Abstract
Burkholderia cepacia complex (BCC) is a group of closely related bacteria with widespread environmental distribution. BCC bacteria are opportunistic pathogens that cause nosocomial infections in patients, especially cystic fibrosis (CF). Multilocus sequence typing (MLST) is used nowadays to differentiate species within the BCC complex. This study collected 41 BCC isolates from vascular access infections (VAIs) and other clinical infections between 2014 and 2020. We preliminarily identified bacterial isolates using standard biochemical procedures and further conducted recA gene sequencing and MLST for species identification. We determined genetic diversity indices using bioinformatics software. We studied 14 isolates retrieved from patients with VAIs and observed that Burkholderia cepacia was the predominant bacterial species, and B. contaminans followed by B. cenocepacia were mainly retrieved from patients with other infections. According to MLST data, we identified that all B. contaminans isolates belonged to ST102, while a wide variety of sequence types (STs) were found in B. cenocepacia isolates. In summary, the high diversity and easy transmission of BCC increase BCC infections, which provides insights into their potential clinical effects in non-CF infections.
1. Introduction
Burkholderia cepacia (formerly Pseudomonas cepacia) was previously known as one bacteria species, but it has expanded to the Burkholderia cepacia complex (BCC), which comprises more than 20 closely related opportunistic pathogenic species [1,2]. BCC is a group of genetically distinct but phenotypically similar bacteria that occur naturally in remarkably diverse ecological niches and possess extraordinary metabolic versatility [3,4,5]. Despite their agricultural potential, BCC is also recognized among opportunistic nosocomial pathogens, with high potential to cause infections in patients with compromised immunity, cystic fibrosis (CF), chronic granulomatous disease, and indwelling medical devices [6,7]. Among the complex species, B. cenocepacia and B. multivorans are the predominant pathogens that account for about 85–97% of all BCC infections in CF [8], causing a severe decline in lung function, possibly further developing into cepacia syndrome. Nevertheless, there is growing clinical interest in B. contaminans, which was previously identified as taxon K, and this interest has been associated with CF patients, especially in Argentina and Spain [9,10]. B. contaminans was first identified from a contaminated Sargasso Sea DNA sample and was designated as Burkholderia SAR-1 metagenome at that time. The species is epidemiologically linked to the contamination of medical devices and pharmaceutical products, such as nasal sprays, dialysis water, intravenous fentanyl, and moist washcloths [11,12,13,14], causing healthcare-associated infections.
Since conventional phenotypic identification of the closely related BCC species is often difficult and not straightforward, 16S rRNA gene sequence analysis was introduced as a useful tool to discriminate BCC bacteria [15,16]. However, as 16S rRNA gene analysis has limited taxonomic resolution that exclusively discriminates BCC at the complex level, 16S rRNA has been surpassed by recA gene sequence analysis as a means to identify BCC at species level [17]; however, recA gene sequence analysis offers limited resolution. Improved resolution in strain identification is offered by multilocus sequence typing (MLST), the gold standard for typing both species and strains within the species, and this provides new insights into the population structure. The ability of MLST to differentiate existing BCC species is greater compared to analysis of the recA gene alone, and it is a valuable approach in offering a high level of strain identification without using polyphasic taxonomy techniques [18].
Here, we conducted a six-year single-institution study to (a) investigate the prevalence of BCC species from vascular access infections and other BCC infections (non-CF), (b) determine the genetic diversity and multilocus sequence of different bacterial species isolated from different infections and specimens, and (c) establish correlation between BCC species, sequence types (STs), and infection types.
2. Materials and Methods
2.1. Ethical Approval
This study was approved by the Institutional Review Board (IRB) of Chang Gung Memorial Hospital (IRB Nos: 201204188B0, 13/05/2013; 2015080482B0, 25/12/2015; and 201801001B1,27/06/2018). Written consent was obtained from patients, and the study was carried out as per approved guidelines.
2.2. Study Setting, Bacterial Isolate Collection, and Identification
This single-institution study was conducted at Chiayi Chang Gung Memorial Hospital, a territory referral hospital in Taiwan, from January 2014 to March 2020. We prospectively collected information on ten patients with BCC vascular access infections (VAIs) requiring tunneled cuffed catheter (TCC) removal. Patients with poor compliance or who refused to join the study were excluded. Informed consent was obtained from all patients with a personal explanation before performing the procedures. The other 26 patients with BCC infections from other diseases, including lung disease, reproductive disease, eye disease, kidney disease, and soft tissue infections between July 2018 and March 2020 were also analyzed for comparison. Bacterial isolates were cultured under laboratory standards. The samples were routinely cultured on blood agar at 37 °C overnight. Strain identification was performed using standard biochemical (phenotypic) procedures. The isolates collected after 2019 were identified using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF).
2.3. Genomic DNA Extraction
A single colony from a clinical isolate was inoculated in tryptic soy broth (TSB) for 16 h, and 1 mL of the overnight culture was harvested by centrifugation at 16,500× g for 5 min. Bacterial cells were suspended in 1 mL of ultrapure water and heated at 100 °C for 15 min. The supernatant containing the DNA was stored at 4 °C until further use.
2.4. Molecular Characterization
Burkholderia cepacia Complex (BCC) Species Identification
We identified and determined the species of BCC isolates by recA gene amplification and sequencing. We performed PCR amplification using specific primers and conditions described by Fehlberg et al. [19]. Cycle sequencing was performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Thermo Fisher Scientific Company, Waltham, MA, USA) and an ABI 3730xl DNA analyzer (Applied Biosystems, Thermo Fisher Scientific Company, Waltham, MA, USA). The recA sequences were further analyzed and aligned to the NCBI nucleotide Basic Local Alignment Search Tool (BLASTn) database.
2.5. Multilocus Sequence Typing (MLST) Locus Amplification and Sequencing
Among BCC isolates, we conducted MLST by amplifying seven housekeeping genes (atpD: ATP synthase beta chain; gltB: glutamate synthase large subunit; gyrB: DNA gyrase subunit B; recA: recombinase A; lepA: GTP binding protein; phaC: acetoacetyl-CoA reductase; and trpB: tryptophan synthase subunit B) using primer sets and PCR amplification conditions as described previously [20]. We designed an alternative forward primer for the undetected trpB gene of BCC: trpE-F2, 5′-AAGGACGCGCTGAACGAAGC-3′. We also used alternative primer sets for undetected atpD, gyrB, lepA, and recA genes, as a previous study described [21]. Briefly, PCR products were sequenced using the BigDye Terminator v3.1 cycle sequencing kit and analyzed with an ABI 3730xl DNA analyzer. We further aligned the sequences from both strands of a given locus of the same isolate, trimmed to the desired length, and edited using BioNumerics software ver. 7.6 (Applied Maths, Sint-Martens-Latem, Belgium).
2.6. Genetic Diversity Indices
The G+C content, number of polymorphic sites, nucleotide diversity (π) (representing the average number of nucleotide differences per site between random pairwise nucleotides), total number of mutations (Eta), number of haplotypes (h), and haplotype diversity (Hd) (defined as the probability that two randomly chosen haplotypes would be different) were calculated using DnaSP Version 6.12.03 [22]. The average nonsynonymous/synonymous substitution rate ratios (dN/dS) were calculated using Molecular Evolutionary Genetics Analysis software, MEGA X (Nei and Gojobori’s method) for inferring the direction and magnitude of natural selection [23].
2.7. Multilocus Sequence Typing (MLST) and Analysis
We assigned the allele numbers, sequence types (STs), and clonal complexes (CCs) of each isolate using BioNumerics software ver. 7.6. We submitted the novel alleles and STs to the PubMLST database (https://pubmlst.org/) for new allele numbers and ST assignment. We also constructed the minimal spanning tree (MST) with MLST data from the seven housekeeping genes using BioNumerics software ver. 7.6 with the categorical coefficient.
3. Results
In total, we collected 41 Burkholderia cepacia complex (BCC) clinical isolates from 36 patients with both VAIs and infections other than VAIs during the six years of the study, starting in January 2014. In five of the patients with VAIs, two BCC isolates from two different types of specimens were collected (i.e., contaminated Hickman catheter tip and blood). The ratio of patients with VAIs to other infections was 1:2.6, with more females than males with VAI and contrary to other infections (Table 1).

Table 1.
Clinical demographics of patients with Burkholderia cepacia complex (BCC) infections.
3.1. Identification and Prevalence of Burkholderia cepacia Complex (BCC)
The majority of Burkholderia species involved in clinical infection in our institution were B. contaminans, followed by B. cepacia and B. cenocepacia (Figure 1). Moreover, based on recA gene alignment to the NCBI database, we classified three isolates as “other BCC” due to the high similarity with Burkholderia spp. The most common species in VAIs was B. cepacia, which was not isolated from other BCC infections (Table 2). B. cepacia isolates were derived from contaminated Hickman catheter tip (n = 8) and blood (n = 3). In contrast, B. contaminans was the predominant species in other BCC infections, followed by B. cenocepacia. B. contaminans isolates were derived from various specimens, with those from the blood (n = 4) and endocervix discharge (n = 4) being predominant, followed by corneal ulcer (n = 2), percutaneous nephrostomy (n = 2), pus (n = 2), Hickman catheter tip (n = 2), and central venous pressure tip (n = 1). B. cenocepacia isolates were mainly derived from respiratory system specimens (4/9), including sputum (n = 3) and bronchoalveolar lavage (n = 1). In this study, 12 cases of multiple-species infections (i.e., isolation of BCC, especially B. contaminans and B. cenocapacia, and other species of bacteria in one specimen) were predominantly found in other BCC infections (11/27), especially from the endocervix discharge of B. contaminans infection.
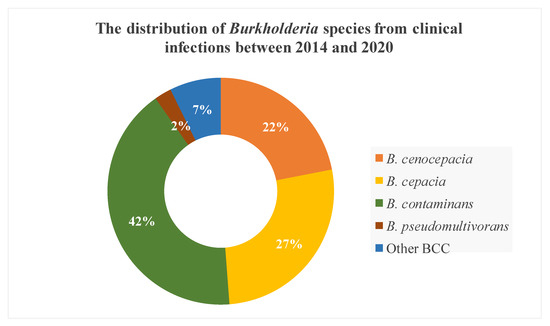
Figure 1.
The species distribution of Burkholderia cepacia complex (BCC) from different clinical infections between 2014 and 2020.

Table 2.
The distribution of Burkholderia species in different clinical infections.
3.2. Genetic Diversity Analysis
Table 3 summarizes data on the genetic diversity of the seven loci, including G+C content, number of alleles, number of polymorphic sites, nucleotide diversity (π), total number of mutations (Eta), number of haplotypes (h), haplotype diversity (Hd), and the ratio of nonsynonymous (dN) to synonymous (dS) substitutions. The number of alleles observed for each DNA fragment ranged from 7 (atpD, recA) to 14 (phaC), and the most frequently detected alleles at each locus were atpD 64 (n = 17), gltB 80 (n = 17), gyrB 76 (n = 17), recA 89 (n = 17), lepA 105 (n = 17), phaC 97 (n = 17), and trpB 70 (n = 17), respectively. The number of different alleles present per locus in B. cenocepacia was highly diverse, including 3 (atpD, recA, lepA), 4 (gltB, phaC), 9 (gyrB), and 5 (trpB). On the contrary, only one allele was present per locus in B. contaminans. The frequency of each allele within each bacterial species is shown in Figure 2.

Table 3.
Nucleotide and allelic diversity of the seven loci in the 41 Burkholderia cepacia complex (BCC) isolates.
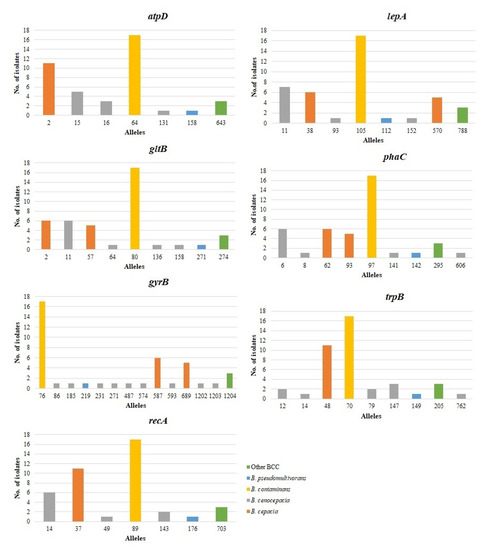
Figure 2.
Frequency of alleles among the seven loci examined. For each locus, the number of times each allele occurred in different Burkholderia cepacia complex (BCC) species is shown.
The average G+C content of the MLST loci ranged from 61.1% for phaC to 69.4% for trpB. The number of polymorphic nucleotide sites (S) varied from 23 (atpD) to 70 (gyrB), in which the gyrB allele possessed the most polymorphic sites at the nucleotide level. The nucleotide diversity indices (π) of the seven loci ranged from 0.01227 (atpD) to 0.04532 (recA). The haplotype diversity (Hd) ranged from 0.748 for atpD to 0.8 for gyrB.
The nonsynonymous to synonymous (dN/dS) ratio measures the level and mode of natural selection acting on protein-coding genes. A dN/dS ratio of <1 indicates negative selection (purifying selection), in which nonsynonymous sites evolved more slowly than synonymous sites during the evolutionary process; a dN/dS >1 indicates positive selection (adaptive selection); and neutral selection is indicated if values are close to 1. The dN/dS ratios for the seven loci varied from 0.01984 (recA) to 9.8871 (gyrB). The dN/dS ratios were extremely high (dN/dS > 1) in four genes (atpD, gltB, gyrB, and trpB), indicating positive selection in the four housekeeping proteins. The dN/dS of recA, phaC, and lepA genes was <1, indicating a purifying selection during the evolutionary process.
3.3. MLST and Population Structure
Of the 41 BCC isolates, we identified 14 STs, including seven new ST types (1725, 1726,1863–1866, 1868) (Table 4). The predominant B. contaminans species in different specimens of VAIs and various clinical infections all belonged to ST102. Intriguingly, all isolates collected from 22 May to 30 July 2019, were ST102 B. contaminans, even those from different types of infections, suggesting that detergent contamination happened during that period. In contrast, the nine B. cenocepacia isolates from different specimens of various clinical infections showed high diversity with nine STs, including six unique STs (1725, 1726, 1854, 1865, 1866, and 1868) and four novel alleles (gyrB 1202–1203; trpB 762; phaC 606). The B. cepacia isolates belonging to ST1723 and ST1724 were only collected from the blood and contaminated catheter tips of patients with VAIs. The three other BCC isolates belonged to the new ST type (ST1863) with four novel alleles (atpD 643; gyrB 1204; recA 703; lepA 788). The MLST minimum spanning tree (MST) of all strains, considering their species and infection cases, is depicted in Figure 3a,b.

Table 4.
The distribution of sequence type (ST) of different species of Burkholderia cepacia complex.
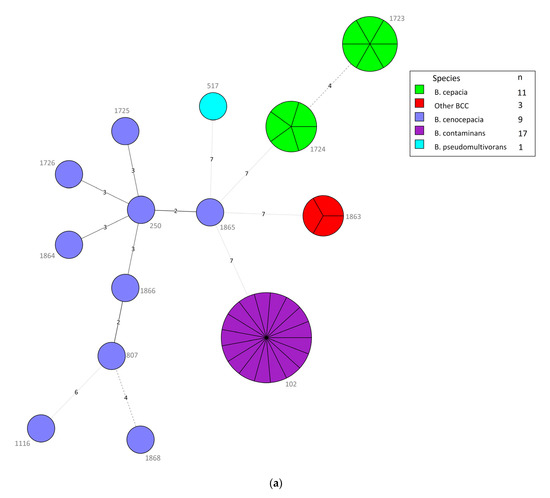
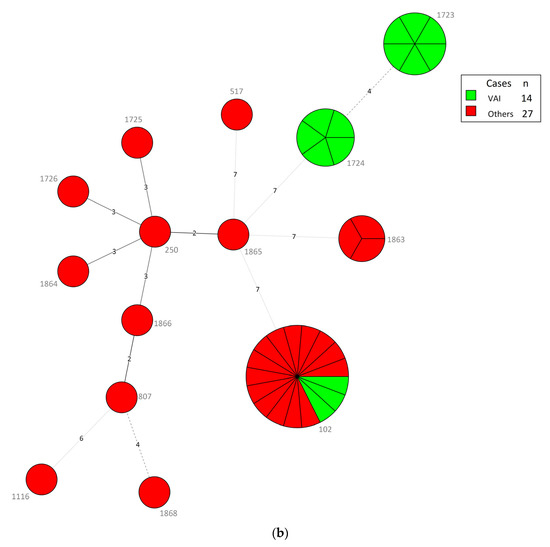
Figure 3.
(a) Minimum spanning tree (MST) constructed with multilocus sequence typing (MLST), using BioNumerics version 7.6, of the 41 Burkholderia cepacia complex (BCC) isolates consisting of different species. Each node within the tree corresponds to a single sequence type (ST). The ST number is labeled next to the node. Each color inside the nodes represents the species of BCC. The size of the node is proportional to the number of isolates found with that profile. The length of branches between each node indicates the number of alleles (out of seven MLST genes) differing between two linked nodes/ST. (b) MST analysis of 41 BCC isolates depending on types of cases (i.e., vascular access infections (VAIs) and other clinical infections). Each color inside the nodes represents the types of cases.
4. Discussion
Bacteria comprising the BCC are ubiquitous in the environment and capable of infecting people with CF. However, its pathogenicity is not limited to CF patients. CF, a common autosomal recessive hereditary disease in the Caucasian population, is uncommon in Asia, with only a few cases published in Taiwan [24,25,26,27]. Our six-year study collected 41 BCC isolates from non-CF patients that were from VAIs among hemodialysis patients and other clinical infections. We noted that VAIs were more common than CF in Taiwan. In previous studies, B. cepacia was the most prevalent species among UK non-CF patients [28], and B. cenocepacia and B. cepacia were the leading bacteremia species in non-CF patients in Taiwan [29]. In this study, the predominant species B. contaminans was collected from different BCC infections, including VAIs and other clinical infections, followed by B. cepacia and B. cenocepacia, which were only collected from VAIs and other clinical infections, respectively. B. contaminans and B. cenocepacia were found in multiple-species infections, especially in endocervix discharge with B. contaminans infections.
MLST is the method of choice for BCC typing due to its ability to differentiate the BCC complex species. In our study, we initially misidentified B. contaminans as B. cepacia and further differentiated them by using MLST. The major sequence type of B. contaminans isolates from VAIs and various clinical infections in our institution belonged to ST102. ST102 may have a global distribution among patients with CF and non-CF; indeed, they are also present in other countries of the European Union, the United States, and Russia, according to data from the PubMLST database (https://pubmlst.org/), which provides a platform to study the epidemiology and biodiversity of BCC bacteria. In contrast to B. contaminans, B. cenocepacia isolates from different infection specimens showed high genetic diversity, in which six of the novel types were revealed in our single institution, suggesting a high capacity of B. cenocepacia to rapidly mutate and adapt to enhance its presence in the patients’ environments or hospital settings.
In a previous study, Kaitwatcharachai et al. [30] reported catheter-related Burkholderia cepacia complex bacteremia in patients undergoing hemodialysis in Thailand, and contaminated chlorhexidine–cetrimide solution was the source of infection. Magalhães et al. [31] reported on a polyclonal bacteremia outbreak involving B. cenocepacia (formerly genomovar III) and B. vietnamiensis infection that occurred in a hemodialysis facility in Brazil. In our study, we found that B. cepacia was the predominant species in VAIs among hemodialysis patients. B. cepacia infection, although previously reported in cystic fibrosis patients, primarily spreads via cross-transmission [32]. Therefore, contamination with B. cepacia—possibly at the skin insertion site, catheter hub, or the contaminated intravenous fluid delivered through the catheter—may have caused the periodic transmission of B. cepacia in our institution.
BCC bacteria possess the ability to survive and proliferate in a water-based environment. BCC bacteria are historically described as major contaminants in both sterile and non-sterile pharmaceuticals, such as intravenous drugs, nasal sprays, and mouthwash, causing numerous nosocomial outbreaks registered during the past years [33]. Therefore, we speculate that the occurrence of the same strain of ST102 B. contaminans in our institution between May 22 and July 30, 2019, may have been caused by contamination in several ways. It is possible that the cause was due to sink drain contamination, in which the pathogen was present in the water or in contaminated products placed in sinks. Alternatively, the use of contaminated chlorhexidine solution for skin disinfection before catheter insertion and during follow-up care may have caused the dissemination of B. contaminans. There is a high probability that contaminated pharmaceuticals, especially detergent, caused the BCC contamination. However, we did not find evidence of BCC contamination in an institutional survey. Nevertheless, easy cross-transmission with BCC, facilitating the rapid spreading of infections between wards, is alarming to us, and further precautions should be taken in the future.
5. Conclusions
This six-year epidemiology surveillance study revealed that B. contaminans, B. cenocepacia, and B. cepacia were the predominant non-CF BCC infections, including VAIs and other clinical infections, at a single institution in Taiwan. The MLST analysis provided important insights into the diversity of the BCC population in our institution. Our study demonstrated the high genetic diversity of B. cenocepacia in different clinical infections, providing its ability to survive in the healthcare setting’s harsh environment and to cause infection in different kinds of diseases. In contrast, single clonal ST102 B. contaminans in different diseases suggested the successful evolution of this strain to survive a wide distribution in clinical environments. Overall, the first insight into BCC’s phylogenetic diversity in healthcare environments will contribute to a greater understanding of their ecological role and evolution and provides insight into the BCC species’ potential clinical impact in non-CF infections.
Author Contributions
Conceptualization, M.Y.W., Y.-H.T., Y.-K.H., and C.C.; Methodology, M.Y.W., Y.-H.T., T.-Y.H., and B.-S.L.; Validation, C.-W.T., M.Y.W., and Y.-K.H.; Formal Analysis, M.Y.W. and Y.-H.T.; Investigation, M.Y.W., Y.-H.T., T.-Y.H., and B.-S.L.; Resources, T.-Y.H. and C.-W.T.; Data Curation, M.Y.W. and Y.-K.H.; Writing—Original Draft Preparation, M.Y.W. and Y.-K.H.; Writing—Review & Editing, Y.-K.H., M.Y.W., Y.-H.T., T.-Y.H., B.-S.L., C.-W.T., and C.C.; Supervision, C.C. and Y.-K.H.; Project Administration, Y.-K.H.; Funding Acquisition, Y.-H.T. and Y.-K.H. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by grants from Chang Gung Memorial Hospital, Chiayi, Taiwan (Grant numbers: CMRPG6J0401, CMRPG6J0402, CMRPG6J0403, CMRPG6H0292, and CMRPG6H0293). The APC was funded by Chang Gung Memorial Hospital, Chiayi, Taiwan.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (IRB) of Chang Gung Memorial Hospital (IRB Nos: 201204188B0, 13/05/2013; IRB2015080482B0, 25/12/2015; and IRB201801001B1, 27/06/2018).
Informed Consent Statement
Written consent was obtained from patients, and the study was carried out as per approved guidelines.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- LiPuma, J.J. Update on the Burkholderia cepacia complex. Curr. Opin. Pulm. Med. 2005, 11, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhou, J.; Zhou, J.; Hu, M.; Zhang, Q.; Kong, N.; Ren, H.; Liang, L.; Yue, J. Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status. Biol. Direct. 2020, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Kunakom, S.; Eustaquio, A.S. Burkholderia as a Source of Natural Products. J. Nat. Prod. 2019, 82, 2018–2037. [Google Scholar] [CrossRef] [PubMed]
- Isles, A.; Maclusky, I.; Corey, M.; Gold, R.; Prober, C.; Fleming, P.; Levison, H. Pseudomonas cepacia infection in cystic fibrosis: An emerging problem. J. Pediatr. 1984, 104, 206–210. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Wong, M.Y.; Huang, T.Y.; Lin, B.S.; Tung, C.W.; Huang, Y.K. Molecular characterization of clinical isolates from vascular access infection: A single-institution study. Microbiologyopen 2020. [Google Scholar] [CrossRef]
- Drevinek, P.; Mahenthiralingam, E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 2010, 16, 821–830. [Google Scholar] [CrossRef]
- Martina, P.; Bettiol, M.; Vescina, C.; Montanaro, P.; Mannino, M.C.; Prieto, C.I.; Vay, C.; Naumann, D.; Schmitt, J.; Yantorno, O.; et al. Genetic diversity of Burkholderia contaminans isolates from cystic fibrosis patients in Argentina. J. Clin. Microbiol. 2013, 51, 339–344. [Google Scholar] [CrossRef]
- Medina-Pascual, M.J.; Valdezate, S.; Carrasco, G.; Villalon, P.; Garrido, N.; Saez-Nieto, J.A. Increase in isolation of Burkholderia contaminans from Spanish patients with cystic fibrosis. Clin. Microbiol. Infect. 2015, 21, 150–156. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Baldwin, A.; Drevinek, P.; Vanlaere, E.; Vandamme, P.; LiPuma, J.J.; Dowson, C.G. Multilocus sequence typing breathes life into a microbial metagenome. PLoS ONE 2006, 1, e17. [Google Scholar] [CrossRef]
- Souza, A.V.; Moreira, C.R.; Pasternak, J.; Hirata, M.L.; Saltini, D.A.; Caetano, V.C.; Ciosak, S.; Azevedo, F.M.; Severino, P.; Vandamme, P.; et al. Characterizing uncommon Burkholderia cepacia complex isolates from an outbreak in a haemodialysis unit. J. Med. Microbiol. 2004, 53, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Christiansen, B.; Caspari, G.; Hogardt, M.; von Thomsen, A.J.; Ott, E.; Mattner, F. Hospital-wide outbreak of Burkholderia contaminans caused by prefabricated moist washcloths. J. Hosp. Infect. 2011, 77, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Moehring, R.W.; Lewis, S.S.; Isaacs, P.J.; Schell, W.A.; Thomann, W.R.; Althaus, M.M.; Hazen, K.C.; Dicks, K.V.; Lipuma, J.J.; Chen, L.F.; et al. Outbreak of bacteremia due to Burkholderia contaminans linked to intravenous fentanyl from an institutional compounding pharmacy. JAMA Intern. Med. 2014, 174, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Segonds, C.; Heulin, T.; Marty, N.; Chabanon, G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 1999, 37, 2201–2208. [Google Scholar] [CrossRef]
- LiPuma, J.J.; Dulaney, B.J.; McMenamin, J.D.; Whitby, P.W.; Stull, T.L.; Coenye, T.; Vandamme, P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 1999, 37, 3167–3170. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Bischof, J.; Byrne, S.K.; Radomski, C.; Davies, J.E.; Av-Gay, Y.; Vandamme, P. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 2000, 38, 3165–3173. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef]
- Fehlberg, L.C.; Andrade, L.H.; Assis, D.M.; Pereira, R.H.; Gales, A.C.; Marques, E.A. Performance of MALDI-ToF MS for species identification of Burkholderia cepacia complex clinical isolates. Diagn. Microbiol. Infect. Dis. 2013, 77, 126–128. [Google Scholar] [CrossRef]
- Spilker, T.; Baldwin, A.; Bumford, A.; Dowson, C.G.; Mahenthiralingam, E.; LiPuma, J.J. Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 2009, 47, 2607–2610. [Google Scholar] [CrossRef]
- Baldwin, A.; Mahenthiralingam, E.; Thickett, K.M.; Honeybourne, D.; Maiden, M.C.; Govan, J.R.; Speert, D.P.; Lipuma, J.J.; Vandamme, P.; Dowson, C.G. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 2005, 43, 4665–4673. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.J.; Lin, S.P.; Lee, H.C.; Chen, C.P.; Chiu, N.C.; Hung, H.Y.; Chern, S.R.; Chuang, C.K. Cystic fibrosis with homozygous R553X mutation in a Taiwanese child. J. Hum. Genet. 2005, 50, 674–678. [Google Scholar] [CrossRef]
- Lin, C.-J.; Chang, S.-P.; Ke, Y.-Y.; Chiu, H.-Y.; Tsao, L.-Y.; Chen, M. Phenotype and genotype of two Taiwanese cystic fibrosis siblings and a survey of delta F508 in East Asians. Pediatr. Neonatol. 2008, 49, 240–244. [Google Scholar] [CrossRef]
- Liu, L.C.; Shyur, S.D.; Chu, S.H.; Huang, L.H.; Kao, Y.H.; Lei, W.T.; Cheng, C.H.; Lo, C.Y.; Chen, C.K.; Fang, L.C. Cystic fibrosis: Experience in one institution. J. Microbiol. Immunol. Infect. 2014, 47, 358–361. [Google Scholar] [CrossRef][Green Version]
- Wang, C.-C.; Lin, H.-C.; Huang, S.-W.; Lu, Y.-C. Early diagnosis of a case of infantile cystic fibrosis and review of literature in Taiwan. J. Med. Sci. 2019, 39. [Google Scholar] [CrossRef]
- Kenna, D.T.D.; Lilley, D.; Coward, A.; Martin, K.; Perry, C.; Pike, R.; Hill, R.; Turton, J.F. Prevalence of Burkholderia species, including members of Burkholderia cepacia complex, among UK cystic and non-cystic fibrosis patients. J. Med. Microbiol. 2017, 66, 490–501. [Google Scholar] [CrossRef]
- Chien, Y.C.; Liao, C.H.; Sheng, W.H.; Chien, J.Y.; Huang, Y.T.; Yu, C.J.; Hsueh, P.R. Clinical characteristics of bacteraemia caused by Burkholderia cepacia complex species and antimicrobial susceptibility of the isolates in a medical centre in Taiwan. Int. J. Antimicrob. Agents 2018, 51, 357–364. [Google Scholar] [CrossRef]
- Kaitwatcharachai, C.; Silpapojakul, K.; Jitsurong, S.; Kalnauwakul, S. An outbreak of Burkholderia cepacia bacteremia in hemodialysis patients: An epidemiologic and molecular study. Am. J. Kidney Dis. 2000, 36, 199–204. [Google Scholar] [CrossRef]
- Magalhães, M.; Doherty, C.; Govan, J.R.W.; Vandamme, P. Polyclonal outbreak of Burkholderia cepacia complex bacteraemia in haemodialysis patients. J. Hosp. Infect. 2003, 54, 120–123. [Google Scholar] [CrossRef]
- Ledson, M.; Gallagher, M.; Corkill, J.; Hart, C.; Walshaw, M. Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia. Thorax 1998, 53, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Tavares, M.; Kozak, M.; Balola, A.; Sá-Correia, I. Burkholderia cepacia Complex Bacteria: A Feared Contamination Risk in Water-Based Pharmaceutical Products. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).