Simple Summary
Schizothorax kozlovi Nikolsky is a unique cold−water fish in the upper reaches of the Yangtze River in China and has high economic value. In our study, genetic diversity and population structure analyses were performed on seven wild populations in the upper reaches of the Yangtze River by GBS. The above results indicate that the populations of S. kozlovi have different degrees of tolerance and selection pressure in response to temperature and altitude. The Wujiang population was genetically differentiated from the Jinsha River and Yalong River populations. The Wujiang intrapopulation has greater genetic diversity and differentiation than the Jinsha River and Yalong River populations, which demonstrates that the Jinsha and Yalong populations require more attention and resources for their protection. The results of this study will increase our understanding of the diversity of S. kozlovi in the upper reaches of the Yangtze River and provide a basis for the conservation and utilization of wild resources.
Abstract
Schizothorax kozlovi Nikolsky is a unique cold−water fish in the upper reaches of the Yangtze River in China and has high economic value. In our study, genetic diversity and population structure analyses were performed on seven wild populations (originating from the Jinsha River, Yalong River, and Wujiang River) in the upper reaches of the Yangtze River by genotyping by sequencing (GBS). The results indicated that a total of 303,970 single−nucleotide polymorphisms (SNPs) were identified from the seven wild populations. Lower genetic diversity was exhibited among the intrapopulations of the three tributaries, and the Wujiang River population had significant genetic differentiation when compared to the Jinsha River and Yalong River populations. Furthermore, the selected SNPs were enriched in cellular processes, environmental adaptation, signal transduction, and related metabolic processes between the Wujiang population and the other two populations. The above results indicate that the populations of S. kozlovi have different degrees of tolerance and selection pressure in response to temperature and altitude. The Wujiang intrapopulation has greater genetic diversity and differentiation than the Jinsha River and Yalong River populations, which demonstrates that the Jinsha and Yalong populations require more attention and resources for their protection. The results of this study will increase our understanding of the diversity of S. kozlovi in the upper reaches of the Yangtze River and provide a basis for the conservation and utilization of wild resources.
1. Introduction
Schizothorax kozlovi (original Schizothoracinae fish) is a cold−water fish that has adapted to the subalpine environment at the edge of the Tibetan Plateau over a long period of time. Additionally, S. kozlovi is a unique fish and is mainly distributed in the upper reaches of the Yangtze River in China, including the Jinsha River, Yalong River, Chishui River, and Wujiang River [1]. Multiple factors have led to the sharp decline in S. kozlovi population resources and the stability and sustainability of population development, such as climate change, overfishing, water pollution, water conservancy, and hydropower project construction [2]. Therefore, analysis of the genetic characteristics of the S. kozlovi wild populations is helpful in monitoring and improving the development level of the aquatic ecological environment in the upper reaches of the Yangtze River. Previous studies of the growth and breeding characteristics [3], diets and resource protection [4], and genetic diversity [5] of S. kozlovi have focused on the upstream Wujiang River. To date, the genetic characteristics of S. kozlovi in the Jinsha and Yalong Rivers have not been reported.
At present, mitochondrial genes are mostly used to study the genetic diversity of Schizothorax species [6,7]. The cytochrome b (cytb) gene has better resolution in the discrimination of Schizothoracinae fish than the cytochrome c oxidase subunit I (COI) gene [7]. However, there are still some confusing clustering relationships, which might have greatly weakened the resolution in the discrimination of Schizothorax species based on the cytb gene [7]. Furthermore, the cytochrome b (1141 bp) gene and the control region (712 bp) were used to analyze the phylogeography of S. waltoni in the Yarlung Tsangpo River (YLTR) on the southern margin of the Qinghai–Tibet Plateau [8]. However, there are some limitations to the use of mitochondrial genes to reveal population evolution, such as the relatively small size of the mitochondrial genome in the whole fish genome and nuclear mitochondrial pseudogene sequence contamination or sequencing errors [7,9].
Genotyping by sequencing (GBS), as one of the simplified genome sequencing approaches, can identify many single−nucleotide polymorphisms (SNPs) to reveal the evolutionary relationship between species and population genetics [10,11,12]. Unlike single−molecule DNA sequencing methods, a highly promising GBS approach can discover tens of thousands of variants and obtain genotypic information at the genomic level [13]. GBS reduces the complexity of genome sequencing through restriction endonucleases that are particularly suitable for species with large genomes, high repetition frequencies, and limited genomic resources [13]. Furthermore, SNPs from GBS are much more abundant, have lower mutation rates, and can be genotyped with lower error rates [14]. In previous studies, GBS has been used to solve a series of evolutionary biological problems, such as fish genetic protection and local adaptation of different populations [15,16].
The present study analyzed the genetic diversity and population structure of seven wild populations of S. kozlovi originating from three tributaries, including the Jinsha River (3 populations), Yalong River (3 populations), and Wujiang River (1 population), in the upper reaches of the Yangtze River by GBS. The results could provide important evidence for germplasm resource protection and the development and utilization of S. kozlovi.
2. Materials and Methods
2.1. Sample Collection
A total of 51 samples of S. kozlovi were randomly selected from seven wild populations at the three tributaries of the upper reaches of the Yangtze River in China (from 2017 to 2019), including the Jinsha River (Luoxu (LX), Zhubalong (ZBL), and Shigu (SG)), Yalong River (Xinlong (XL), Yajiang (YJ), and Muli (ML)), and Wujiang River (Zongjihe (ZJH)) (Figure 1 and Table 1). Otherwise, Schizopygopsis malacanthus and Gymnodiptychus pachycheilus were collected from the Yalong River and sequenced as outgroups by GBS to construct phylogenetic trees. S. kozlovi, S. malacanthus, and G. pachycheilus species were identified based on a reference book [17]. Each sample included composite samples of a pair of fins on one side and back muscles and was stored in 75% alcohol and a −20 °C refrigerator until use. Genomic DNA was extracted by the traditional phenol–chloroform extraction method.
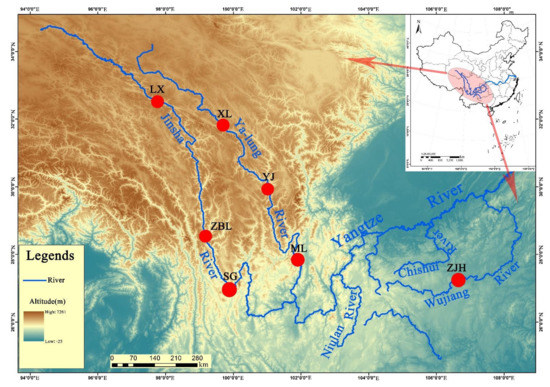
Figure 1.
Map of sampling sites. LX, Luoxu; ZBL, Zhubalong; SG, Shigu; XL, Xinlong; YJ, Yajiang; ML, Muli; ZJH, Zongjihe.

Table 1.
Specimens for GBS.
2.2. Library Preparation
Genomic DNA (gDNA) was extracted from frozen tissue (TIANcombi DNA Lyse & Det PCR Kit, Tiangen Biotech, Beijing, China). The quality of gDNA was evaluated by 1% agarose gel electrophoresis, and the concentration was tested using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Boston, MA, USA). The optimal concentration (50 ng/μL) of all gDNA samples was utilized to construct the sequencing library by superGBS technology [18].
2.3. Sequencing and Genotyping
All gDNA samples were sequenced by the GBS method [18], which was performed by the OE Biotech Co., Ltd. (Shanghai, China). Briefly, gDNA samples were digested by the restriction enzymes pstI−HF and mspI (NEB, Beverly, MA, USA). The digested gDNA fragments were linked by T4 ligase plus to connect barcode adapters at both ends of the fragment. Then, an improved magnetic bead recovery system was used to recover 300–700 bp fragments. The recovered fragments were amplified by PCR using high−fidelity enzymes. Furthermore, Qubit was used to determine the concentration of PCR products, and only concentrations greater than 5 ng/μL were used. Finally, PCR amplification fragments were sequenced on an Illumina Nova, PE150 (Illumina, Inc., San Diego, CA, USA). The raw reads were obtained after removing the potential adaptor sequences by using STACKS [19] software. The clean reads were obtained by FASTP [20] software after filtering the redundant reads. The genome sequence of Oxygymnocypris stewartia [21] was used as the reference genome. Then, the clean reads were mapped to the reference genome to analyze SNP markers by BWA [22].
2.4. Genetic Diversity and Population Structure Analysis
The parameters of genetic diversity were analyzed by VCFTOOLS [23], including the expected heterozygosity (HE), polymorphism information content (PIC), observed heterozygosity (HO), and nucleotide diversity (π). ARLEQUIN 3.5.1.3 [24] was used to calculate F−statistics (Fst). Population structure was determined according to the K value corresponding to the minimum value of the CV error (cross−validation error) using ADMIXTURE v1.3.0 (Alexander, CA, USA) [25], and sometimes the K value was set from 2–7. Population gene exchange and differentiation analyses for the three tributaries (Jinsha River, Yalong River, and Wujiang River) were performed by Treemix V1.12 (Pickrell, Chicago, IL, USA) [26].
2.5. Reconstructing the Phylogenetic Tree
The neighbor−joining (NJ) method [27] was utilized to construct the phylogenetic tree and differentiate populations. S. malacanthus and G. pachycheilus were selected as outgroups to construct the phylogenetic trees. The distance matrix was calculated by TreeBest [28] software, and the reliability of the NJ tree was tested by the bootstrap method (repeated 1000 times) [29]. Then, the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 6 May 2021) was used to visualize the phylogenetic tree. Plink2 [30] software was used for the principal component analysis (PCA) of the SNP markers obtained, and the two feature vectors with the greatest influence were obtained.
2.6. Functional Enrichment of the Selected SNPs
The selected SNPs were screened by Bayescan v2.1 (Foll, Grenoble, France) [31] software. The parameter was set to PR_odds 10 (the prior probability of the neutral model was set to 10), which was consistent with the default software parameter. SNPs with p-values less than 0.05 after FDR (Benjamini-Hochberg) correction in the resulting data were identified as selected sites. For functional enrichment analysis, all selected SNPs were mapped to terms in the GO and KEGG databases. Then, significantly enriched GO terms and KEGG pathways were searched for among the selected SNPs using p < 0.05 as the threshold. GO terms were cataloged into three subgroups, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs).
3. Results
3.1. GBS Data and SNP Discovery
From the Illumina HiSeq sequencing platform, 41.01 Gb raw reads were obtained from 51 samples (The original sequencing data are provided in the Supplementary Materials). Then, after removing the tags with insufficient sequencing depth, a total of 39.0 Gb of clean reads were retained and used for subsequent analysis. The Q20 and Q30 values were ≥95.93% and ≥89.66, respectively. The average GC content was approximately 45%. A 94.04% similarity was observed when the simplified genome sequences of S. kozlovi were compared to the reference genome sequences of O. stewartia.
3.2. Clustering Analysis of Differential SNPs
A total of 303,970 SNPs were obtained by mutation detection and filtering (Dataset S1 and Dataset S2). The results show that the types of DNA base mutations included transition and transversion. Transition (AT→GC or GC→AT) was the main mutation type (Figure 2A). Furthermore, most SNPs were located in intergenic and intron regions and were next derived from the exon, downstream, and upstream regions (Figure 2B).
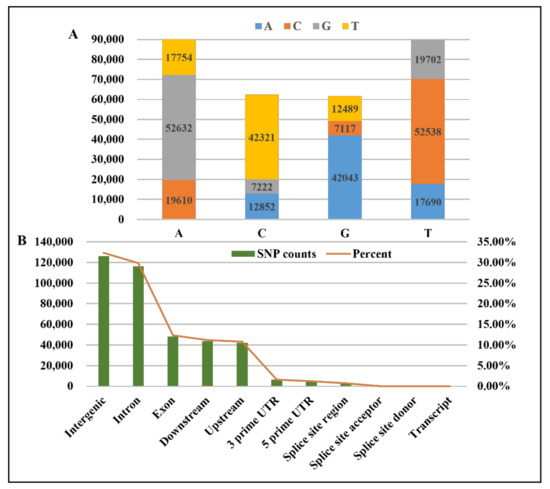
Figure 2.
The DNA base mutation and location in the genome of differential SNPs. (A) DNA base mutation of differential SNPs; (B) location in the genome of differential SNPs.
3.3. Analysis of Population Genetic Diversity
According to SNP data, the values of HE, HO, π, and PIC were 0.04901–0.07821, 0.09175–0.10054, 0.07452–0.09202, and 0.03676–0.06128, respectively (Table 2). The average values of HE, HO, π, and PIC of the Wujiang population were greater than those of the Jinsha and Yalong populations. The results showed that the genetic diversity of the Wujiang population was higher than that of the Jinsha and Yalong populations.

Table 2.
Genetic diversity parameters among S. kozlovi populations.
The Fst value and genetic distance among populations were 0.0038–0.1890 and 0.0038–0.2095, respectively. The Fst value and the genetic distance values were 0.1791–0.1824 (p < 0.05) and 0.1974–0.2073 between the Wujiang (ZJH) and Jinsha populations, respectively. Furthermore, the Fst values and the genetic distance values were 0.1580–0.1890 and 0.1720–0.2095 between the Wujiang (ZJH) and Yalong populations, respectively. Thus, there was a large genetic differentiation between Wujiang (ZJH) and the other two populations, with the highest Fst value (0.15 < Fst < 0.25) and genetic distance (Table 3). The ML population of the Yalong River had moderate genetic differentiation from the three Jinsha populations. The genetic diversity within the Jinsha and Yalong populations was low. However, the genetic distance within Jinsha populations was the smallest.

Table 3.
Genetic distance within and between S. kozlovi populations (above diagonal) and genetic fixation index (Fst) (below diagonal) of populations.
3.4. Phylogenetic Analysis
The NJ phylogenetic tree was constructed based on 303,970 SNPs (Figure 3). According to the outgroups S. malacanthus (SC69) and G. pachycheilus (SC70), the Jinsha and Yalong populations could not be separated. Specifically, Wujiang (ZJH) and the other two populations were clustered into different branches. The results show that Wujiang (ZJH) and the other two populations had significant differentiation.
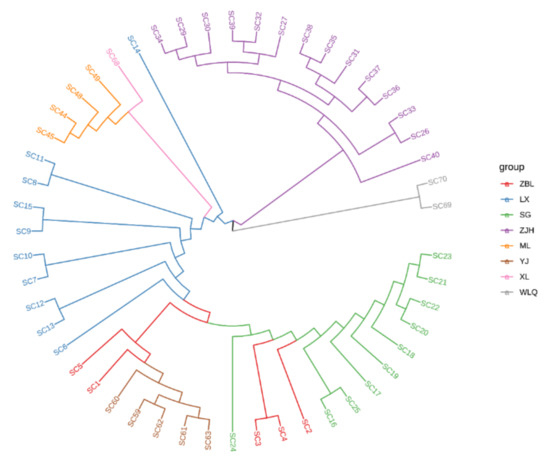
Figure 3.
NJ phylogenetic tree of S. kozlovi populations. ZBL, SC1−SC5; LX, SC6−SC15; SG, SC16−SC25; ZJH, SC26−SC40; ML, SC41−SC51; YJ, SC52−SC67; XL, SC68; WLQ, SC69 and SC70. ZBL, Zhubalong; LX, Luoxu; SG, Shigu; XL, Xinlong; YJ, Yajiang; ML, Muli; ZJH, Zongjihe. The Jinsha River includes LX, ZBL, and SG. The Yalong River contains XL, YJ, and ML. The Wujiang River includes ZJH.
3.5. PCA
According to the 303,970 SNPs, the PCA results showed that the contribution rates of the first principal component (PC1) and the second principal component (PC3) were 47.15% and 21.05%, respectively (Figure 4A). The Jinsha and Yalong populations were clustered into one group, and the Wujiang (ZJH) population was clustered separately. When the K value was 2 based on the CV error, the Wujiang (ZJH) population and two other populations (Jinsha River and Yalong River) could be identified (Figure 4B). Furthermore, the Wujiang (ZJH) population was the first independent population, and gene flow existed between the Jinsha River and Yalong River populations throughout history (Figure 4C). The above results suggest that the Wujiang (ZJH) population of S. kolzovi had genetic differentiation from the Jinsha and Yalong populations.
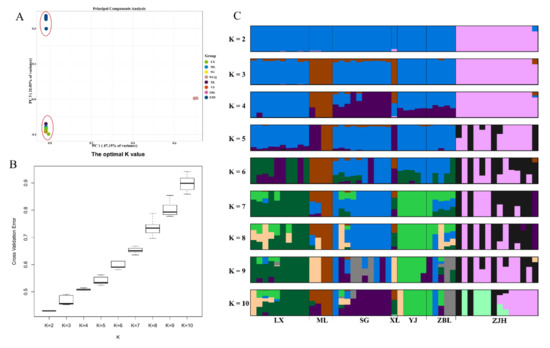
Figure 4.
Principal component analysis and the error rate of the S. kolzovi admixture K value. (A) Principal component analysis for S. kolzovi based on 303,970 SNPs; (B) the error rate of the S. kolzovi admixture K value by cross−validation; (C) the clustering results of S. kolzovi. LX, Luoxu; ZBL, Zhubalong; SG, Shigu; XL, Xinlong; YJ, Yajiang; ML, Muli; ZJH, Zongjihe.
3.6. Functional Annotations of the Selected SNPs
The functional annotations of the selected SNPs were analyzed based on KEGG signaling pathways and GO terms. The differential KEGG pathways (Dataset S3) and GO terms (Dataset S4) of the selected SNPs between the two groups were screened by p-values ≤ 0.05. A total of 30, 18, and 23 significant KEGG pathways were identified in the YL vs. WJ, JS vs. WJ, and JS vs. YL groups, respectively (Figure 5A). Then, eight common pathways were found in the YL vs. WJ, JS vs. WJ, and JS vs. YL groups, including cellular process (apoptosis−fly (4 genes), cell cycle (11 genes), and p53 signaling pathway (6 genes)), environmental adaptation (circadian rhythm (2 genes)), and signal transduction (MAPK signaling pathway (22 genes), Notch signaling pathway (8 genes), Hippo signaling pathway (7 genes), and TGF−beta signaling pathway (7 genes)) (Figure 5B). The Hippo signaling pathway (7 genes) and Notch signaling pathway (4 genes) were found in the JSJ vs. WJ and JS vs. YL groups. Specifically, the Notch signaling pathway was common in the YL vs. WJ, JS vs. WJ, and JS vs. YL groups (Figure 5B).
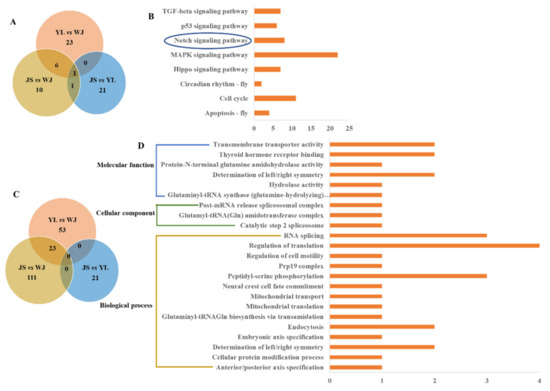
Figure 5.
Functional annotations of the selected SNPs based on KEGG pathways and GO terms. (A) Venn diagram of significant KEGG pathways; (B) six common signaling pathways; (C) Venn diagram of significant GO terms; (D) 23 common GO terms. YL, Yalong River; JS, Jinsha River; WJ, Wujiang River. The Notch signaling pathway was common in the YL vs. WJ, JS vs. WJ, and JS vs. YL groups.
There were 134 significant GO terms in the JS vs. WJ groups, and 23 common terms were identified in the YL vs. WJ and JS vs. WJ groups (Figure 5C). Then, the 23 common terms were classified into molecular functions (6 terms), cellular components (3 terms), and biological processes (14 terms) (Figure 5D). Several important GO terms were screened, such as mitochondrial translation, mitochondrial transport, regulation of translation, RNA splicing, peptidyl−serine phosphorylation, thyroid hormone receptor binding, and transmembrane transporter activity.
4. Discussion
To date, GBS technology has not been widely utilized for the analysis of Schizothoracinae fish. In our study, 41.01 Gb of raw data and 39.0 Gb of clean data were obtained from 51 samples on average. The average value of each sample was 0.76 Gb, which was larger than the 0.63 Gb of Cynoglossus semilaevis [32]. The sequencing data were of high quality, with an average mapping rate of 94.02%, Q20 ≥ 95.93%, and Q30 ≥ 89.66%. The Q20 and Q30 values of GBS data from S. kozlovi were greater than those of GBS data from C. semilaevis (Q20 ≥ 93.74% and Q30 ≥ 87.01%) [32]. Furthermore, the 303.970 SNPs from the present study were far more abundant than those from the mitochondrial control region [33] and the amplified fragment length polymorphism (AFLP) marker [2] of S. kozlovi. These data objectively reflect the advantages of high efficiency and high recovery of GBS technology. Otherwise, SNPs obtained from GBS technology were characterized by high density and uniformity in distribution on the genome [34]. Therefore, compared with traditional analytical methods, the results of this study can more comprehensively and accurately reveal the genetic characteristics of the species of interest and provide a reference for the development of related research in the future.
As an important indicator of evolutionary potential, a higher level of genetic diversity signifies stronger adaptability to environmental change [35,36]. Polymorphic information content (PIC) can reflect the degree of population genetic variation, such as a highly polymorphic locus with PIC > 0.5, a weakly polymorphic locus with PIC < 0.25, and a moderately polymorphic locus with 0.25 < PIC < 0.5 [37]. Then, the levels of average heterozygosity and genetic diversity were positively correlated with the capacity to adapt to the environment [38]. The average values of observed heterozygosity (HO 0.09578) and expected heterozygosity (HE 0.06743) of the three river populations were lower than those of Schizothorax wangchiachii (HO 0.2007 and HE 0.3160) and Schizothorax lissolabiatus (HO 0.2695 and HE 0.2892) [39]. The above data show that the Yalong River, Jinsha River, and Wujiang River intrapopulations showed lower polymorphism and genetic diversity. The reason might be related to the decrease in the wild population of S. kozlovi due to changes in the aquatic environment and human fishing. The populations in the upper reaches of the Jinsha River and the middle reaches of the Yalong River are overfished in terms of their growth and recruitment. The population resources of S. kozlovi in the upper reaches of the Wujiang River are also in a state of overexploitation [2,5], resulting in a decrease in the number of wild S. kozlovi year by year. In addition, the number of parents of S. kozlovi decreased following a decrease in the number of alleles in the offspring population and the occurrence of a low degree of genetic variation [3]. These factors may result in low heterozygosity, serious inbreeding, and decreased genetic diversity of S. kozlovi.
The level of genetic differentiation can be determined by Fst values [40]. According to the classification criteria of genetic differentiation−based Fst values, Wujiang vs. Yalong and Wujiang vs. Jinsha had relatively high genetic differentiation (0.15 < Fst < 0.25), which indicates that genetic differentiation of the Wujiang population had occurred. In addition to the Jinsha River in the upper reaches of the Yangtze River, S. kozlovi is also distributed in the upper reaches of some water systems of the Yunnan–Guizhou Plateau. The geographical distance between the Yunnan–Guizhou Plateau and the population of the upper Yangtze River is large, which indicates that the geographical distance has a great influence on the degree of genetic differentiation of the natural populations of S. kozlovi. Geographic proximity suggests more gene exchange and a lower degree of genetic differentiation. Similar results have also been verified in Schizothorax prenanti [41], Schizothorax biddulphi [42], and Schizothorax molesworthi [43]. Furthermore, the gene exchange of S. kozlovi distributed in the upper reaches of the Yangtze River was hindered to some extent by hydropower development. Thus, the Wujiang population had large genetic differentiation and low gene exchange with the Jiansha and Yalong River populations. The above result suggests that scientific management measures should be taken to increase the number of wild populations and the genetic diversity of S. kozlovi.
Temperature is a key environmental factor affecting species distribution. The temperature selection strategies of Salvelinus fontinalis in different water layers reduced the overlap of the same ecological niche through intraspecific temporal and spatial isolation, which led to the maximization of the use of feed resources in the environment and enhanced the overall ability of the population to adapt to the environment [44]. This difference in intraspecific temperature selection behavior was also found in other fish species [44,45,46]. For example, the small tributaries were rich in bait and had suitable temperatures, with growth faster for Oncorhynchus mykiss [47]. However, O. mykiss could form a competitive monopoly in the heat−sheltered area with lower−than−average water temperature through cluster behavior in the main stream of a river [47]. The above behavior of O. mykiss enhanced the overall environmental adaptability of the population and maximized the population’s growth rate [47]. The temperature and altitude of different habitats reflect the differences in their adaptation to the environment [48]. Previous studies have shown that the genetic isolation of Diptychus maculates is significantly related to temperature, and the large variation in environmental factors leads to weak gene flow in the species [48]. There was information that showed that the average temperature of the low−altitude upper Wujiang River in the past ten years (approximately 15 °C) was significantly higher than that of the high−altitude upper Jinsha River (approximately 2.5 °C) and middle Yalong River (0.64 °C) [49,50,51]. Therefore, these results suggest that temperature may be one of the reasons for the genetic differentiation of S. kozlovi populations between the low−altitude upper Wujiang River and the high−altitude upper Jinsha River and Yalong River.
Loci with high genetic differentiation values were interpreted as having undergone natural selection, which could reflect the selection pressure from certain factors. To date, several molecular pathways have been correlated with the high−altitude adaptation of fish. Several types of functional selection genes are involved in the regulation of temperature changes by processes such as signal transduction, energy metabolism, and membrane [44]. Specifically, transcriptome research on Gymnocypris namensis [52] and Gymnocypris selincuoensis [53] demonstrated similar results. In our study, the common molecular pathways of selected SNPs among the YL vs. WJ, JS vs. WJ, and JS vs. YL groups of S. kozlovi populations were enriched in the cellular process (3 signaling pathways), environmental adaptation (circadian rhythm), signal transduction (4 signaling pathways), and related metabolism GO terms (such as mitochondrial translation and mitochondrial transport). The temperature−related SNPS found in this study are mainly involved in the functions of genes, including mitochondrial translation and transport, energy metabolism, etc. It is possible that temperature is a stimulus to S. kozlovi, causing various physiological reactions, and thus gene transcription has become very active. High mitochondrial gene concentration has been studied in other cold−adapted ectotherms. Windisch demonstrated that the mitochondrial ribosomal protein in Pachycara brachycephalum has a high transcription rate under cold conditions, which has a positive effect on the adaptation of Antarctic fish to cold environments [54]. In addition, increased energy requirements under temperature stress are usually required to maintain normal body function, thus allocating less energy to growth, storage, and reproduction [55]. In aquatic ectotherms such as mussels, extremely high temperatures will lead to a reduction in aerobic range or induce a reduced metabolic rate [56]. Thus, these results signify that adaptive differentiation−related regions have been generated in the genome of S. kozlovi populations and are driven by differences in environmental factors such as temperature, leading to differences in physiological regulation among different populations.
5. Conclusions
In summary, our study analyzed the genetic diversity and population differentiation in three different populations of S. kozlovi, including those from the Jinsha River, Yalong River, and Wujiang River, based on GBS technology. Based on the analysis of selected SNPs, low genetic diversity existed in each population. However, there was higher genetic differentiation between the Wujiang River population and the other two populations (Jinsha River and Yalong River). Furthermore, the most selected SNP markers were enriched in cellular processes, environmental adaptation, signal transduction, and related metabolic processes. Although the Xinlong population had only 1 sample, it did not affect the overall analysis results of the Yalong River population. Our research helps to reveal the diversity level of S. kozlovi in the upper reaches of the Yangtze River and provides the molecular basis for the formulation of biological protection measures.
Supplementary Materials
The original sequencing data can be downloaded: https://dataview.ncbi.nlm.nih.gov/object/PRJNA853098?reviewer=438d0dvi55oepdgk94bpi56253. The following supporting information can be downloaded: https://www.mdpi.com/article/10.3390/ani12172181/s1. A total of 303,970 SNPs were obtained by mutation detection and filtering (Dataset S1 and Dataset S2). The differential KEGG pathways (Dataset S3) and GO terms (Dataset S4) of the selected SNPs between the two groups were screened by p-values ≤ 0.05. A total of 30, 18, and 23 significant KEGG pathways were identified in the Y.L. vs. W.J., J.S. vs. W.J. and J.S. vs. Y.L. groups, respectively.
Author Contributions
Conceptualization, H.C. and L.Y.; methodology, J.H. and Z.H.; software, M.Z.; formal analysis, J.H., Y.P. and Z.H.; writing—original draft preparation, J.H., Z.H., D.Y. and Z.M.; writing—review and editing, S.Y. (Song Yang), S.Y. (Shiyong Yang), F.D. and Q.Z.; funding acquisition, Z.H. and T.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China [grant numbers 31972777, 2019; 31402286, 2015].
Institutional Review Board Statement
The study was conducted in accordance with the guidelines of the Animal Research and Ethics Committees of Sichuan Agricultural University and followed the animal experiments guidelines of Sichuan Agricultural University.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be available upon request.
Acknowledgments
We thank Songpei Zhang for their technical assistance and Jie Luo and Yongxiang Chen for the fish management and sample collection, and Ziting Tang for the revising the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, W.; Zeng, S.; Wang, X.; Zhan, H.X.; Zhou, L.; Xiong, P.; Xiang, Y. Genetic diversity in farmed Schizothorax kozlovi based on mitochondrial DNA D-loop and Cyt b gene sequence. Southwest China J. Agric. Sci. 2022, 35, 475–480. [Google Scholar]
- An, D.D.; Dai, Y.G.; Zou, X.J. AFLP Analysis of Genetic Structure and Diversity in Population of Schizothorax kolzovi from the Wujiang River. Prog. Fishery Sci. 2021, 42, 39–45. [Google Scholar]
- Chen, Y.X.; Luo, Q.S. The fecunidty of Schizothorax kozlovi from Wu River. Zool. Res. 1995, 4, 324–325. [Google Scholar]
- Zhang, X.J.; Dai, Y.G. Feeding habits and resources protection of Schizothorax kozlovi. J. Hydrol. 2011, 32, 110–114. [Google Scholar]
- Li, Z.L.; Hu, S.Y.; Chen, Y.X.; Zhao, H.T. Age Structure and Growth Characteristics of Schizothorax kozlovi in the Upper Wujiang River. J. Hydrol. 2015, 2, 75–80. [Google Scholar]
- Chen, D.; Fu, C.; Lei, Z.; Li, Y.; Zhao, T.; Fan, H.; Hu, T.; Wang, Q.; Zong, H. Complete mitochondrial genome and phylogenetic analysis of Schizothorax sinensis (Teleostei: Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 2021, 6, 1118–1119. [Google Scholar] [CrossRef]
- Ma, Q.; He, K.; Wang, X.; Jiang, J.; Zhang, X.; Song, Z. Better Resolution for Cytochrome b than Cytochrome c Oxidase Subunit I to Identify Schizothorax Species (Teleostei: Cyprinidae) from the Tibetan Plateau and Its Adjacent Area. DNA Cell Biol. 2020, 39, 579–598. [Google Scholar] [CrossRef]
- Guo, X.-Z.; Zhang, G.-R.; Wei, K.-J.; Ji, W.; Yan, R.-J.; Wei, Q.-W.; Gardner, J.P.A. Phylogeography of the threatened tetraploid fish, Schizothorax waltoni, in the Yarlung Tsangpo River on the southern Qinghai-Tibet Plateau: Implications for conservation. Sci. Rep. 2019, 9, 2704. [Google Scholar] [CrossRef]
- Hassanin, A.; Ropiquet, A.; Couloux, A.; Cruaud, C. Evolution of the Mitochondrial Genome in Mammals Living at High Altitude: New Insights from a Study of the Tribe Caprini (Bovidae, Antilopinae). J. Mol. Evol. 2009, 68, 293–310. [Google Scholar] [CrossRef]
- Wang, N.; Yuan, Y.; Wang, H.; Yu, D.; Liu, Y.; Zhang, A.; Gowda, M.; Nair, S.K.; Hao, Z.; Lu, Y.; et al. Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Sci. Rep. 2020, 10, 16308. [Google Scholar] [CrossRef]
- Han, Y.P. Genotyping-by-Sequencing-Based Genome-Wide Association Studies of Fusarium Wilt Resistance in Radishes (Raphanus sativus L.). Genes 2021, 12, 858. [Google Scholar]
- Jadhav, M.P.; Gangurde, S.S.; Hake, A.A.; Yadawad, A.; Mahadevaiah, S.S.; Pattanashetti, S.K.; Gowda, M.V.C.; Shirasawa, K.; Varshney, R.K.; Pandey, M.K.; et al. Genotyping-by-Sequencing Based Genetic Mapping Identified Major and Con-sistent Genomic Regions for Productivity and Quality Traits in Peanut. Front. Plant Sci. 2021, 12, 668020. [Google Scholar] [CrossRef] [PubMed]
- Szarmach, S.J.; Brelsford, A.; Witt, C.C.; Toews, D.P. Comparing divergence landscapes from reduced-representation and whole genome resequencing in the yellow-rumped warbler (Setophaga coronata) species complex. Mol. Ecol. 2021, 30, 5994–6005. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.; Sebastin, R.; Lee, G.A.; Lee, K.J.; Cho, G.T. Genome-Wide SNP Markers for Genotypic and Phenotypic Differentiation of Melon (Cucumis melo L.) Varieties Using Genotyping-by-Sequencing. Int. J. Mol. Sci. 2021, 22, 6722. [Google Scholar]
- Boulanger, E.; Benestan, L.; Guerin, P.; Dalongeville, A.; Mouillot, D.; Manel, S. Climate differently influences the genomic patterns of two sympatric marine fish species. J. Anim. Ecol. 2021, 91, 1180–1195. [Google Scholar] [CrossRef]
- Huo, B.; Liu, X.; Chen, S.; Liu, J.; Zhou, Q.; Shen, J.; Li, D.; Tang, R.; Chen, J.; Zhou, X. Population Structure, Genetic Diversity and Differentiation of Triplophysa tenuis in Xinjiang Tarim River. Front. Genet. 2022, 13, 860678. [Google Scholar] [CrossRef]
- Ding, R.H. The Fishes of Sichuan; Sichuan Science and Technology Press: Chengdu, China, 1994. [Google Scholar]
- Qin, P.; Davis, G.; Dipnarayan, S.; Stephan, S.; Debkanta, C.; Wang, X.; Mathews, D.M.; Malmberg, R.L.; Devos, K.M. UGbS-Flex, a novel bioinformatics pipeline for imputation-free SNP discovery in polyploids without a reference genome: Finger millet as a case study. BMC Plant Biol. 2018, 18, 117. [Google Scholar]
- Julian, C.; Paul, A.H.; Susan, B.; Angel, A.; William, A.C. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Liu, H.-P.; Xiao, S.-J.; Wu, N.; Wang, D.; Liu, Y.-C.; Zhou, C.-W.; Liu, Q.-Y.; Yang, R.-B.; Jiang, W.-K.; Liang, Q.-Q.; et al. The sequence and de novo assembly of Oxygymnocypris stewartii genome. Sci. Data 2019, 6, 190009. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef]
- Li, J. Methodology A fast neighbor joining method. Genet. Mol. Res. 2015, 14, 8733–8743. [Google Scholar] [CrossRef]
- Fabian, S.; Mateus, P.; Matthieu, M.; Miguel, P.; Alex, B. TreeFam v9: A new website, more species and orthology-on-the-fly. Nucleic Acids. Res. 2014, 42, D922–D925. [Google Scholar]
- Noma, H.; Nagashima, K.; Maruo, K.; Gosho, M.; Furukawa, T.A. Bartlett-type corrections and bootstrap adjustments of likelihood-based inference methods for network meta-analysis. Stat. Med. 2017, 37, 1178–1190. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Foll, M.; Gaggiotti, O.E. A Genome-Scan Method to Identify Selected Loci Appropriate for Both Dominant and Codominant Markers: A Bayesian Perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Liu, H.; Zhang, B.; Chen, C.X.; Dai, Y.Y.; He, X.X.; Wang, Q.S. Analysis of genetic diversity of wild and cultivated populations of Cynoglossus semilaevis based on genotyping-by-sequencing technology. Trans. Oceanol. Limnol. 2020, 1, 107–114. [Google Scholar]
- Chen, Y.X. The Genetic Characterization and Population Genetic Diversity of Schizothorax Kozlovi Nikolsky. Ph.D. Thesis, Sichuan Agricultural University, Ya’an, China, 2013. [Google Scholar]
- Gao, L.; Bao, X.B.; Yu, S.Q.; Li, Z.; Li, Y.F.; He, C.B. Analysis of the genetic characteristics of Umbonium thomasi populations along the Yellow and Bohai Seas using GBS. J. Fish. Sci. China 2020, 27, 204–212. [Google Scholar]
- Bernatchez, L. On the maintenance of genetic variation and adaptation to environmental change: Considerations from popu-lation genomics in fishes. J. Fish Biol. 2016, 89, 2519–2556. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Huang, Y.; Jiang, J.; Guisan, A. Genetic diversity in frogs linked to past and future climate changes on the roof of the world. J. Anim. Ecol. 2019, 88, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Saugstad, L.F. From genetics to epigenetics. Nutr. Health 2006, 18, 285–300. [Google Scholar] [CrossRef]
- Jin, F.P.; Li, G.H.; Leng, Y.; Wu, J.X.; Zuo, P.X.; Lei, C.Y. Genetic diversity analysis of four Schizothoras lissolabiatus Sao popu-lations in the middle and Upper Rearches the Lancang River. Acta Hydrobiol. Sin. 2021, 45, 60–68. [Google Scholar]
- Kitada, S.; Nakamichi, R.; Kishino, H. Understanding population structure in an evolutionary context: Population-specific FST and pairwise FST. G3 Genes|Genomes|Genet. 2021, 11, jkab316. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Hu, B.J.; Ye, X.Y.; Liu, G.J.; Zheng, S.M.; Hua, Y.E. Genetic diversity of the prenant’s Schizothoracin (Schizothorax prenanti) based on partial mtDNA CYTB sequences. Acta Hydrobiol. Sin. 2017, 41, 609–616. [Google Scholar]
- Ayelhan, H.S.; Guo, Y.; Wei, M.; Yang, T.; Karjan, A. Genetic diversity and population differentiation of Schizothorax biddulphi based on mtDNA control region sequences. J. Fish. Sci. China 2016, 23, 944–954. [Google Scholar]
- Dan, Y.U.; Zhang, Z.; Zhang, J.; Lin, P.C.; Xiong, S.R.; Tang, F.L.; Liu, H.Z. Genetic diversity and population demography of Schizothorax molesworthi from the MOTUO area of lower Reaches of the Yarlung Zangbo River and Lohit River. Acta Hydrobiol. Sin. 2019, 43, 923–930. [Google Scholar]
- Yi, S.K. The Phylogeny, Population Structure and Adaptive Evolution of Misgurnus Species in China. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2017. [Google Scholar]
- Folio, D.M.; Gil, J.; Caudron, A.; Labonne, J. Genotype-by-environment interactions drive the maintenance of genetic variation in a Salmo trutta L. hybrid zone. Evol. Appl. 2021, 14, 2698–2711. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.R.; Reid, B.N.; Fitzpatrick, S.W. Genome-wide diversity and habitat underlie fine-scale phenotypic differentiation in the rainbow darter (Etheostoma caeruleum). Evol. Appl. 2020, 14, 498–512. [Google Scholar] [CrossRef] [PubMed]
- Petty, J.T.; Thorne, D.; Huntsman, B.; Mazik, P.M. The temperature–productivity squeeze: Constraints on brook trout growth along an Appalachian river continuum. Hydrobiologia 2013, 727, 151–166. [Google Scholar] [CrossRef]
- Yang, T.Y.; Meng, W.; Guo, B.C. Population Genomic Analysis of Two Endemic Schizothoracins Reveals Their Genetic Dif-ferences and Underlying Selection Associated with Altitude and Temperature. Animals 2020, 10, 447. [Google Scholar] [CrossRef]
- Lu, L.U.; Wang, Q.; Wang, G.; Liu, Y.; Liu, C. Trend of climate change over the recent 60 years and its hydrological responses for Jinsha river basin. J. North China Univ. Water Resour. Electr. Power 2016, 37, 16–21. [Google Scholar]
- Huang, Y.Y.; Zhao, H.L.; Jiang, Y.Z.; Xin, L.U. Runoff and its influencing factors in the upper reaches of the Yalong River. Arid Land Geogr. 2018, 41, 127–133. [Google Scholar]
- Zhu, X.S. Preliminary Study on Water Area Changes and Climate Effects in Wujiang River Basin in Guizhou. Ph.D. Thesis, Guizhou Normal University, Guiyang, China, 2020. [Google Scholar]
- Luo, H.; Liu, H.; Zhang, J.; Hu, B.; Zhou, C.; Xiang, M.; Yang, Y.; Zhou, M.; Jing, T.; Li, Z.; et al. Full-length transcript sequencing accelerates the transcriptome research of Gymnocypris namensis, an iconic fish of the Tibetan Plateau. Sci. Rep. 2020, 10, 9668. [Google Scholar] [CrossRef]
- Feng, X.; Jia, Y.; Zhu, R.; Chen, K.; Chen, Y. Characterization and analysis of the transcriptome in Gymnocypris selincuoensis on the Qinghai-Tibetan Plateau using single-molecule long-read sequencing and RNA-seq. DNA Res. 2019, 26, 353–363. [Google Scholar] [CrossRef]
- Windisch, H.S.; Frickenhaus, S.; John, U.; Knust, R.; Prtner, H.O.; Lucassen, M. Stress response or beneficial temperature ac-climation: Transcriptomic signatures in antarctic fish (Pachycara brachycephalum). Mol. Ecol. 2014, 23, 3469–3482. [Google Scholar] [CrossRef]
- Pörtner, H.-O. Oxygen- and capacity-limitation of thermal tolerance: A matrix for integrating climate-related stressor effects in marine ecosystems. J. Exp. Biol. 2010, 213, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Múgica, M.; Sokolova, I.; Izagirre, U.; Marigómez, I. Season-dependent effects of elevated temperature on stress biomarkers, energy metabolism and gamete development in mussels. Mar. Environ. Res. 2015, 103, 1–10. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).