Optimization of Fermentation Conditions and Product Identification of a Saponin-Producing Endophytic Fungus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Chemicals
2.2. Identification of the Strain NSJ105
2.3. Liquid Culture of Endophytic Fungus
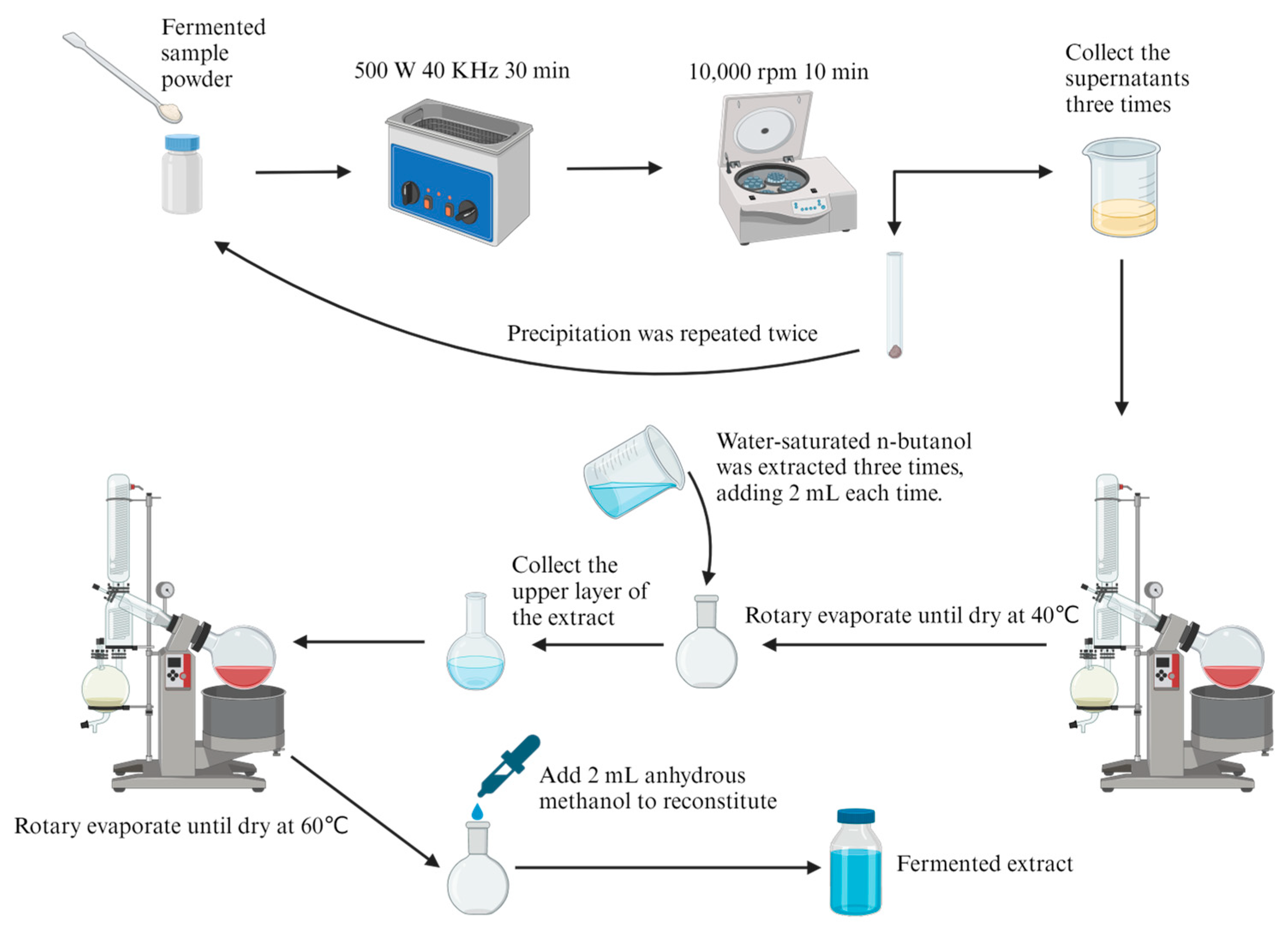
2.4. Extraction of Total Saponins
2.5. Determination of Total Saponins
2.6. Design of Fermentation Conditions
2.7. Regression Analysis and Verification of Fermentation Protocols
2.8. Identification and Analysis of Fermentation Extracts
2.9. Statistical Analysis
3. Results
3.1. Identification of the Strain NSJ105
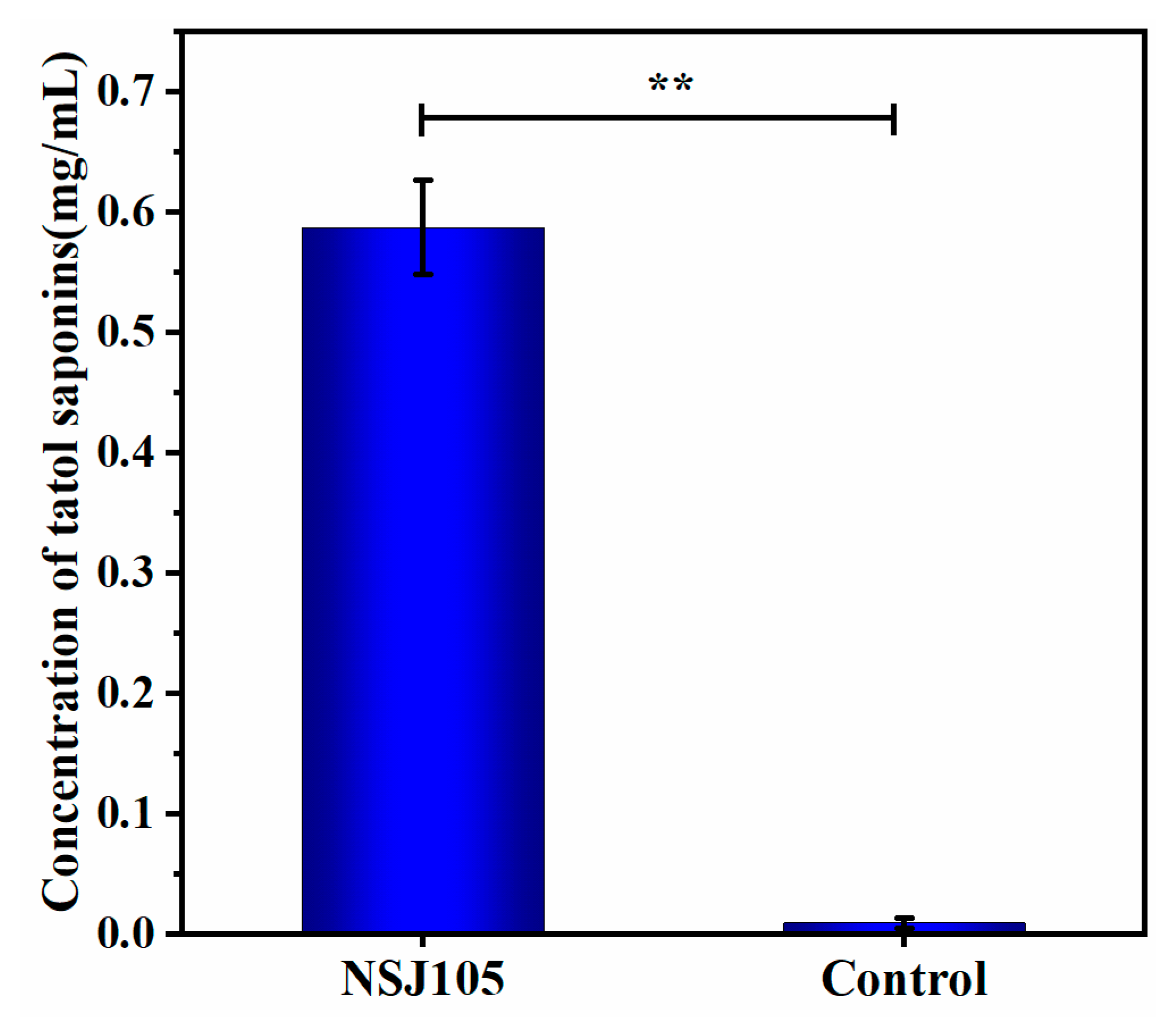
3.2. Liquid Fermentation In Vitro of NSJ105
3.3. Total Saponins Concentration by Uniform Design Test
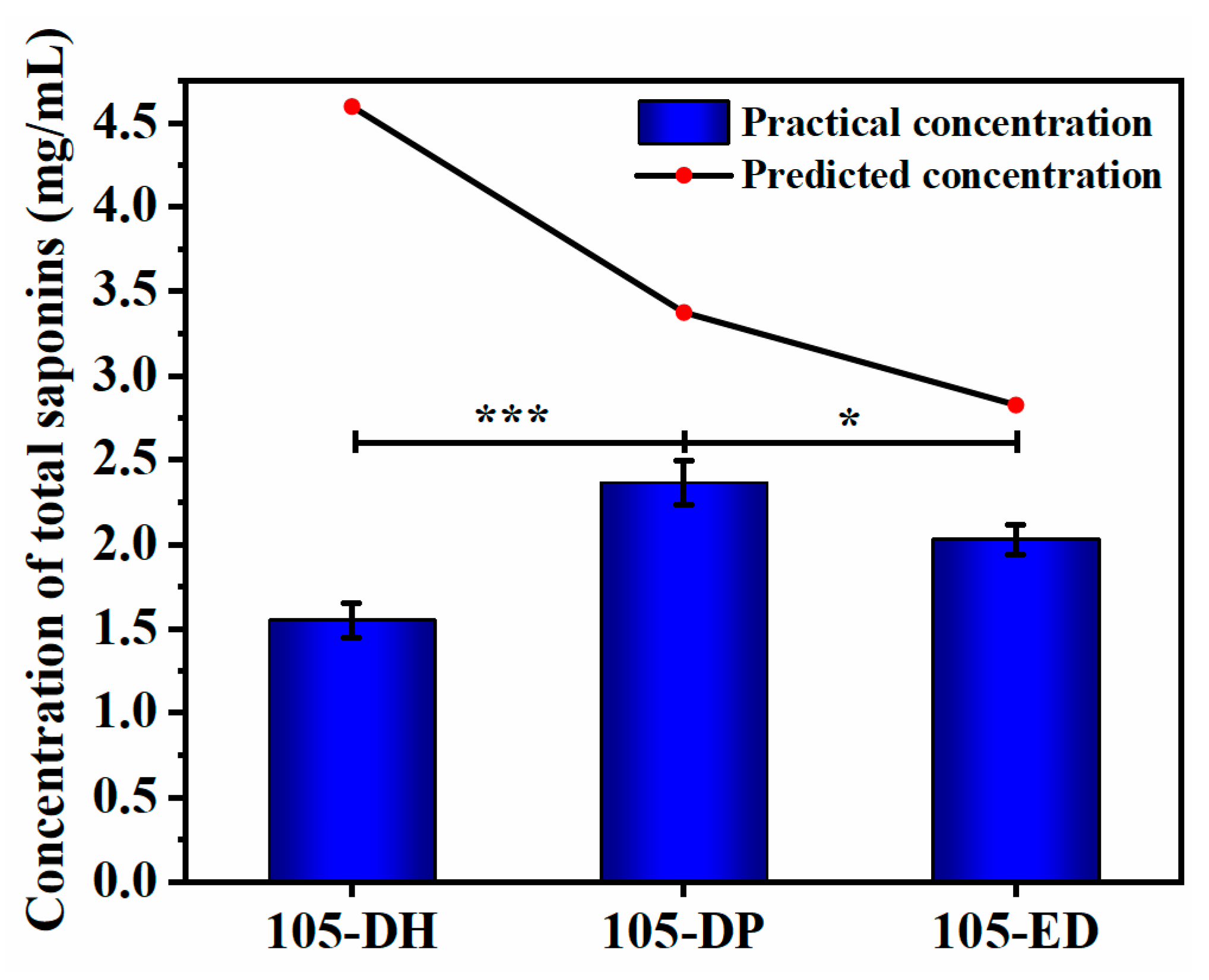
3.4. Optimization of Fermentation Conditions
3.5. Identification and Analysis of Fermentation Products
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Liu, J.; Zuo, T.T.; Hu, Y.; Li, Z.; Wang, H.D.; Xu, X.Y.; Yang, W.Z.; Guo, D.A. Advances and challenges in ginseng research from 2011 to 2020: The phytochemistry, quality control, metabolism, and biosynthesis. Nat. Prod. Rep. 2022, 39, 875–909. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.K.; Kwon, S.W.; Park, J.H. Chemical diversity of ginseng saponins from Panax ginseng. J. Ginseng Res. 2015, 39, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.W.; Wang, C.Z.; Yuan, C.S. Ginsenosides from American ginseng: Chemical and pharmacological diversity. Phytochemistry 2011, 72, 689–699. [Google Scholar] [CrossRef]
- You, L.; Cha, S.; Kim, M.Y.; Cho, J.Y. Ginsenosides are active ingredients in Panax ginseng with immunomodulatory properties from cellular to organismal levels. J. Ginseng Res. 2022, 46, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Y.; Zeng, J.Z.; Wong, A.S.T. Chemical Structures and Pharmacological Profiles of Ginseng Saponins. Molecules 2019, 24, 2443. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; Cho, J.Y.; Kim, B.; Bae, B.S.; Kim, J.H. Red ginseng (Panax ginseng) decreases isoproterenol-induced cardiac injury via antioxidant properties in porcine. J. Med. Food 2014, 17, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qu, C.Y.; Li, J.X.; Wang, Y.F.; Li, W.; Wang, C.Z.; Wang, D.S.; Song, J.; Sun, G.Z.; Yuan, C.S. Hypoglycemic and Hypolipidemic Effects of Malonyl Ginsenosides from American Ginseng (Panax quinquefolius L.) on Type 2 Diabetic Mice. ACS Omega 2021, 6, 33652–33664. [Google Scholar] [CrossRef]
- Mucalo, I.; Jovanovski, E.; Rahelic, D.; Bozikov, V.; Romic, Z.; Vuksan, V. Effect of American ginseng (Panax quinquefolius L.) on arterial stiffness in subjects with type-2 diabetes and concomitant hypertension. J. Ethnopharmacol. 2013, 150, 148–153. [Google Scholar] [CrossRef]
- Yue, P.Y.; Mak, N.K.; Cheng, Y.K.; Leung, K.W.; Ng, T.B.; Fan, D.T.; Yeung, H.W.; Wong, R.N. Pharmacogenomics and the Yin/Yang actions of ginseng: Anti-tumor, angiomodulating and steroid-like activities of ginsenosides. Chin. Med. 2007, 2, 6. [Google Scholar] [CrossRef]
- Chung, S.I.; Kang, M.Y.; Lee, S.C. In Vitro and In Vivo Antioxidant Activity of Aged Ginseng (Panax ginseng). Prev. Nutr. Food Sci. 2016, 21, 24–30. [Google Scholar] [CrossRef][Green Version]
- Piao, X.; Zhang, H.; Kang, J.P.; Yang, D.U.; Li, Y.; Pang, S.; Jin, Y.; Yang, D.C.; Wang, Y. Advances in Saponin Diversity of Panax ginseng. Molecules 2020, 25, 3452. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Chaudhary, N.; Seo, H.J.; Kim, M.Y.; Shin, T.S.; Kim, J.D. Immunomodulatory effect of tea saponin in immune T-cells and T-lymphoma cells via regulation of Th1, Th2 immune response and MAPK/ERK2 signaling pathway. Immunopharmacol. Immunotoxicol. 2014, 36, 202–210. [Google Scholar] [CrossRef]
- Li, M.; Ma, M.; Wu, Z.; Liang, X.; Zheng, Q.; Li, D.; An, T.; Wang, G. Advances in the biosynthesis and metabolic engineering of rare ginsenosides. Appl. Microbiol. Biotechnol. 2023, 107, 3391–3404. [Google Scholar] [CrossRef] [PubMed]
- Kusari, S.; Hertweck, C.; Spiteller, M. Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 2012, 19, 792–798. [Google Scholar] [CrossRef]
- Wei, G.; Chen, Z.; Wang, B.; Wei, F.; Zhang, G.; Wang, Y.; Zhu, G.; Zhou, Y.; Zhao, Q.; He, M.; et al. Endophytes isolated from Panax notoginseng converted ginsenosides. Microb. Biotechnol. 2021, 14, 1730–1746. [Google Scholar] [CrossRef]
- Li, R.; Duan, W.; Ran, Z.; Chen, X.; Yu, H.; Fang, L.; Guo, L.; Zhou, J. Diversity and correlation analysis of endophytes and metabolites of Panax quinquefolius L. in various tissues. BMC Plant Biol. 2023, 23, 275. [Google Scholar]
- Nisa, H.; Kamili, A.N.; Nawchoo, I.A.; Shafi, S.; Shameem, N.; Bandh, S.A. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015, 82, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, H.Y.; You, X.L.; Li, Y.H. Diversity of endophytic fungi from roots of Panax ginseng and their saponin yield capacities. Springerplus 2013, 2, 107. [Google Scholar] [CrossRef]
- Xu, W.; Lyu, W.; Duan, C.; Ma, F.; Li, X.; Li, D. Preparation and bioactivity of the rare ginsenosides Rg3 and Rh2: An updated review. Fitoterapia 2023, 167, 105514. [Google Scholar] [CrossRef]
- Chu, L.L.; Bae, H. Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. 2022, 46, 1–10. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Yan, X.; Wang, J.; Zhu, L.; Wang, J.; Li, S.; Kim, Y.M. Oxidative stress, growth inhibition, and DNA damage in earthworms induced by the combined pollution of typical neonicotinoid insecticides and heavy metals. Sci. Total Environ. 2021, 754, 141873. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Zhang, Y.; Lin, T.; Gao, Y.; Cai, Y.; Zhou, C.; Yang, L.; Liu, B.; Dong, S.; et al. Optimization of the proportions of advantageous components in the hypolipidemic “bioequivalent substance system” of Jiang-Zhi-Ning and its mechanism of action. Pharm. Biol. 2023, 61, 1374–1386. [Google Scholar] [CrossRef]
- Peng, W.; Zhong, J.; Yang, J.; Ren, Y.; Xu, T.; Xiao, S.; Zhou, J.; Tan, H. The artificial neural network approach based on uniform design to optimize the fed-batch fermentation condition: Application to the production of iturin A. Microb. Cell Fact. 2014, 13, 54. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, J.; Gao, X.; Wang, K.; Wang, W.; Wang, Q.; Yan, P. Isolation of a novel deep-sea Bacillus circulus strain and uniform design for optimization of its anti-aflatoxigenic bioactive metabolites production. Bioengineered 2019, 10, 13–22. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Wang, C.Q.; Yi, L.W.; Zhao, L.; Zhou, Y.Z.; Guo, F.; Huo, Y.S.; Zhao, D.Q.; Xu, F.; Wang, X.; Cai, S.Q. 177 Saponins, Including 11 New Compounds in Wild Ginseng Tentatively Identified via HPLC-IT-TOF-MS(n), and Differences among Wild Ginseng, Ginseng under Forest, and Cultivated Ginseng. Molecules 2021, 26, 3371. [Google Scholar] [CrossRef]
- Hong, C.E.; Kim, J.U.; Lee, J.W.; Lee, S.W.; Jo, I.H. Diversity of bacterial endophytes in Panax ginseng and their protective effects against pathogens. 3 Biotech. 2018, 8, 397. [Google Scholar] [CrossRef]
- Yan, H.; Jin, H.; Fu, Y.; Yin, Z.; Yin, C. Production of Rare Ginsenosides Rg3 and Rh2 by Endophytic Bacteria from Panax ginseng. J. Agric. Food Chem. 2019, 67, 8493–8499. [Google Scholar] [CrossRef]
- Bu, Q.T.; Zhang, W.Y.; Chen, Q.C.; Zhang, C.Z.; Gong, X.J.; Liu, W.C.; Li, W.; Zheng, Y.N. Anti-diabetic effect of ginsenoside Rb(3) in alloxan-induced diabetic mice. Med. Chem. 2012, 8, 934–941. [Google Scholar]
- Zhu, J.R.; Tao, Y.F.; Lou, S.; Wu, Z.M. Protective effects of ginsenoside Rb(3) on oxygen and glucose deprivation-induced ischemic injury in PC12 cells. Acta Pharmacol. Sin. 2010, 31, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Shi, J.S.; Jiang, Z.L. Inhibitory influence of ginsenoside Rb(3) on activation of strychnine-sensitive glycine receptors in hippocampal neurons of rat. Brain Res. 2005, 1037, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, L.; Ao, W.; Chen, Y.; Li, S.; Huang, Z.; Yu, D.; Dong, Y.; Gu, J.; Zeng, H. Bioinformatics study of the potential therapeutic effects of ginsenoside Rf in reversing nonalcoholic fatty liver disease. Biomed. Pharmacother. 2022, 149, 112879. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.C.; Zhang, H.; Zhang, L.X.; Liu, Z.; Sun, G.Z.; Lei, J.; Qin, Y.X.; Zheng, Y.N.; Li, X.; Pan, H.Y. Biotransformation of ginsenoside Rf to Rh1 by recombinant beta-glucosidase. Molecules 2009, 14, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gu, L.J.; Zhang, D.L.; Wang, Z.; Wang, C.Y.; Li, Z.; Sung, C.K. Microbial conversion of rare ginsenoside Rf to 20(S)-protopanaxatriol by Aspergillus niger. Biosci. Biotechnol. Biochem. 2010, 74, 96–100. [Google Scholar] [CrossRef][Green Version]


| Factors | Potato Concentration (g/L) | Glucose Concentration (g/L) | Inoculation Volume (%) | pH | Incubation Temperature (°C) | Incubation Time (d) |
|---|---|---|---|---|---|---|
| N1 | 100 | 10 | 4 | 6.0 | 28 | 10 |
| N2 | 200 | 35 | 4 | 7.0 | 22 | 6 |
| N3 | 200 | 15 | 2 | 7.0 | 22 | 12 |
| N4 | 200 | 35 | 2 | 5.0 | 28 | 10 |
| N5 | 300 | 25 | 4 | 4.0 | 28 | 12 |
| N6 | 100 | 30 | 2 | 4.0 | 22 | 8 |
| N7 | 300 | 20 | 2 | 6.0 | 28 | 6 |
| N8 | 100 | 25 | 8 | 7.0 | 28 | 8 |
| N9 | 100 | 20 | 8 | 5.0 | 22 | 12 |
| N10 | 300 | 10 | 4 | 5.0 | 22 | 8 |
| N11 | 300 | 30 | 8 | 6.0 | 22 | 10 |
| N12 | 200 | 15 | 8 | 4.0 | 28 | 6 |
| Time (min) | Water (%) | Acetonitrile (%) |
|---|---|---|
| 0 | 77 | 23 |
| 13 | 54 | 46 |
| 33 | 32 | 68 |
| 45 | 32 | 68 |
| 55 | 0 | 100 |
| 60 | 0 | 100 |
| 63 | 77 | 23 |
| Protocols | Multifactor and Interaction Term Stepwise Regression (105-DH) | Multifactor and Square Term Stepwise Regression (105-DP) | Quadratic Polynomial Stepwise Regression (105-ED) |
|---|---|---|---|
| Potato concentration (g/L) | 103.5 | 97.3 | 202.3 |
| Glucose concentration (g/L) | 35.5 | 20.6 | 10.6 |
| Inoculation (%) | 8.2 | 2.1 | 2.0 |
| pH | 2.0 | 2.1 | 7.7 |
| Incubation temperature (°C) | 21.2 | 29.2 | 28.8 |
| Incubation time (d) | 6 | 6 | 6 |
| Predicted concentration (mg/mL) | 3.385 | 2.485 | 2.082 |
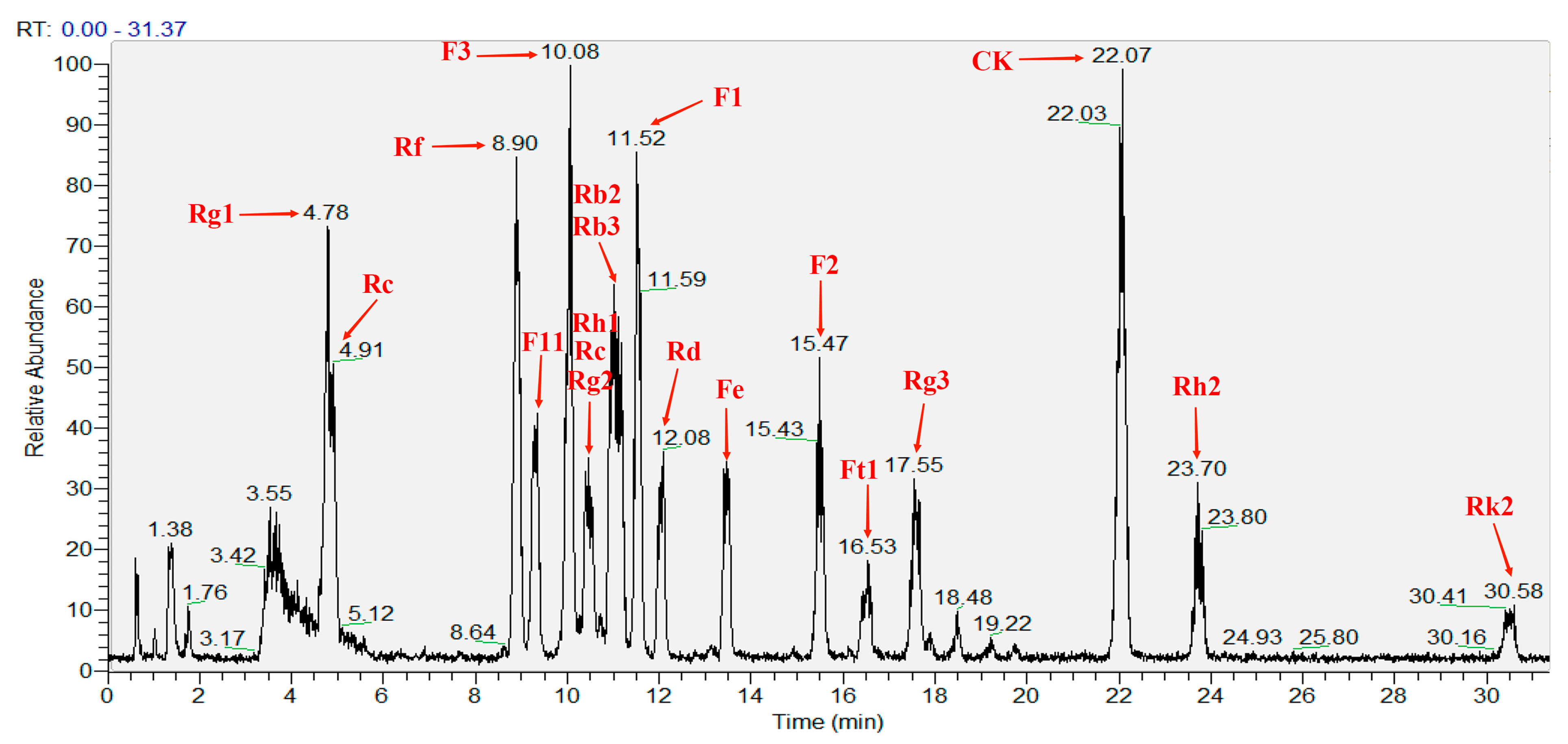
| Standard Name | RT (min) | Area | M/Z (M + Na) | Calculate Mass Errors (ppm) |
|---|---|---|---|---|
| Ginsenoside Rg1 | 4.78 | 210,621,828 | 823.4778 | 3.76 |
| Ginsenoside Re | 4.91 | 185,905,150 | 969.5354 | 3.51 |
| Ginsenoside Rf | 8.90 | 326,583,879 | 823.4778 | 3.76 |
| Pseudo-ginsenoside F11 | 9.34 | 193,324,327 | 823.4777 | 3.89 |
| Ginsenoside F3 | 10.08 | 359,421,580 | 793.4682 | 2.65 |
| Ginsenoside Rh1 | 10.45 | 33,692,890 | 661.4218 | 9.37 |
| Ginsenoside Rc | 10.52 | 56,417,603 | 1101.5771 | 3.54 |
| Ginsenoside Rg2 | 10.72 | 39,293,309 | 807.4778 | 10.03 |
| Ginsenoside Rb2 | 11.01 | 223,210,191 | 1101.5769 | 3.72 |
| Ginsenoside Rb3 | 11.11 | 189,872,269 | 1101.5764 | 4.18 |
| Ginsenoside F1 | 11.52 | 313,614,257 | 661.4257 | 3.48 |
| Ginsenoside Rd | 12.08 | 158,247,318 | 969.5354 | 3.51 |
| Noto-ginsenoside Fe | 13.47 | 159,923,021 | 939.5243 | 4.15 |
| Ginsenoside F2 | 15.47 | 185,211,392 | 807.4832 | 3.34 |
| Noto-ginsenoside Ft1 | 16.53 | 81,671,445 | 939.5245 | 3.94 |
| Ginsenoside Rg3 | 17.55 | 186,817,422 | 807.4833 | 3.22 |
| Ginsenoside CK | 22.07 | 466,725,112 | 645.4313 | 2.79 |
| Ginsenoside Rh2 | 23.70 | 115,834,711 | 645.4308 | 3.56 |
| Ginsenoside Rk2 | 30.58 | 65,261,096 | 627.4207 | 3.03 |
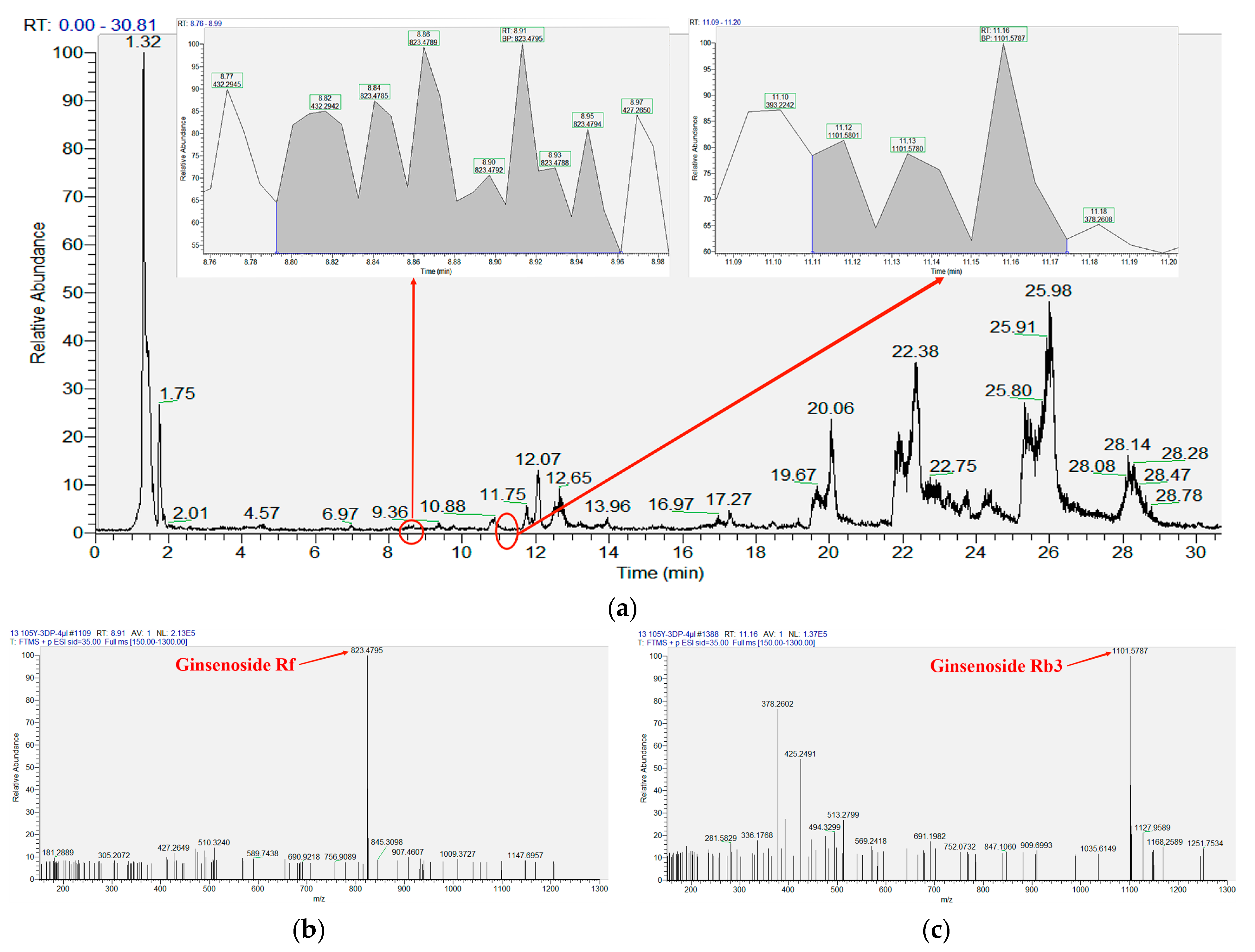
| Compound Name | RT (min) | M/Z (M + Na) | Calculate Mass Errors (ppm) | Area | Concentration (mg/L) |
|---|---|---|---|---|---|
| Ginsenoside Rf | 8.91 | 823.4795 | 1.70 | 2,145,591 | 0.66 |
| Ginsenoside Rb3 | 11.16 | 1101.5787 | 2.09 | 352,600 | 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Wang, J.; Gao, Y.; Gao, X.; Yan, P. Optimization of Fermentation Conditions and Product Identification of a Saponin-Producing Endophytic Fungus. Microorganisms 2023, 11, 2331. https://doi.org/10.3390/microorganisms11092331
Chen Q, Wang J, Gao Y, Gao X, Yan P. Optimization of Fermentation Conditions and Product Identification of a Saponin-Producing Endophytic Fungus. Microorganisms. 2023; 11(9):2331. https://doi.org/10.3390/microorganisms11092331
Chicago/Turabian StyleChen, Qiqi, Jingying Wang, Yuhang Gao, Xiujun Gao, and Peisheng Yan. 2023. "Optimization of Fermentation Conditions and Product Identification of a Saponin-Producing Endophytic Fungus" Microorganisms 11, no. 9: 2331. https://doi.org/10.3390/microorganisms11092331
APA StyleChen, Q., Wang, J., Gao, Y., Gao, X., & Yan, P. (2023). Optimization of Fermentation Conditions and Product Identification of a Saponin-Producing Endophytic Fungus. Microorganisms, 11(9), 2331. https://doi.org/10.3390/microorganisms11092331










