Transcriptional Terminators Allow Leak-Free Chromosomal Integration of Genetic Constructs in Cyanobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Plasmid Construction
2.3. Strain Construction
2.4. Assays
3. Results
3.1. Screening Rho-independent Terminators
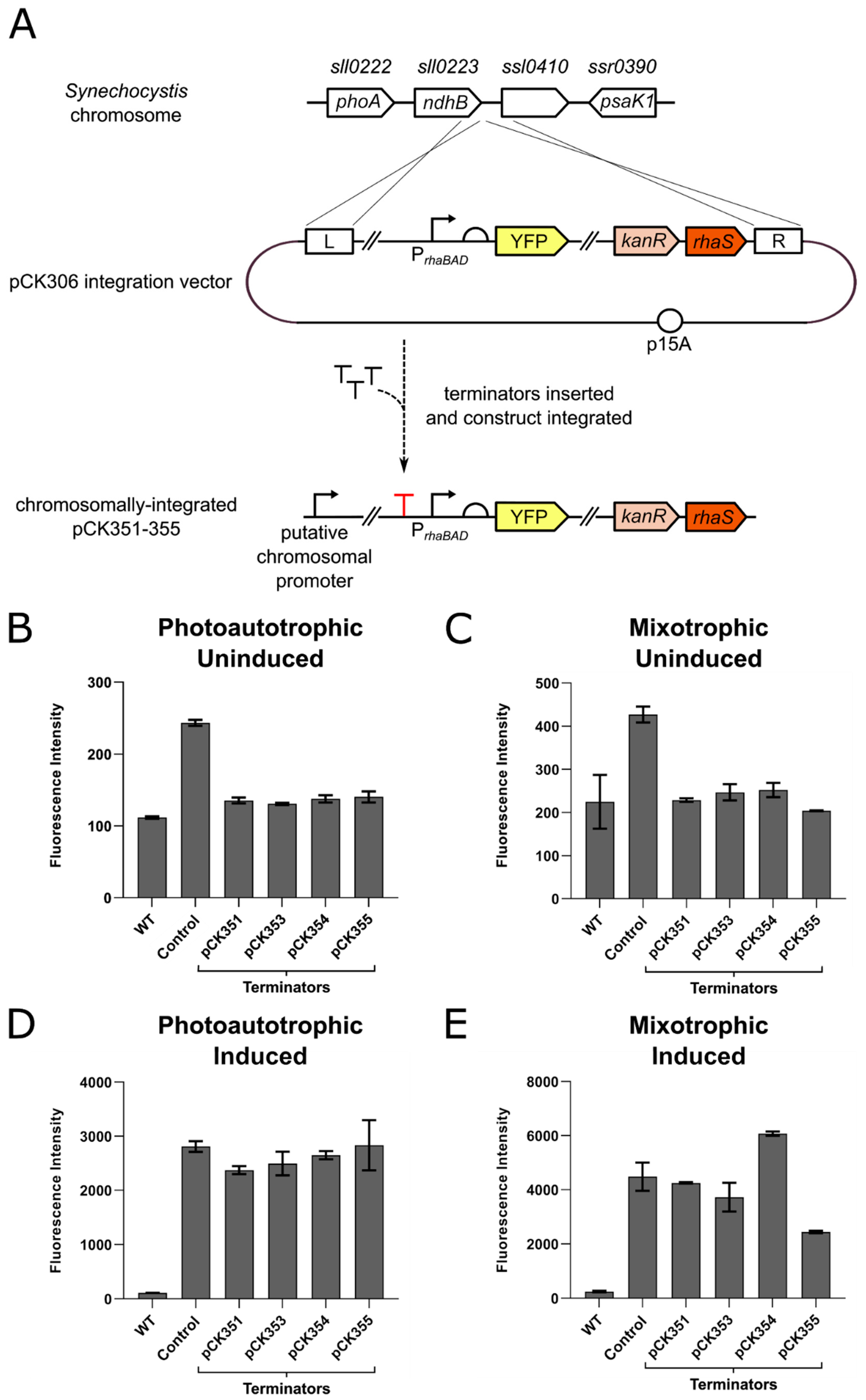
3.2. Strong Terminators Insulate Expression Constructs Integrated in the Synechocystis Chromosome from Transcriptional Read-Through
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Berla, B.M.; Saha, R.; Immethun, C.M.; Maranas, C.D.; Moon, T.S.; Pakrasi, H.B. Synthetic biology of cyanobacteria: Unique challenges and opportunities. Front. Microbiol. 2013, 4, 246. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-H.; Camsund, D.; Lindblad, P.; Heidorn, T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010, 38, 2577–2593. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.L.; Taylor, G.M.; Hitchcock, A.; Torres-Méndez, A.; Heap, J.T. A Rhamnose-Inducible System for Precise and Temporal Control of Gene Expression in Cyanobacteria. ACS Synth. Biol. 2018, 7, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Ramey, C.J.; Barón-Sola, Á.; Aucoin, H.R.; Boyle, N.R. Genome Engineering in Cyanobacteria: Where We Are and Where We Need To Go. ACS Synth. Biol. 2015, 4, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, V.; Jain, I.H.; O’Shea, E.K. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 2011, 12, R47. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988, 167, 3–27. [Google Scholar] [PubMed]
- Kelly, C.L.; Liu, Z.; Yoshihara, A.; Jenkinson, S.F.; Wormald, M.R.; Otero, J.; Estévez, A.; Kato, A.; Marqvorsen, M.H.S.; Fleet, G.W.J.; et al. Synthetic Chemical Inducers and Genetic Decoupling Enable Orthogonal Control of the rhaBAD Promoter. ACS Synth. Biol. 2016, 5, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Liu, P.; Nielsen, A.A.K.; Brophy, J.A.N.; Clancy, K.; Peterson, T.; Voigt, C.A. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 2013, 10, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Geerts, D.; Bovy, A.; de Vrieze, G.; Borrias, M.; Weisbeek, P. Inducible expression of heterologous genes targeted to a chromosomal platform in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology 1995, 141, 831–841. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miyake, K.; Abe, K.; Ferri, S.; Nakajima, M.; Nakamura, M.; Yoshida, W.; Kojima, K.; Ikebukuro, K.; Sode, K. A green-light inducible lytic system for cyanobacterial cells. Biotechnol. Biofuels 2014, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.H.; Frigaard, N.-U. Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 2014, 21, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Jeng, S.T.; Gardner, J.F.; Gumport, R.I. Transcription termination by bacteriophage T7 RNA polymerase at rho-independent terminators. J. Biol. Chem. 1990, 265, 3823–3830. [Google Scholar] [PubMed]
- Cambray, G.; Guimaraes, J.C.; Mutalik, V.K.; Lam, C.; Mai, Q.-A.; Thimmaiah, T.; Carothers, J.M.; Arkin, A.P.; Endy, D. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013, 41, 5139–5148. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Pakrasi, H.B. Exploring native genetic elements as plug-in tools for synthetic biology in the cyanobacterium Synechocystis sp. PCC 6803. Microb. Cell Fact. 2018, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, J.; Georg, J.; Scholz, I.; Sharma, C.M.; Dienst, D.; Bantscheff, J.; Voss, B.; Steglich, C.; Wilde, A.; Vogel, J.; et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 2011, 108, 2124–2129. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.M.; Mordaka, P.M.; Heap, J.T. Start-Stop Assembly: A functionally scarless DNA assembly system optimized for metabolic engineering. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
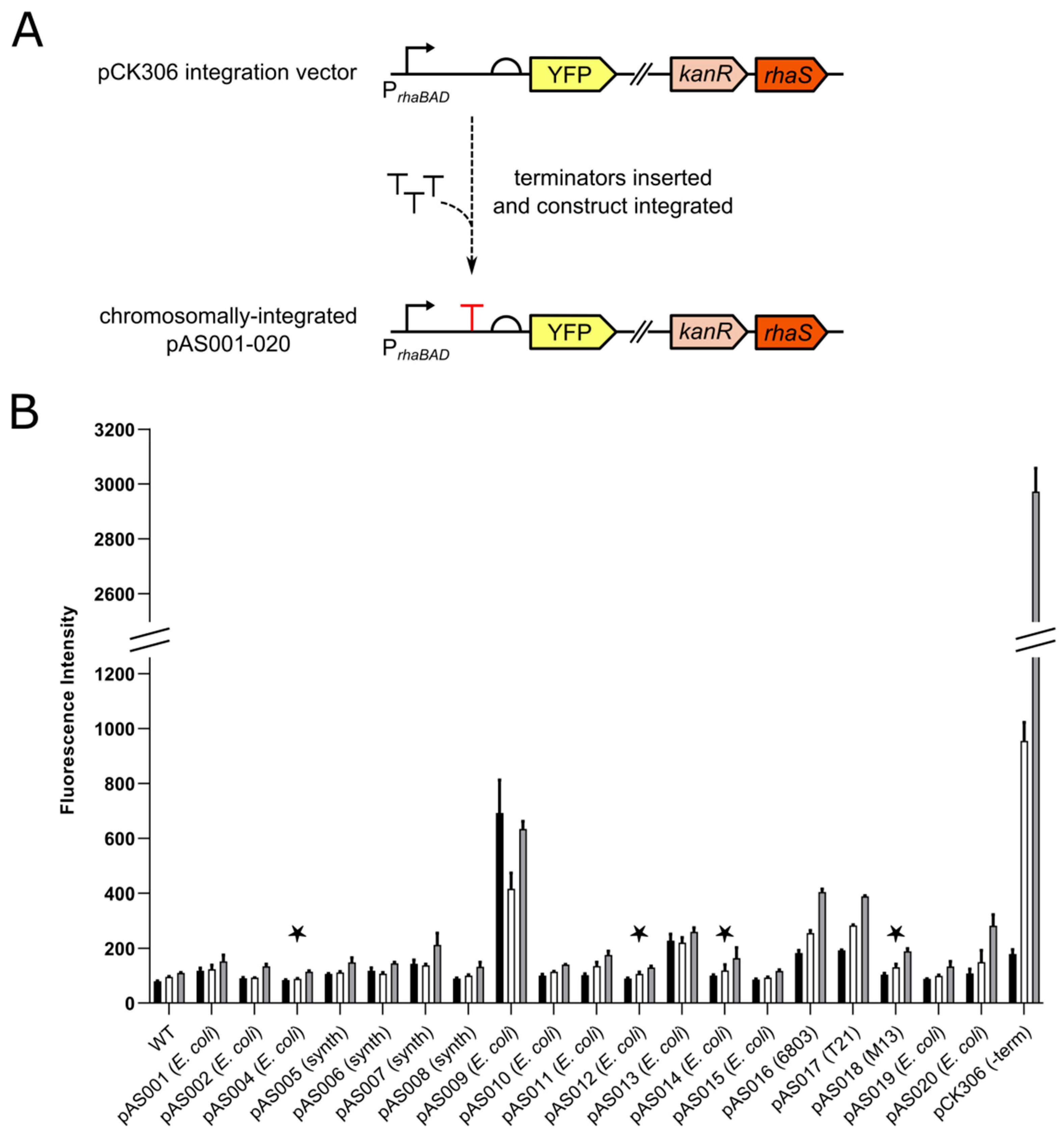
| Terminator | Plasmid | Length (bp) | Sequence (5′–3′) | ΔG (kcal/mol) | Origin | References |
|---|---|---|---|---|---|---|
| ECK120029600 | pAS001 | 90 | TTCAGCCAAAAAACTTAAGACCGCCGGTCTTGTCCACTACCTTGCAGTAATGCGGTGGACAGGATCGGCGGTTTTCTTTTCTCTTCTCAA | −42.00 | E. coli K12 | [8] |
| ECK120033737; thrL attenuator | pAS002 | 57 | GGAAACACAGAAAAAAGCCCGCACCTGACAGTGCGGGCTTTTTTTTTCGACCAAAGG | −25.00 | E. coli K12 | [8,12] |
| ECK120034435 | pAS004; pCK351 | 57 | CTCGGTACCAAATTCCAGAAAAGAGACGCTGAAAAGCGTCTTTTTTCGTTTTGGTCC | −27.90 | E. coli K12 | [8] |
| L3S2P21 | pAS005 | 61 | CTCGGTACCAAATTCCAGAAAAGAGGCCTCCCGAAAGGGGGGCCTTTTTTCGTTTTGGTCC | −37.90 | Synthetic | [8] |
| L3S2P56 | pAS006 | 57 | CTCGGTACCAAATTTTCGAAAAAAGACGCTGAAAAGCGTCTTTTTTCGTTTTGGTCC | −28.80 | Synthetic | [8] |
| L3S2P51 | pAS007 | 57 | CTCGGTACCAAAAAAAAAAAAAAAGACGCTGAAAAGCGTCTTTTTTCGTTTTGGTCC | −24.90 | Synthetic | [8] |
| L3S1P56 | pAS008 | 52 | TTTTCGAAAAAAGGCCTCCCAAATCGGGGGGCCTTTTTTATTGATAACAAAA | −23.40 | Synthetic | [8] |
| Bba_B0015; rrnB terminator | pAS009 | 130 | CCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTCTACTAGAGTCACACTGGCTCACCTTCGGGTGGGCCTTTCTGCGTTTATA | −72.10 | E. coli K12 | [2] |
| ECK120035133 | pAS010 | 43 | ACTGATTTTTAAGGCGACTGATGAGTCGCCTTTTTTTTGTCT | −15.40 | E. coli K12 | [8] |
| ECK120017009 | pAS011 | 44 | GATCTAACTAAAAAGGCCGCTCTGCGGCCTTTTTTCTTTTCACT | −16.20 | E. coli K12 | [8] |
| ECK120015170 | pAS012; pCK353 | 47 | ACAATTTTCGAAAAAACCCGCTTCGGCGGGTTTTTTTATAGCTAAAA | −20.10 | E. coli K12 | [8] |
| ECK120033736 | pAS013 | 53 | AACGCATGAGAAAGCCCCCGGAAGATCACCTTCCGGGGGCTTTTTTATTGCGC | −37.80 | E. coli K12 | [8] |
| ECK120010799 | pAS014; pCK354 | 60 | GTTATGAGTCAGGAAAAAAGGCGACAGAGTAATCTGTCGCCTTTTTTCTTTGCTTGCTTT | −33.60 | E. coli K12 | [8] |
| BBa_B0010; rrnB terminator | pAS015 | 80 | CCAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTCGTTTTATCTGTTGTTTGTCGGTGAACGCTCTC | −42.90 | E. coli K12 | [9] |
| ΩgroEL | pAS016 | 89 | GGTTTAGTAGACCGACTACCACTTTTCTCATAAAATCCCAGGGAGGTTTCGGCCTCCCTTTTTTTCACTTGCTAAGCTCTCTTTCGTTT | −20.80 | Synechocystis sp. PCC 6803 | [11] |
| T21 | pAS017 | 74 | ATTGAGCAAGTAGCAACACTATTCGCATAAGCTGCCGTTAGTGACTCTTAAGTTGCAACGGTGGCTTTTTTTAT | −25.40 | Bacteriophage T21 | [13] |
| M13 Central | pAS018 | 85 | AAAGCAAGCTGATAAACCGATACAATTAAAGGCTCCTTTTGGAGCCTTTTTTTTTGGAGATTTTCAACATGAAAAAATTATTATT | −18.60 | Bacteriophage M13 | [13] |
| ilvBN terminator | pAS019; pCK355 | 36 | AAGACCCCCGCACCGAAAGGTCCGGGGGTTTTTTTT | −24.40 | E. coli K12 | [8,13] |
| ECK120010793 | pAS020 | 34 | TACGTAAAAACCCGCTTCGGCGGGTTTTTACTTT | −24.40 | E. coli K12 | [8,13] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelly, C.L.; Taylor, G.M.; Šatkutė, A.; Dekker, L.; Heap, J.T. Transcriptional Terminators Allow Leak-Free Chromosomal Integration of Genetic Constructs in Cyanobacteria. Microorganisms 2019, 7, 263. https://doi.org/10.3390/microorganisms7080263
Kelly CL, Taylor GM, Šatkutė A, Dekker L, Heap JT. Transcriptional Terminators Allow Leak-Free Chromosomal Integration of Genetic Constructs in Cyanobacteria. Microorganisms. 2019; 7(8):263. https://doi.org/10.3390/microorganisms7080263
Chicago/Turabian StyleKelly, Ciarán L., George M. Taylor, Aistė Šatkutė, Linda Dekker, and John T. Heap. 2019. "Transcriptional Terminators Allow Leak-Free Chromosomal Integration of Genetic Constructs in Cyanobacteria" Microorganisms 7, no. 8: 263. https://doi.org/10.3390/microorganisms7080263
APA StyleKelly, C. L., Taylor, G. M., Šatkutė, A., Dekker, L., & Heap, J. T. (2019). Transcriptional Terminators Allow Leak-Free Chromosomal Integration of Genetic Constructs in Cyanobacteria. Microorganisms, 7(8), 263. https://doi.org/10.3390/microorganisms7080263





