Abstract
Coronaviruses in general are a zoonotic pathogen with significant cross-species transmission. They are widely distributed in nature and have recently become a major threat to global public health. Vaccines are the preferred strategy for the prevention of coronaviruses. However, the rapid rate of virus mutation, large number of prevalent strains, and lag in vaccine development contribute to the continuing frequent occurrence of coronavirus diseases. There is an urgent need for new antiviral strategies to address coronavirus infections effectively. Antiviral drugs are important in the prevention and control of viral diseases. Members of the genus coronavirus are highly similar in life-cycle processes such as viral invasion and replication. These, together with the high degree of similarity in the protein sequences and structures of viruses in the same genus, provide common targets for antiviral drug screening of coronaviruses and have led to important advances in recent years. In this review, we summarize the pathogenic mechanisms of coronavirus, common drugs targeting coronavirus entry into host cells, and common drug targets against coronaviruses based on biosynthesis and on viral assembly and release. We also describe the common targets of antiviral drugs against coronaviruses and the progress of antiviral drug research. Our aim is to provide a theoretical basis for the development of antiviral drugs and to accelerate the development and utilization of commonly used antiviral drugs in China.
1. Introduction
Antiviral drugs are widely recognized as a valuable approach for preventing and controlling viral diseases, particularly in the acute treatment of new outbreaks. These drugs can significantly alleviate the disease, reduce mortality, and serve as an effective alternative or supplement to vaccines. Currently, there are several main types of antiviral drugs available, including inhibitors of viral adhesion, internalization, and release; drugs that restrict polymerase or protease activity; and nucleoside/nucleotide reverse transcriptase inhibitors and integrase inhibitors [1]. Most of these antiviral drugs target a particular protein of a particular virus, which offers the advantage of high specificity and minimal harm to the host. However, the downside is that the mutable nature of viruses can easily lead to drug resistance, owing to mutations in the viral drug target, which also means that existing drugs are of limited help in controlling viral infections. In addition, although some antiviral drugs effectively suppress viral infections, they often come with drawbacks such as high toxicity, low potency, adverse drug reactions, and off-target side effects [2]. Development of virus-specific drugs first requires addressing the basic biology of the virus to find a suitable target, followed by medicinal chemistry studies, compound screening, optimization, animal studies, and ultimately clinical trials. This process can be lengthy, cumbersome, and difficult. Most of the drugs that have been developed reduce clinical symptoms and symptom duration but cannot completely remove the virus. The drugs must be administered regularly, which greatly limits their clinical application [3]. This, coupled with the limited number of disease proteins suitable for drug development, has resulted in the slow progress of research on antiviral drugs. There are still no effective antiviral drugs available for many viral infections, which has been a common and critical problem in this field.
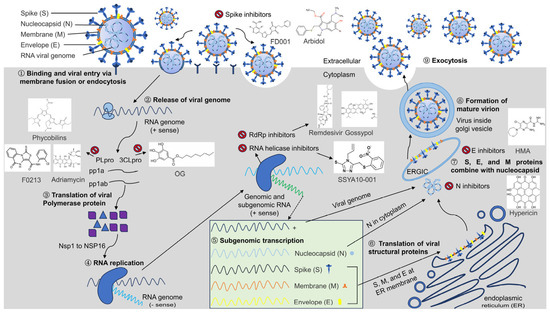
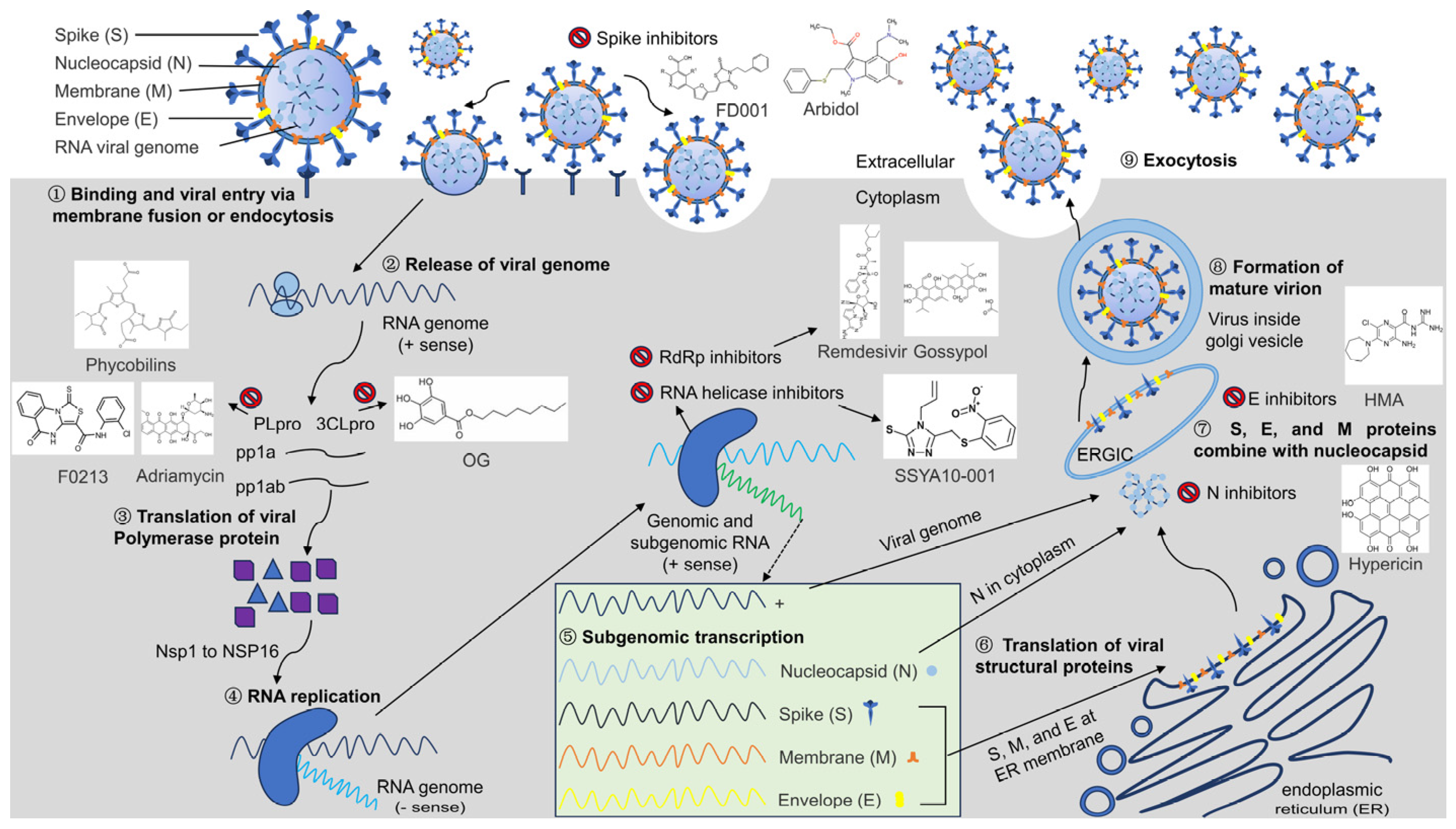
The discovery of antiviral drug targets has been advanced with the revelation of viral life-cycle mechanisms, such as viral invasion, genome transcription, replication, assembly, and release; mechanistic revelations of virus–host interaction mechanisms; and the rapid development of antiviral drug screening and development technologies. Currently, anti-coronaviral drug targets mainly focus on three aspects: viral invasion, replication, and release [4]. In terms of viral invasion, the design of antiviral drugs mainly revolves around interfering with the fusion of the viral vesicle membrane with the host cell membrane or inhibiting host receptor proteins. For example, antiviral drugs inhibit the coupling of viruses to relevant host receptors by binding to specific viral proteins, inhibiting viral entry into target cells in the form of receptor binding [5,6], or inhibiting viral replication by specifically inhibiting the contact, adhesion, and fusion of viral lipid vesicle membranes to host cell membranes [7,8]. In the context of viral replication, the design of antiviral drugs is centered around interfering with the function of viral replication-associated proteases. For example, some antiviral drugs can inhibit viral replication by targeting viral replication-related enzymes. In terms of viral assembly and release, antiviral drugs inhibit virus formation by inhibiting the modification of cytosolic proteins by viral proteases. These targeting efforts have revealed small-molecule compounds, chemically synthesized drugs, natural products, peptides, and other antiviral drugs, which have enhanced antiviral research and opened the door to the development of antiviral drugs with broad-spectrum activity.
In addition to vaccines, research on antiviral drugs is one of the key strategies for coronavirus prevention and control. Although different viruses replicate in different ways, there are several common stages in the viral replication cycle. These include viral adherence, invasion, exsiccation, biosynthesis, assembly, and release [9,10]. Based on key replication mechanisms, such as the viral life cycle, a series of antiviral drug studies have been performed by blocking any one, or a combination of, the aforementioned viral life-cycle stages, resulting in effective drugs. However, coronaviruses have proven to be challenging. Their positive-strand RNA can be used both directly as a translation template and as a template for the synthesis of negative-stranded RNA, which in turn replicates the daughter positive-stranded RNA, thereby generating a large number of copies of the viral genome that are used to generate progeny viruses that infect new host cells. The replication of one virion can lead to 100 to 1000 progeny. In addition, coronaviruses have one of the largest known genomes among RNA viruses. This large genome instills a high mutation frequency during replication [11]. This aspect, coupled with the increased risk of cross-species transmission of coronaviruses due to increased human–animal interactions, has constrained the rapid development of coronavirus antiviral drugs. In seeking to overcome this constraint, the utilization of common antiviral targets, as well as the development of multi-combination antiviral drugs based on common targets, which has been promising for other viruses, is a potentially valuable direction for coronaviruses. Identification and exploitation of common coronavirus drug targets is central to this research.
In this review, we focus on the life cycle of coronavirus-infected hosts and combine the research results of scholars from various countries to consider the common antiviral drug targets of coronaviruses. The aim is to provide reference data for the clinical treatment of coronavirus infections and the timely development of broad-spectrum antiviral drugs.
6. Conclusions and Prospects
Infectious diseases are among the major public health challenges. Outbreaks of viral infections in humans and animals have prompted a great deal of research on the pathogens of diseases worldwide [76]. There have been some encouraging advances in co-opted antiviral agents and a variety of approaches for designing anti-coronavirus strategies based on the mechanism of viral infection have been reported. However, many infectious diseases remain untreated [77]. In addition, emerging viral outbreaks pose a continuing threat to human health and life. Efficient and broad-spectrum inhibitors against viruses are powerful assets enabling rapid responses in the early stages of outbreaks and for preventative measures in the first instances of infection [78]. Therefore, the development of commensal antiviral drugs based on viral commensal mechanisms is of particular importance for dealing with emerging and outbreaks of infectious diseases [79].
Common antiviral drugs usually need to target critical proteins belonging to different viruses. Thus, in developing common antivirals, the basic biology of viruses must first be addressed to find suitable targets [80]. Based on the key aspects of the whole life cycle of infection, replication, and release of different RNA viruses, antiviral drug research has identified multiple common targets. These include S proteins targeting the entry into host cells, proteases targeting viral biosynthesis, and viral structural proteins targeting viral assembly and release, which can simultaneously inhibit the same or different coronaviruses. Thus, the combination of co-targeted drugs may enhance the effectiveness of antiviral drugs against each coronavirus type. In the future, if, after sequence identification, a new-onset virus is found to be from a known genus of viruses with a high degree of homology to existing viruses of the same genus, it may be treated by a stockpile of cocktail therapies targeting viruses of that genus [81]. The sequences and structures of proteins performing the same function are often highly similar across all viruses or in the same genus, and most antiviral drugs that are designed to target a conserved viral target have good antiviral efficacy against that virus as well as potential inhibitory effects on other viruses in the same genus or family [82]. Therefore, broad-spectrum viral inhibitors with conserved targets could be the first to be tried in cases of new outbreaks of viruses from known viral genera but with low homology to their homologs. In addition, based on the discovery of these new drug targets against the viral life cycle and their regulatory mechanisms, new drug targets with common antiviral effects and their precise structures and regulatory mechanisms can also be explored in future studies based on the characteristic molecular events in the replication process of other viruses.
Coronaviruses require the cooperative action of many host factors and cellular metabolic pathways to successfully infect host cells and effectively reproduce. Therefore, some critical host factors, such as ACE2, CD147, furin, cathepsin L, TMPRSS2, HSP90, HS, DC-SIGN/L-SIGN, SA, and TfR1, have also shown potential as antiviral targets [83,84,85]. For example, ACE2, CD147, and other functional receptors of coronaviruses can mediate the entry of viruses into host cells by binding to S proteins. Therefore, some antiviral drugs targeting functional receptors such as ACE2 can inhibit coronavirus replication in vitro by interfering with the binding of the virus to the receptor and affecting the invasion of the virus, thus exerting broad-spectrum antiviral effects. Similar to this mechanism of action, antiviral targets such as Furin, TMPRSS2, and other host cytokine proteases that mediate viral invasion can block the protein hydrolysis cleavage sites between the S1/S2 subunits of the viral S-protein, which are required for viral cell entry. Blocking the proteases necessary for these cleavages can affect the viral life cycle and pathogenicity, thereby inhibiting viral replication. Drugs targeting host cells can effectively inhibit the rapid replication of viral nucleic acids and combat viral drug-resistant mutations. However, broad-spectrum antiviral drugs targeting the host may be harmful to the host, and the in vitro inhibitory activity and in vivo therapeutic efficacy may not be fully consistent. In response to the host factors during viral infection and proliferation, the common principles, targeting, and safety of their inhibition of viral replication can be explored in the future to find new targets that are critical for viral replication but not for host cells. In this way, broad-spectrum and highly effective multi-targeted chemotherapy modalities can be designed to support chemical interventions.
The development of coronaviral antiviral drugs for multiple targets and multiple antiviral combinations, as well as broad-spectrum anti-coronaviral drugs for a common target, is a future research direction. Clarifying the mechanism of action of coronaviral antiviral drugs will further enhance and improve the drug design strategy for coronaviral antiviral drugs, facilitate screening for more efficient antiviral drugs, further enrich the potential of antiviral drug family to combat the current coronavirus pandemic and prevent future outbreaks of coronaviruses and the potential spillover of zoonotic coronaviruses, and provide basic support for the research and development of original RNA viral infectious disease. The collective knowledge also provides basic support for the development of original therapeutic drugs for RNA viral infectious diseases.
Author Contributions
Literature search, writing—original draft preparation, J.W.; Literature search, writing—review and editing, Q.Z.; Funding acquisition, literature search, writing—review and editing, X.X.; Funding acquisition, writing—review and editing, D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant no. U23A20236) and the Heilongjiang Provincial Natural Science Foundation (grant nos. ZD2023C006 and LH2023C084).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541. [Google Scholar] [CrossRef]
- Ton, A.T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 2020, 39, e2000028. [Google Scholar] [CrossRef]
- Thomasy, S.M.; Maggs, D.J. A review of antiviral drugs and other compounds with activity against feline herpesvirus type 1. Vet. Ophthalmol. 2016, 19 (Suppl. 1), 119–130. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, L.V. Overview of Targets and Potential Drugs of SARS-CoV-2 According to the Viral Replication. J. Proteome Res. 2021, 20, 49–59. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef]
- Wang, X.; Cao, R.; Zhang, H.; Liu, J.; Xu, M.; Hu, H.; Li, Y.; Zhao, L.; Li, W.; Sun, X.; et al. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020, 6, 28. [Google Scholar] [CrossRef]
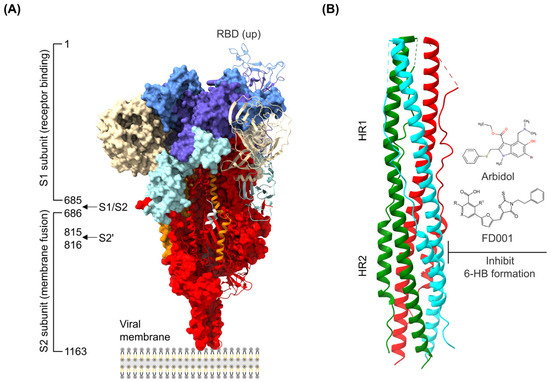
- Wang, C.; Xia, S.; Zhang, P.; Zhang, T.; Wang, W.; Tian, Y.; Meng, G.; Jiang, S.; Liu, K. Discovery of Hydrocarbon-Stapled Short alpha-Helical Peptides as Promising Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Fusion Inhibitors. J. Med. Chem. 2018, 61, 2018–2026. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 2249. [Google Scholar] [CrossRef]
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef]
- de Groot, R.J.; Baker, S.C.; Baric, R.S.; Brown, C.S.; Drosten, C.; Enjuanes, L.; Fouchier, R.A.; Galiano, M.; Gorbalenya, A.E.; Memish, Z.A.; et al. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013, 87, 7790–7792. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Y.; Zhai, J.; Xue, M.; Zheng, C.; Yu, L. Future trajectory of SARS-CoV-2: Constant spillover back and forth between humans and animals. Virus Res. 2023, 328, 199075. [Google Scholar] [CrossRef]
- Silva, L.R.; da Silva Santos-Júnior, P.F.; de Andrade Brandão, J.; Anderson, L.; Bassi, Ê.J.; Xavier de Araújo-Júnior, J.; Cardoso, S.H.; da Silva-Júnior, E.F. Druggable targets from coronaviruses for designing new antiviral drugs. Bioorg. Med. Chem. 2020, 28, 115745. [Google Scholar] [CrossRef] [PubMed]
- Uma Reddy, B.; Routhu, N.K.; Kumar, A. Multifaceted roles of plant derived small molecule inhibitors on replication cycle of SARS-CoV-2. Microb. Pathog. 2022, 168, 105512. [Google Scholar] [CrossRef] [PubMed]
- Drożdżal, S.; Rosik, J.; Lechowicz, K.; Machaj, F.; Szostak, B.; Przybyciński, J.; Lorzadeh, S.; Kotfis, K.; Ghavami, S.; Łos, M.J. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer Chemother. 2021, 59, 100794. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Castillo, G.; Mora-Díaz, J.C.; Breuer, M.; Singh, P.; Nelli, R.K.; Giménez-Lirola, L.G. Molecular mechanisms of human coronavirus NL63 infection and replication. Virus Res. 2023, 327, 199078. [Google Scholar] [CrossRef]
- Upadhyay, M.; Gupta, S. Endoplasmic reticulum secretory pathway: Potential target against SARS-CoV-2. Virus Res. 2022, 320, 198897. [Google Scholar] [CrossRef]
- Pu, J.; He, X.; Xu, W.; Wang, C.; Lan, Q.; Hua, C.; Wang, K.; Lu, L.; Jiang, S. The Analogs of Furanyl Methylidene Rhodanine Exhibit Broad-Spectrum Inhibitory and Inactivating Activities against Enveloped Viruses, including SARS-CoV-2 and Its Variants. Viruses 2022, 14, 489. [Google Scholar] [CrossRef]
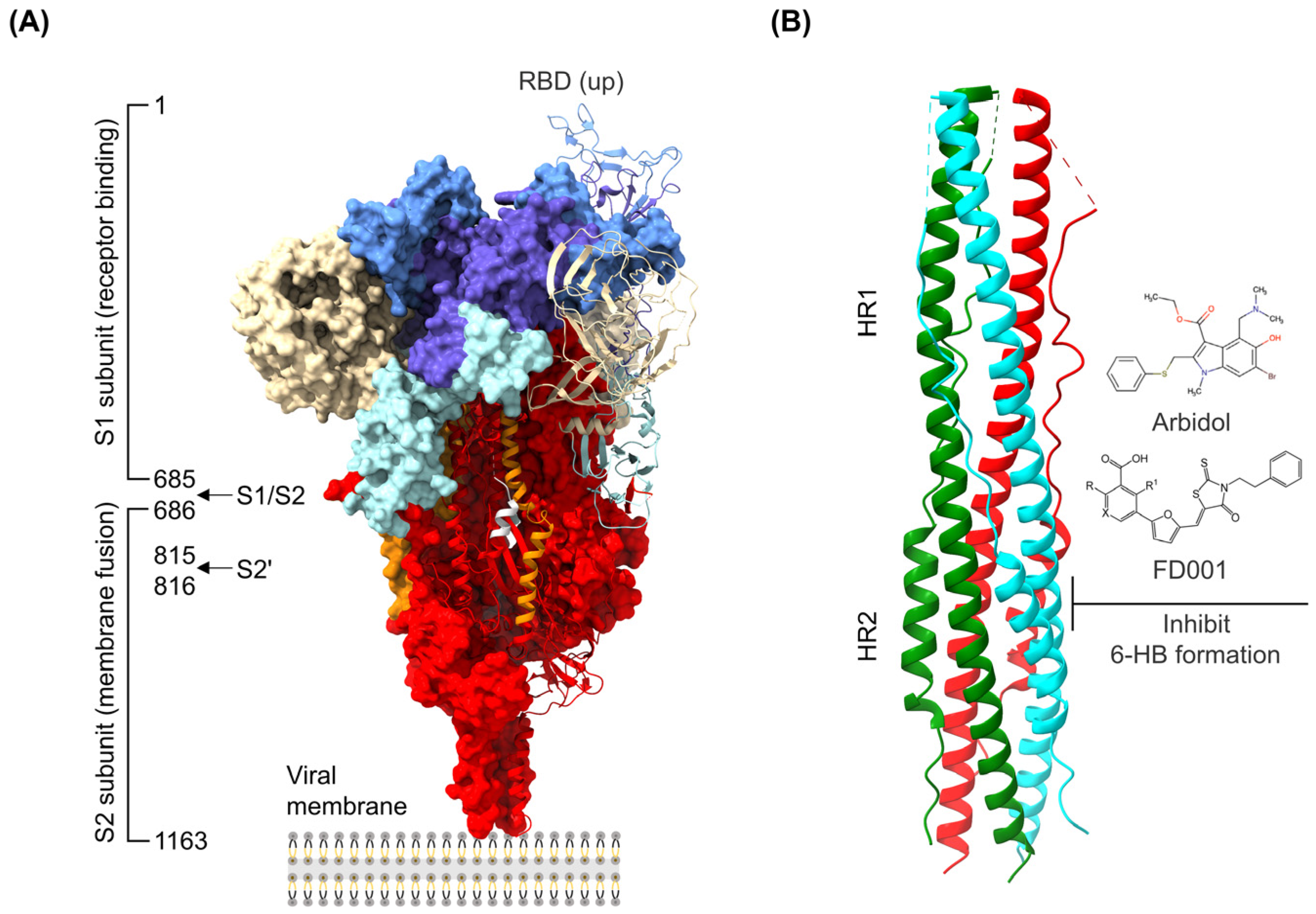
- Vankadari, N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents 2020, 56, 105998. [Google Scholar] [CrossRef]
- Pendyala, B.; Patras, A.; Dash, C. Phycobilins as Potent Food Bioactive Broad-Spectrum Inhibitors Against Proteases of SARS-CoV-2 and Other Coronaviruses: A Preliminary Study. Front. Microbiol. 2021, 12, 645713. [Google Scholar] [CrossRef]
- Alaofi, A.L.; Shahid, M.; Raish, M.; Ansari, M.A.; Syed, R.; Kalam, M.A. Identification of Doxorubicin as Repurposing Inhibitory Drug for MERS-CoV PLpro. Molecules 2022, 27, 7553. [Google Scholar] [CrossRef]
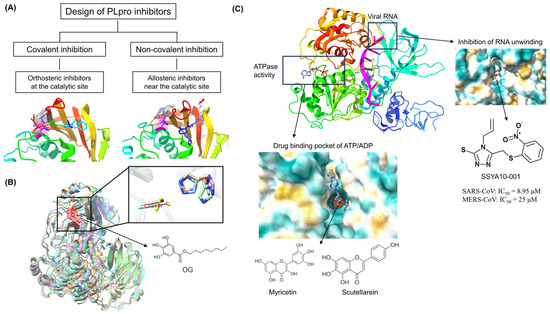
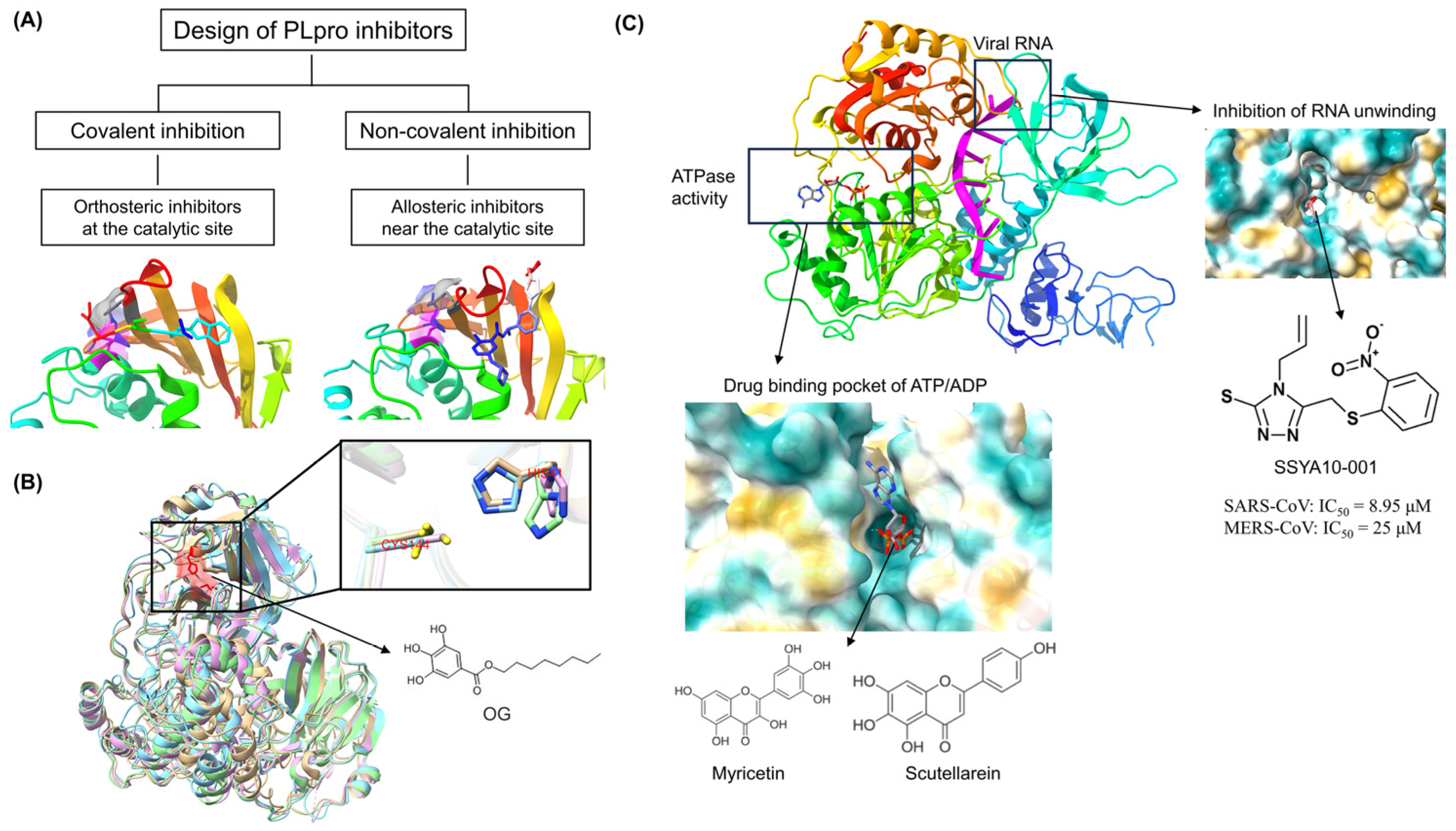
- Yuan, S.; Gao, X.; Tang, K.; Cai, J.P.; Hu, M.; Luo, P.; Wen, L.; Ye, Z.W.; Luo, C.; Tsang, J.O.; et al. Targeting papain-like protease for broad-spectrum coronavirus inhibition. Protein Cell 2022, 13, 940–953. [Google Scholar] [CrossRef]
- Lin, Y.; Zang, R.; Ma, Y.; Wang, Z.; Li, L.; Ding, S.; Zhang, R.; Wei, Z.; Yang, J.; Wang, X. Xanthohumol Is a Potent Pan-Inhibitor of Coronaviruses Targeting Main Protease. Int. J. Mol. Sci. 2021, 22, 12134. [Google Scholar] [CrossRef]
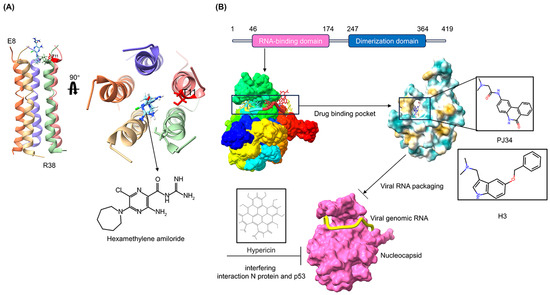
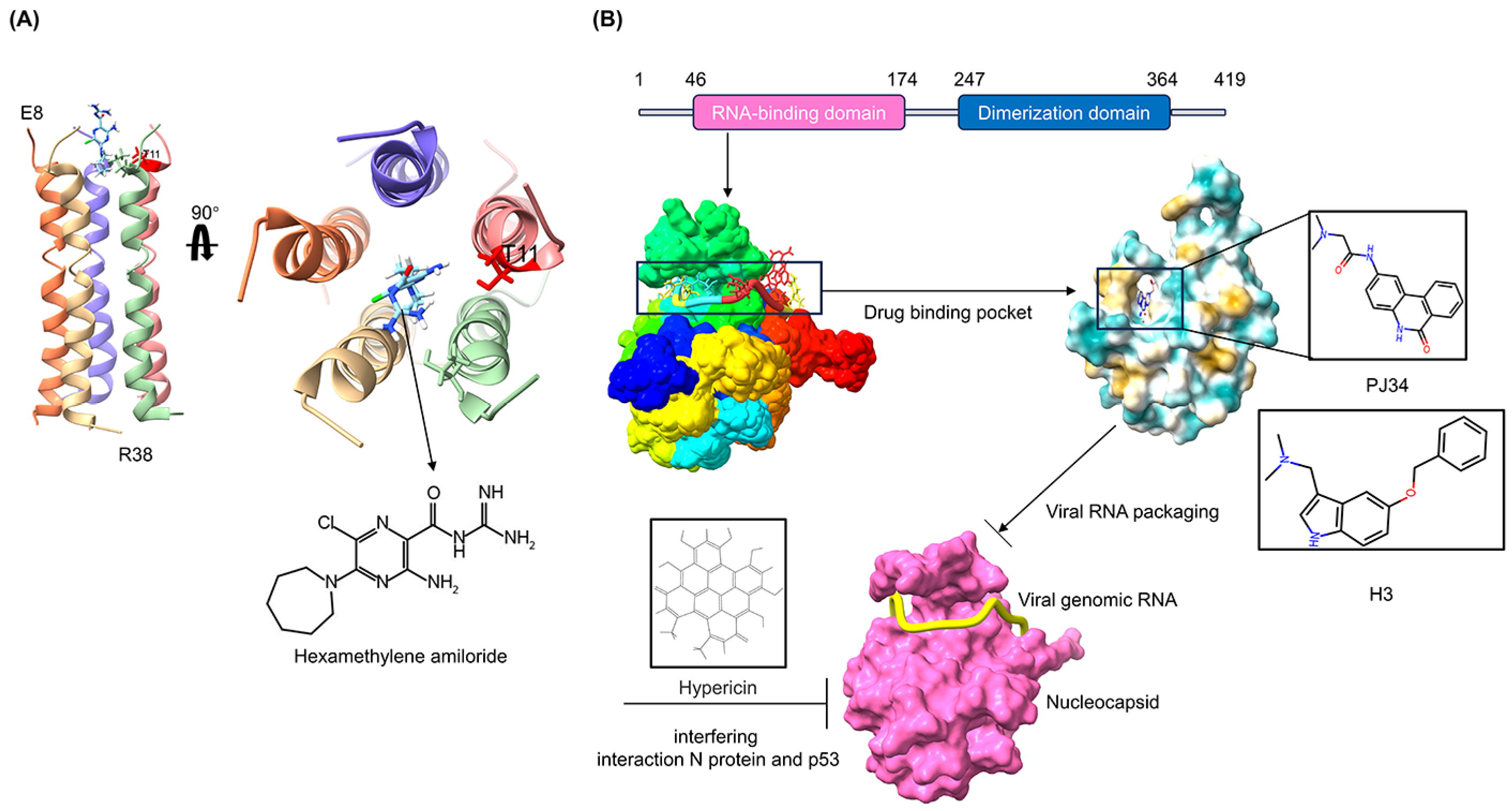
- Zhang, Y.; Chen, H.; Zou, M.; Oerlemans, R.; Shao, C.; Ren, Y.; Zhang, R.; Huang, X.; Li, G.; Cong, Y. Hypericin Inhibit Alpha-Coronavirus Replication by Targeting 3CL Protease. Viruses 2021, 13, 1825. [Google Scholar] [CrossRef]
- Wang, P.; Bai, J.; Liu, X.; Wang, M.; Wang, X.; Jiang, P. Tomatidine inhibits porcine epidemic diarrhea virus replication by targeting 3CL protease. Vet. Res. 2020, 51, 136. [Google Scholar] [CrossRef]
- Su, M.; Yin, B.; Xing, X.; Li, Z.; Zhang, J.; Feng, S.; Li, L.; Zhao, F.; Yang, X.; Yu, S.; et al. Octyl gallate targeting the 3C-like protease exhibits highly efficient antiviral activity against swine enteric coronavirus PEDV. Vet. Microbiol. 2023, 281, 109743. [Google Scholar] [CrossRef] [PubMed]
- Mahgoub, R.E.; Mohamed, F.E.; Ali, B.R.; Ferreira, J.; Rabeh, W.M.; Atatreh, N.; Ghattas, M.A. Discovery of pyrimidoindol and benzylpyrrolyl inhibitors targeting SARS-CoV-2 main protease (M(pro)) through pharmacophore modelling, covalent docking, and biological evaluation. J. Mol. Graph. Model. 2024, 127, 108672. [Google Scholar] [CrossRef]
- Runfeng, L.; Yunlong, H.; Jicheng, H.; Weiqi, P.; Qinhai, M.; Yongxia, S.; Chufang, L.; Jin, Z.; Zhenhua, J.; Haiming, J.; et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Wen, Z.; Wang, C.; Liu, W.; Zhang, Y.; Liu, J.; Ding, T.; Shuai, L.; Zhong, G.; et al. Gossypol Broadly Inhibits Coronaviruses by Targeting RNA-Dependent RNA Polymerases. Adv. Sci. 2022, 9, e2203499. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Singh, K.; Kassim, A.; Coleman, C.M.; Elliott, R.; Weiss, S.R.; Frieman, M.B.; Sarafianos, S.G. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob. Agents Chemother. 2014, 58, 4894–4898. [Google Scholar] [CrossRef]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef]
- Su, M.; Shi, D.; Xing, X.; Qi, S.; Yang, D.; Zhang, J.; Han, Y.; Zhu, Q.; Sun, H.; Wang, X.; et al. Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication. J. Virol. 2021, 95, e0018721. [Google Scholar] [CrossRef]
- Guo, L.; Lin, S.; Chen, Z.; Cao, Y.; He, B.; Lu, G. Targetable elements in SARS-CoV-2 S2 subunit for the design of pan-coronavirus fusion inhibitors and vaccines. Signal Transduct. Target. Ther. 2023, 8, 197. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Stincarelli, M.A.; Quagliata, M.; Di Santo, A.; Pacini, L.; Fernandez, F.R.; Arvia, R.; Rinaldi, S.; Papini, A.M.; Rovero, P.; Giannecchini, S. SARS-CoV-2 inhibitory activity of a short peptide derived from internal fusion peptide of S2 subunit of spike glycoprotein. Virus Res. 2023, 334, 199170. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Osipiuk, J.; Azizi, S.A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Melo-Filho, C.C.; Bobrowski, T.; Martin, H.J.; Sessions, Z.; Popov, K.I.; Moorman, N.J.; Baric, R.S.; Muratov, E.N.; Tropsha, A. Conserved coronavirus proteins as targets of broad-spectrum antivirals. Antivir. Res. 2022, 204, 105360. [Google Scholar] [CrossRef]
- Shen, Z.; Ratia, K.; Cooper, L.; Kong, D.; Lee, H.; Kwon, Y.; Li, Y.; Alqarni, S.; Huang, F.; Dubrovskyi, O.; et al. Design of SARS-CoV-2 PLpro Inhibitors for COVID-19 Antiviral Therapy Leveraging Binding Cooperativity. J. Med. Chem. 2022, 65, 2940–2955. [Google Scholar] [CrossRef]
- Xiong, M.; Su, H.; Zhao, W.; Xie, H.; Shao, Q.; Xu, Y. What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 2021, 41, 1965–1998. [Google Scholar] [CrossRef]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900-3909. [Google Scholar] [CrossRef]
- Zhang, W.F.; Stephen, P.; Thériault, J.F.; Wang, R.; Lin, S.X. Novel Coronavirus Polymerase and Nucleotidyl-Transferase Structures: Potential to Target New Outbreaks. J. Phys. Chem. Lett. 2020, 11, 4430–4435. [Google Scholar] [CrossRef]
- Tian, L.; Qiang, T.; Liang, C.; Ren, X.; Jia, M.; Zhang, J.; Li, J.; Wan, M.; YuWen, X.; Li, H.; et al. RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021, 213, 113201. [Google Scholar]
- Abou Baker, D.H.; Hassan, E.M.; El Gengaihi, S. An overview on medicinal plants used for combating coronavirus: Current potentials and challenges. J. Agric. Food Res. 2023, 13, 100632. [Google Scholar] [CrossRef]
- Reina, J. Remdesivir, the antiviral hope against SARS-CoV-2. Rev. Española Quimioter. 2020, 33, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Şimşek Yavuz, S.; Ünal, S. Antiviral treatment of COVID-19. Turk. J. Med. Sci. 2020, 50, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Guo, X.; Hu, T.; Wei, D.; Ma, X.; Wu, J.; Huang, B.; Shen, J. Significant Inhibition of Porcine Epidemic Diarrhea Virus In Vitro by Remdesivir, Its Parent Nucleoside and β-D-N(4)-hydroxycytidine. Virol. Sin. 2021, 36, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Barauskas, O.; Kim, C.; Babusis, D.; Murakami, E.; Kornyeyev, D.; Lee, G.; Stepan, G.; Perron, M.; Bannister, R.; et al. Off-Target In Vitro Profiling Demonstrates that Remdesivir Is a Highly Selective Antiviral Agent. Antimicrob. Agents Chemother. 2021, 65, e02237-02220. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yin, W.; Xu, H.E. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 47–53. [Google Scholar] [CrossRef]
- Tsai, S.C.; Lu, C.C.; Bau, D.T.; Chiu, Y.J.; Yen, Y.T.; Hsu, Y.M.; Fu, C.W.; Kuo, S.C.; Lo, Y.S.; Chiu, H.Y.; et al. Approaches towards fighting the COVID-19 pandemic (Review). Int. J. Mol. Med. 2021, 47, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Lee, P.I.; Hsueh, P.R. Treatment options for COVID-19: The reality and challenges. J. Microbiol. Immunol. Infect. Wei Mian Yu Gan Ran Za Zhi 2020, 53, 436–443. [Google Scholar] [CrossRef]
- Lu, C.C.; Chen, M.Y.; Lee, W.S.; Chang, Y.L. Potential therapeutic agents against COVID-19: What we know so far. J. Chin. Med. Assoc. JCMA 2020, 83, 534–536. [Google Scholar] [CrossRef]
- Persaud, K.E.; Sahu, R.R.; Neary, M.C.; Kapdi, A.R.; Lakshman, M.K. Two short approaches to the COVID-19 drug β-D-N(4)-hydroxycytidine and its prodrug molnupiravir. Org. Biomol. Chem. 2024, 22, 735–740. [Google Scholar] [CrossRef]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573.e1513. [Google Scholar] [CrossRef]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z.; et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef]
- Yazdi, A.K.; Pakarian, P.; Perveen, S.; Hajian, T.; Santhakumar, V.; Bolotokova, A.; Li, F.; Vedadi, M. Kinetic Characterization of SARS-CoV-2 nsp13 ATPase Activity and Discovery of Small-Molecule Inhibitors. ACS Infect. Dis. 2022, 8, 1533–1542. [Google Scholar] [CrossRef]
- Zeng, J.; Weissmann, F.; Bertolin, A.P.; Posse, V.; Canal, B.; Ulferts, R.; Wu, M.; Harvey, R.; Hussain, S.; Milligan, J.C.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 2021, 478, 2405–2423. [Google Scholar] [CrossRef]
- Squeglia, F.; Romano, M.; Ruggiero, A.; Maga, G.; Berisio, R. Host DDX Helicases as Possible SARS-CoV-2 Proviral Factors: A Structural Overview of Their Hijacking Through Multiple Viral Proteins. Front. Chem. 2020, 8, 602162. [Google Scholar] [CrossRef]
- Shannon, A.; Chazot, A.; Feracci, M.; Falcou, C.; Fattorini, V.; Selisko, B.; Good, S.; Moussa, A.; Sommadossi, J.P.; Ferron, F.; et al. An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex. Nucleic Acids Res. 2024, 52, 1325–1340. [Google Scholar] [CrossRef]
- Singh, I.; Li, F.; Fink, E.A.; Chau, I.; Li, A.; Rodriguez-Hernández, A.; Glenn, I.; Zapatero-Belinchón, F.J.; Rodriguez, M.L.; Devkota, K.; et al. Structure-Based Discovery of Inhibitors of the SARS-CoV-2 Nsp14 N7-Methyltransferase. J. Med. Chem. 2023, 66, 7785–7803. [Google Scholar] [CrossRef]
- Asthana, A.; Corona, A.; Shin, W.J.; Kwak, M.J.; Gaughan, C.; Tramontano, E.; Jung, J.U.; Schobert, R.; Jha, B.K.; Silverman, R.H.; et al. Analogs of the Catechol Derivative Dynasore Inhibit HIV-1 Ribonuclease H, SARS-CoV-2 nsp14 Exoribonuclease, and Virus Replication. Viruses 2023, 15, 1539. [Google Scholar] [CrossRef]
- Kottur, J.; White, K.M.; Rodriguez, M.L.; Rechkoblit, O.; Quintana-Feliciano, R.; Nayar, A.; García-Sastre, A.; Aggarwal, A.K. Structures of SARS-CoV-2 N7-methyltransferase with DOT1L and PRMT7 inhibitors provide a platform for new antivirals. PLoS Pathog. 2023, 19, e1011546. [Google Scholar] [CrossRef]
- El Omari, K.; Li, S.; Kotecha, A.; Walter, T.S.; Bignon, E.A.; Harlos, K.; Somerharju, P.; De Haas, F.; Clare, D.K.; Molin, M.; et al. The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins. Nat. Commun. 2019, 10, 846. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, R.; Lee, I.; Zhang, W.; Sun, J.; Wang, W.; Meng, X. Characterization of the SARS-CoV-2 E Protein: Sequence, Structure, Viroporin, and Inhibitors. Protein Sci. A Publ. Protein Soc. 2021, 30, 1114–1130. [Google Scholar] [CrossRef]
- Borkotoky, S.; Banerjee, M. A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem). J. Biomol. Struct. Dyn. 2021, 39, 4111–4121. [Google Scholar] [CrossRef]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef]
- Peng, Y.; Du, N.; Lei, Y.; Dorje, S.; Qi, J.; Luo, T.; Gao, G.F.; Song, H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020, 39, e105938. [Google Scholar] [CrossRef]
- Cubuk, J.; Alston, J.J.; Incicco, J.J.; Singh, S.; Stuchell-Brereton, M.D.; Ward, M.D.; Zimmerman, M.I.; Vithani, N.; Griffith, D.; Wagoner, J.A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021, 12, 1936. [Google Scholar] [CrossRef]
- Totura, A.L.; Bavari, S. Broad-spectrum coronavirus antiviral drug discovery. Expert Opin. Drug Discov. 2019, 14, 397–412. [Google Scholar] [CrossRef]
- Geraghty, R.J.; Aliota, M.T.; Bonnac, L.F. Broad-Spectrum Antiviral Strategies and Nucleoside Analogues. Viruses 2021, 13, 667. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93, 1128. [Google Scholar] [CrossRef]
- Mazzon, M.; Ortega-Prieto, A.M.; Imrie, D.; Luft, C.; Hess, L.; Czieso, S.; Grove, J.; Skelton, J.K.; Farleigh, L.; Bugert, J.J.; et al. Identification of Broad-Spectrum Antiviral Compounds by Targeting Viral Entry. Viruses 2019, 11, 176. [Google Scholar] [CrossRef]
- Sabbah, D.A.; Hajjo, R.; Bardaweel, S.K.; Zhong, H.A. An Updated Review on SARS-CoV-2 Main Proteinase (M(Pro)): Protein Structure and Small-Molecule Inhibitors. Curr. Top. Med. Chem. 2021, 21, 442–460. [Google Scholar] [CrossRef]
- Lu, L.; Su, S.; Yang, H.; Jiang, S. Antivirals with common targets against highly pathogenic viruses. Cell 2021, 184, 1604–1620. [Google Scholar] [CrossRef]
- Jones, J.C.; Yen, H.L.; Adams, P.; Armstrong, K.; Govorkova, E.A. Influenza antivirals and their role in pandemic preparedness. Antivir. Res. 2023, 210, 105499. [Google Scholar] [CrossRef]
- Muralidar, S.; Ambi, S.V.; Sekaran, S.; Krishnan, U.M. The emergence of COVID-19 as a global pandemic: Understanding the epidemiology, immune response and potential therapeutic targets of SARS-CoV-2. Biochimie 2020, 179, 85–100. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y.Q.; Xu, L.D.; Xiao, L.; Feng, Y.; Wang, B.; Huang, Y.W. Role of heat shock protein 90 as an antiviral target for swine enteric coronaviruses. Virus Res. 2023, 329, 199103. [Google Scholar] [CrossRef]
- Hu, X.; Cui, J.; Chen, J.; Du, S.; Wang, X.; Zhang, Y.; Qian, J.; Chen, H.; Wei, F.; Cai, Q.; et al. Identification of hACE2-interacting sites in SARS-CoV-2 spike receptor binding domain for antiviral drugs screening. Virus Res. 2022, 321, 198915. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).