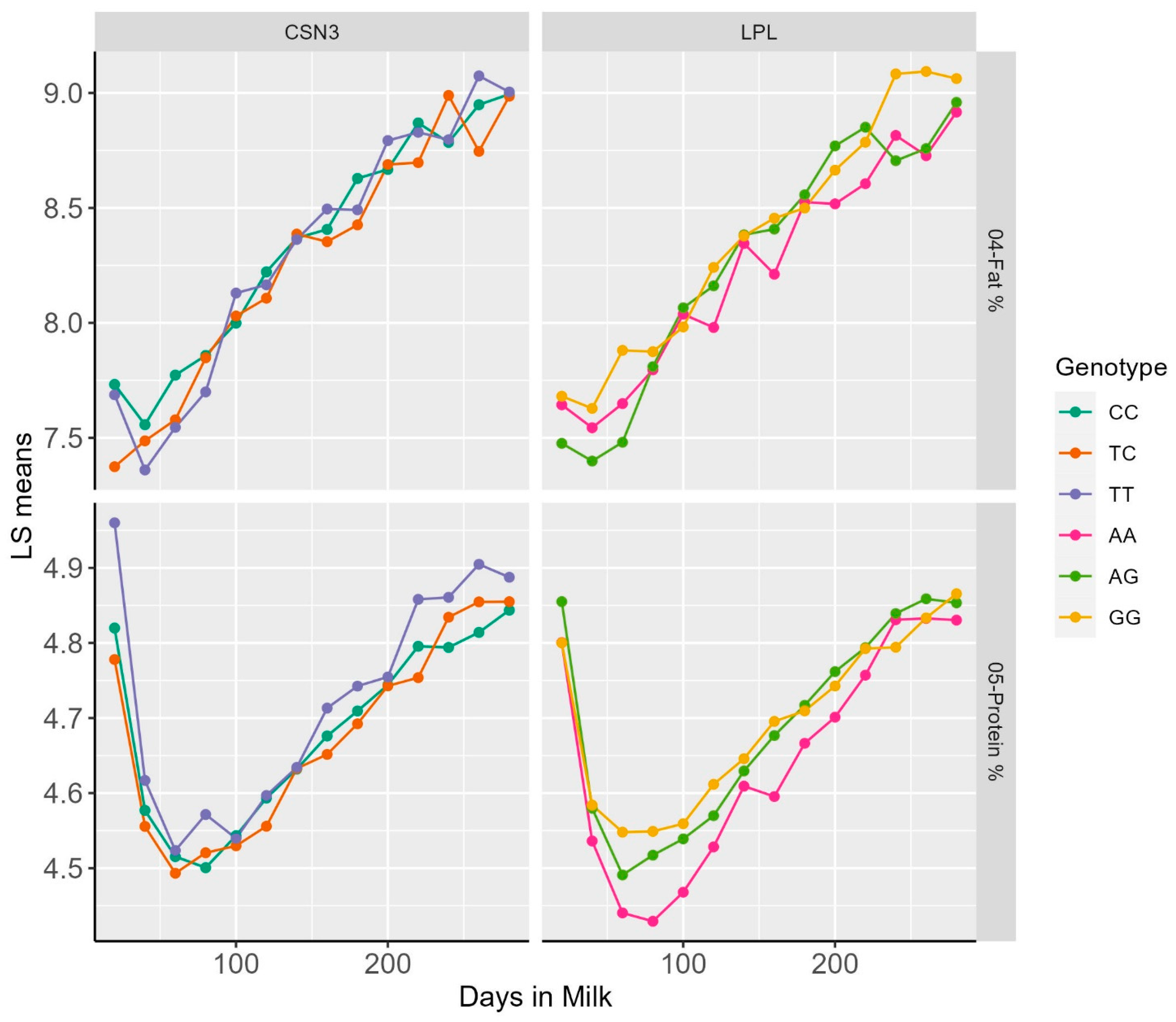
Representative examples of DIM classes and least square means for dPP and dFP for the LPL, as well as dPP and SCS for CSN3, are reported within the lactation patterns of different genotypes (Figure 1).
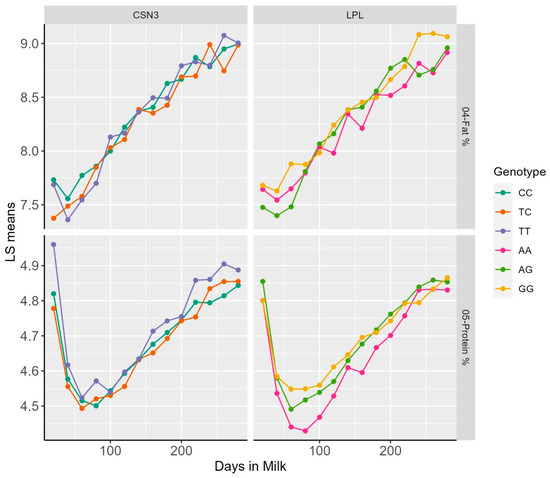
Figure 1.
Plot of LS means of FP and PP over Days in milk within genotypic classes for the polymorphisms in the LPL and CSN3 genes.
4. Conclusions
The genetic improvement of dairy traits is among the main goals of the Italian Mediterranean river buffalo association of breeders. In the present study, we have extended to a larger population the investigation on four polymorphisms that have been previously associated with dairy traits in a limited number of samples, often from a single farm. Three out of four SNPs—CSN1S1, CSN3 and LPL—were associated with at least one of the analyzed traits (dMY, dPY, dFY, dPP, dFP, SCS and Urea) using both an allelic and a genotypic model. In particular, CSN1S1 (AJ005430:c.578C>T) showed favorable associations with all yield traits (dMY, dPY, dFY) and SCS, whereas CSN3 positively associated with SCS and Urea. Favorable effects on dMY, dFP and dPP were observed for LPL. Conversely, SCD did not show any association with milk traits. Overall, our results provide important indications for the preselection of young buffalo bulls for dairy traits, but they also highlight the importance of confirmation studies in larger populations to validate previous associations and enable more efficient setup of gene-assisted breeding programs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani14101414/s1, Figure S1: Genotyping by duplex ACRS-PCR (CSN1S1 and CSN3) and PCR-RFLP (LPL and SCD) for the four investigated SNPs (CSN1S1 = AJ005430:c.578C>T; CSN3 = HQ677596:c.536C>T; LPL = AWWX01438720.1:g14229A>G and SCD = FM876222:g.133A>C); Figure S2: Minor allele frequencies (MAF) detected in the eight investigated herds for the four SNPs (CSN1S1 = AJ005430:c.578C>T; CSN3 = HQ677596:c.536C>T; SCD = FM876222:g.133A>C; LPL = AWWX01438720.1:g14229A>G); Table S1: Within-herd genotypic frequency, observed and expected heterozygosity and p-values for Hardy–Weinberg tests for the four investigated SNPs; Table S2: p-values for fixed effect included in the statistical model for the seven analyzed traits.
Author Contributions
A.P. conceived, designed and performed the experiments. A.P. and G.G. analyzed the data. A.P. and G.G. wrote the manuscript. A.P. and G.C. contributed reagents, materials, and analysis tools. G.C., Y.Z. and Q.L. revised the article critically for important intellectual content. A.P., G.G., Y.Z., Q.L. and G.C. gave final approval of the version to be published. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Italian Ministry of University and Research (D.D. 1332) by the project PON01_00486 (Sequenziamento del genoma bufalino per il miglioramento quali-quantitativo delle produzioni agro-alimentari–GENOBU). The University of Turin, Grant Number PAUA_RIC_N_COMP_21_01 (Miglioramento genetico della bufala Mediterranea Italiana) and Agritech National Research Center of the European Union Next-Generation EU (Piano Nazionale di Ripresa e Resilienza -PNRR- Missione 4 Componente 2, Investimento 1.4-D.D. 1032 17/06/2022, CN00000022) also funded the study.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Bioethics Committee of the University of Torino (Prot. N. 0239749 del 17 May 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors thank the Italian Association of Animal Breeders (AIA) and Alessia Tondi for providing data.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAO. Food and Agriculture Organization of the United Nations. Statistics of Live Animals. 2021. Available online: http://www.fao.org/faostat/en/#data/QA (accessed on 6 May 2024).
- Zhang, Y.; Colli, L.; Barker, J. Asian water buffalo: Domestication, history and genetics. Anim. Genet. 2020, 51, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Iannuzzi, L.; King, W.; Di Berardino, D. Chromosome evolution in domestic bovids as revealed by chromosome banding and FISH-mapping techniques. Cytogenet. Genome Res. 2009, 126, 49–62. [Google Scholar] [CrossRef]
- MacGregor, R. The Domestic Buffalo. Vet. Rec. 1941, 53, 443–450. [Google Scholar]
- Pauciullo, A.; Versace, C.; Perucatti, A.; Gaspa, G.; Li, L.-Y.; Yang, C.-Y.; Zheng, H.-Y.; Liu, Q.; Shang, J.-H. Oocyte aneuploidy rates in river and swamp buffalo types (Bubalus bubalis) determined by Multi-color Fluorescence In Situ Hybridization (M-FISH). Sci. Rep. 2022, 12, 8440. [Google Scholar] [CrossRef]
- Tong, F.; Wang, T.; Gao, N.L.; Liu, Z.; Cui, K.; Duan, Y.; Wu, S.; Luo, Y.; Li, Z.; Yang, C. The microbiome of the buffalo digestive tract. Nat. Commun. 2022, 13, 823. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Shi, W.; Jiang, J.; Li, Z.; Fu, P.; Yang, C.; Rehman, S.u.; Pauciullo, A.; Liu, Q.; Shi, D. Comparative metabolomics analysis of milk components between Italian Mediterranean buffaloes and Chinese Holstein cows based on LC-MS/MS technology. PLoS ONE 2022, 17, e0262878. [Google Scholar] [CrossRef]
- Low, W.Y.; Tearle, R.; Bickhart, D.M.; Rosen, B.D.; Kingan, S.B.; Swale, T.; Thibaud-Nissen, F.; Murphy, T.D.; Young, R.; Lefevre, L. Chromosome-level assembly of the water buffalo genome surpasses human and goat genomes in sequence contiguity. Nat. Commun. 2019, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Iamartino, D.; Nicolazzi, E.L.; Van Tassell, C.P.; Reecy, J.M.; Fritz-Waters, E.R.; Koltes, J.E.; Biffani, S.; Sonstegard, T.S.; Schroeder, S.G.; Ajmone-Marsan, P.; et al. Design and validation of a 90K SNP genotyping assay for the water buffalo (Bubalus bubalis). PLoS ONE 2017, 12, e0185220. [Google Scholar] [CrossRef]
- Cesarani, A.; Biffani, S.; Garcia, A.; Lourenco, D.; Bertolini, G.; Neglia, G.; Misztal, I.; Macciotta, N.P.P. Genomic investigation of milk production in Italian buffalo. Ital. J. Anim. Sci. 2021, 20, 539–547. [Google Scholar] [CrossRef]
- Deng, T.; Liang, A.; Liang, S.; Ma, X.; Lu, X.; Duan, A.; Pang, C.; Hua, G.; Liu, S.; Campanile, G. Integrative analysis of transcriptome and GWAS data to identify the hub genes associated with milk yield trait in buffalo. Front. Genet. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Deng, T.; Zhou, Y.; Ye, T.; Zhou, Z.; Zhang, S.; Shao, B.; Wei, P.; Sun, H.; Khan, F. Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus Bubalis). Anim. Genet. 2019, 50, 207–216. [Google Scholar] [CrossRef]
- Liu, J.; Liang, A.; Campanile, G.; Plastow, G.; Zhang, C.; Wang, Z.; Salzano, A.; Gasparrini, B.; Cassandro, M.; Yang, L. Genome-wide association studies to identify quantitative trait loci affecting milk production traits in water buffalo. J. Dairy Sci. 2018, 101, 433–444. [Google Scholar] [CrossRef]
- Pauciullo, A.; Cosenza, G.; Steri, R.; Coletta, A.; Jemma, L.; Feligini, M.; Di Berardino, D.; Macciotta, N.P.; Ramunno, L. An association analysis between OXT genotype and milk yield and flow in Italian Mediterranean river buffalo. J. Dairy Res. 2012, 79, 150–156. [Google Scholar] [CrossRef]
- Pauciullo, A.; Cosenza, G.; Steri, R.; Coletta, A.; La Battaglia, A.; Di Berardino, D.; Macciotta, N.P.; Ramunno, L. A single nucleotide polymorphism in the promoter region of river buffalo stearoyl CoA desaturase gene (SCD) is associated with milk yield. J. Dairy Res. 2012, 79, 429–435. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pauciullo, A.; Ramunno, L.; Macciotta, N.P.; Gaspa, G.; Coletta, A.; Apicella, E.; Gallo, D.; Cosenza, G. Genetic variability detected at the lactoferrin locus (LTF) in the Italian Mediterranean river buffalo. Anim. Prod. Sci. 2016, 56, 102–107. [Google Scholar] [CrossRef][Green Version]
- Hosseini, S.M.; Tingzhu, Y.; Pasandideh, M.; Liang, A.; Hua, G.; Schreurs, N.M.; Raza, S.H.A.; Salzano, A.; Campanile, G.; Gasparrini, B. Genetic Association of PPARGC1A Gene Single Nucleotide Polymorphism with Milk Production Traits in Italian Mediterranean Buffalo. BioMed Res. Int. 2021, 2021, 3653157. [Google Scholar] [CrossRef]
- Li, J.; Liang, A.; Li, Z.; Du, C.; Hua, G.; Salzano, A.; Campanile, G.; Gasparrini, B.; Yang, L. An association analysis between PRL genotype and milk production traits in Italian Mediterranean river buffalo. J. Dairy Res. 2017, 84, 430–433. [Google Scholar] [CrossRef] [PubMed]
- Bonfatti, V.; Giantin, M.; Gervaso, M.; Coletta, A.; Dacasto, M.; Carnier, P. Effect of CSN1S1-CSN3 (αS1-κ-casein) composite genotype on milk production traits and milk coagulation properties in Mediterranean water buffalo. J. Dairy Sci. 2012, 95, 3435–3443. [Google Scholar] [CrossRef]
- Cosenza, G.; Iannaccone, M.; Auzino, B.; Macciotta, N.; Kovitvadhi, A.; Nicolae, I.; Pauciullo, A. Remarkable genetic diversity detected at river buffalo prolactin receptor (PRLR) gene and association studies with milk fatty acid composition. Anim. Genet. 2018, 49, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, G.; Pauciullo, A.; Macciotta, N.; Apicella, E.; Steri, R.; La Battaglia, A.; Jemma, L.; Coletta, A.; Di Berardino, D.; Ramunno, L. Mediterranean river buffalo CSN1S1 gene: Search for polymorphisms and association studies. Anim. Prod. Sci. 2015, 55, 654–660. [Google Scholar] [CrossRef]
- Pauciullo, A.; Martorello, S.; Carku, K.; Versace, C.; Coletta, A.; Cosenza, G. A novel duplex ACRS-PCR for composite CSN1S1–CSN3 genotype discrimination in domestic buffalo. Ital. J. Anim. Sci. 2021, 20, 1264–1269. [Google Scholar] [CrossRef]
- Zicarelli, L.; Di Palo, R.; Napolano, R.; Tonhati, H.; De Carlo, E.; Gagliardi, R.; Di Luccia, A.; la Gatta, B. Influence of αS1-casein and κ-casein polymorphism on the curd yield of Italian Mediterranean buffalo (Bubalus bubalis L.) milk. Int. Dairy J. 2020, 100, 104559. [Google Scholar] [CrossRef]
- Correddu, F.; Serdino, J.; Manca, M.G.; Cosenza, G.; Pauciullo, A.; Ramunno, L.; Macciotta, N.P. Use of multivariate factor analysis to characterize the fatty acid profile of buffalo milk. J. Food Compos. Anal. 2017, 60, 25–31. [Google Scholar] [CrossRef]
- Cosenza, G.; Gallo, D.; Auzino, B.; Gaspa, G.; Pauciullo, A. Complete CSN1S2 Characterization, Novel Allele Identification and Association With Milk Fatty Acid Composition in River Buffalo. Front. Genet. 2021, 11, 622494. [Google Scholar] [CrossRef]
- Cosenza, G.; Macciotta, N.P.; Nudda, A.; Coletta, A.; Ramunno, L.; Pauciullo, A. A novel polymorphism in the oxytocin receptor encoding gene (OXTR) affects milk fatty acid composition in Italian Mediterranean river buffalo. J. Dairy Res. 2017, 84, 170–180. [Google Scholar] [CrossRef]
- Gu, M.; Cosenza, G.; Gaspa, G.; Iannaccone, M.; Macciotta, N.; Chemello, G.; Di Stasio, L.; Pauciullo, A. Sequencing of lipoprotein lipase gene in the Mediterranean river buffalo identified novel variants affecting gene expression. J. Dairy Sci. 2020, 103, 6374–6382. [Google Scholar] [CrossRef]
- Gu, M.; Cosenza, G.; Iannaccone, M.; Macciotta, N.; Guo, Y.; Di Stasio, L.; Pauciullo, A. The single nucleotide polymorphism g. 133A> C in the stearoyl CoA desaturase gene (SCD) promoter affects gene expression and quali-quantitative properties of river buffalo milk. J. Dairy Sci. 2019, 102, 442–451. [Google Scholar] [CrossRef]
- Gu, M.; Cosenza, G.; Nicolae, I.; Bota, A.; Guo, Y.; Di Stasio, L.; Pauciullo, A. Transcript analysis at DGAT1 reveals different mRNA profiles in river buffaloes with extreme phenotypes for milk fat. J. Dairy Sci. 2017, 100, 8265–8276. [Google Scholar] [CrossRef] [PubMed]
- Pauciullo, A.; Cosenza, G.; D’avino, A.; Colimoro, L.; Nicodemo, D.; Coletta, A.; Feligini, M.; Marchitelli, C.; Di Berardino, D.; Ramunno, L. Sequence analysis and genetic variability of stearoyl CoA desaturase (SCD) gene in the Italian Mediterranean river buffalo. Mol. Cell. Probes 2010, 24, 407–410. [Google Scholar] [CrossRef]
- Goossens, M.; Kan, Y.Y. DNA analysis in the diagnosis of hemoglobin disorders. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1981; Volume 76, pp. 805–817. [Google Scholar]
- Ali, A.; Shook, G. An optimum transformation for somatic cell concentration in milk. J. Dairy Sci. 1980, 63, 487–490. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Costa, A.; Negrini, R.; De Marchi, M.; Campanile, G.; Neglia, G. Phenotypic characterization of milk yield and quality traits in a large population of water buffaloes. Animals 2020, 10, 327. [Google Scholar] [CrossRef]
- Rosati, A.; Van Vleck, L.D. Estimation of genetic parameters for milk, fat, protein and mozzarella cheese production for the Italian river buffalo Bubalus bubalis population. Livest. Prod. Sci. 2002, 74, 185–190. [Google Scholar] [CrossRef]
- AIA. Italian Breeders Association (Associazione Italiana Allevatori, AIA) Bollettino Online. Statistiche ufficiali. 2022. Available online: http://bollettino.aia.it/Contenuti.aspx?CD_GruppoStampe=RS&CD_Specie=C4 (accessed on 6 May 2024).
- Chen, Y.; Atashi, H.; Vanderick, S.; Mota, R.; Soyeurt, H.; Hammami, H.; Gengler, N. Genetic analysis of milk urea concentration and its genetic relationship with selected traits of interest in dairy cows. J. Dairy Sci. 2021, 104, 12741–12755. [Google Scholar] [CrossRef]
- Samoré, A.; Romani, C.; Rossoni, A.; Frigo, E.; Pedron, O.; Bagnato, A. Genetic parameters for casein and urea contentin the Italian Brown Swiss dairy cattle. Ital. J. Anim. Sci. 2007, 6, 201–203. [Google Scholar] [CrossRef]
- Ariyarathne, H.B.; Correa-Luna, M.; Blair, H.T.; Garrick, D.J.; Lopez-Villalobos, N. Genetic parameters for efficiency of crude protein utilisation and its relationship with production traits across lactations in grazing dairy cows. New Zealand J. Agric. Res. 2021, 64, 62–82. [Google Scholar] [CrossRef]
- Lopez-Villalobos, N.; Correa-Luna, M.; Burke, J.; Sneddon, N.; Schutz, M.; Donaghy, D.; Kemp, P. Genetic parameters for milk urea concentration and milk traits in New Zealand grazing dairy cattle. NZJ Anim. Sci. Prod 2018, 78, 56–61. [Google Scholar]
- Badaoui, B.; Serradilla, J.; Tomas, A.; Urrutia, B.; Ares, J.; Carrizosa, J.; Sanchez, A.; Jordana, J.; Amills, M. Identification of two polymorphisms in the goat lipoprotein lipase gene and their association with milk production traits. J. Dairy Sci. 2007, 90, 3012–3017. [Google Scholar] [CrossRef]
- Brzáková, M.; Rychtářová, J.; Čítek, J.; Sztankóová, Z. A candidate gene association study for economically important traits in Czech dairy goat breeds. Animals 2021, 11, 1796. [Google Scholar] [CrossRef]
- Crepaldi, P.; Nicoloso, L.; Coizet, B.; Milanesi, E.; Pagnacco, G.; Fresi, P.; Dimauro, C.; Macciotta, N.P.P. Associations of acetyl-coenzyme A carboxylase α, stearoyl-coenzyme A desaturase, and lipoprotein lipase genes with dairy traits in Alpine goats. J. Dairy Sci. 2013, 96, 1856–1864. [Google Scholar] [CrossRef]
- Marchitelli, C.; Contarini, G.; De Matteis, G.; Crisà, A.; Pariset, L.; Scatà, M.C.; Catillo, G.; Napolitano, F.; Moioli, B. Milk fatty acid variability: Effect of some candidate genes involved in lipid synthesis. J. Dairy Res. 2013, 80, 165–173. [Google Scholar] [CrossRef]
- Legarra, A.; Garcia-Baccino, C.A.; Wientjes, Y.C.; Vitezica, Z.G. The correlation of substitution effects across populations and generations in the presence of nonadditive functional gene action. Genetics 2021, 219, iyab138. [Google Scholar] [CrossRef]
- Jollès, P.; Loucheux-Lefebvre, M.-H.; Henschen, A. Structural relatedness of κ-casein and fibrinogen γ-chain. J. Mol. Evol. 1978, 11, 271–277. [Google Scholar] [CrossRef]
- Luyendyk, J.P.; Schoenecker, J.G.; Flick, M.J. The multifaceted role of fibrinogen in tissue injury and inflammation. Blood J. Am. Soc. Hematol. 2019, 133, 511–520. [Google Scholar] [CrossRef]
- Córdova-Dávalos, L.E.; Jiménez, M.; Salinas, E. Glycomacropeptide bioactivity and health: A review highlighting action mechanisms and signaling pathways. Nutrients 2019, 11, 598. [Google Scholar] [CrossRef]
- Olsen, W.; Liang, N.; Dallas, D.C. Macrophage-Immunomodulatory Actions of Bovine Whey Protein Isolate, Glycomacropeptide, and Their In Vitro and In Vivo Digests. Nutrients 2023, 15, 4942. [Google Scholar] [CrossRef]
- Qu, Y.; Park, S.H.; Dallas, D.C. The Role of Bovine Kappa-Casein Glycomacropeptide in Modulating the Microbiome and Inflammatory Responses of Irritable Bowel Syndrome. Nutrients 2023, 15, 3991. [Google Scholar] [CrossRef]
- Viale, E.; Tiezzi, F.; Maretto, F.; De Marchi, M.; Penasa, M.; Cassandro, M. Association of candidate gene polymorphisms with milk technological traits, yield, composition, and somatic cell score in Italian Holstein-Friesian sires. J. Dairy Sci. 2017, 100, 7271–7281. [Google Scholar] [CrossRef]
- Čítek, J.; Brzáková, M.; Hanusová, L.; Hanuš, O.; Večerek, L.; Samková, E.; Jozová, E.; Hoštičková, I.; Trávníček, J.; Klojda, M. Somatic cell score: Gene polymorphisms and other effects in Holstein and Simmental cows. Anim. Biosci. 2022, 35, 13. [Google Scholar] [CrossRef]
- Prinzenberg, E.-M.; Brandt, H.; Bennewitz, J.; Kalm, E.; Erhardt, G. Allele frequencies for SNPs in the αS1-casein gene (CSN1S1) 5′ flanking region in European cattle and association with economic traits in German Holstein. Livest. Prod. Sci. 2005, 98, 155–160. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).