Abstract
Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov. strain T was isolated from a terrestrial, high-altitude hot spring in Tajikistan. This strain is an obligate anaerobic rod and their cells occur singly, in pairs, or as short chains under the optimal growth conditions of a temperature of 65 °C and pH 6.5, with peptone, glucose, and galactose as the preferred substrates. The minimum generation time of this strain is 150 min. Strain T can efficiently degrade feather keratin at 65–75 °C; this unusual feature is also exhibited by a few other members of the Fervidobacterium genus. The total genome size of this bacterial strain is 2,002,515 base pairs, with a C + G content of 39.0%. The maximum digital DNA–DNA hybridization (dDDH) value of 76.9% was observed on comparing the genome of this strain with that of Fervidobacterium pennivorans type strain DSM9078. This study describes the physiological and genomic properties of strain T, with an emphasis on its keratinolytic power and differences from other members of the genus Fervidobacterium.
1. Introduction
The phylum Thermotogae includes extremely thermophilic anaerobic bacteria, with optimum growth temperatures up to 80 °C [1]. They represent one of the deepest bacterial phylogenetic branches based on a 16S rRNA gene sequence analysis. The physiological characteristics of the members of this phylum are similar to those of the early microbes, who were subjected to extreme environmental conditions during the early stages of life on Earth [2]. All members of Thermotogae are Gram-negative rod-shaped cells surrounded by a characteristic sheath-like envelope or “toga” [3,4]. These bacteria have a fermentative metabolism and have been found in a variety of geothermal environments such as oil reservoirs, hydrothermal vents, and terrestrial hot springs [5]. Since the discovery of the first species in the genus, Fervidobacterium [6], six more species of this genus have been isolated from terrestrial hot springs worldwide [7,8,9,10,11,12]. Representatives of the genus Fervidobacterium can grow on a range of complex substrates such as cellulose, starch, or proteinaceous compounds; however, only a few members have been described as being capable of breaking down keratin, including Fervidobacterium islandicum strain AW-1, Fervidobacterium thailandense, and Fervidobacterium pennivorans [9,10,11]. A total of 287 Thermotogae genome sequences, including metagenome assemblies, are currently publicly available in the GenBank database (https://www.ncbi.nlm.nih.gov/data-hub/taxonomy/200918/; accessed on 2 August 2022), thereby representing a huge resource of hyper-stable enzymes with potential biotechnological applications.
Keratin is a structural protein present in feathers, hair, skin, wool, horns, etc. and is one of the most abundant polymers on Earth after cellulose and chitin [13]. The presence of a large number of intramolecular cysteine disulfide bonds and inter- and intra-molecular polar (i.e., hydrogen and ionic bonds) and nonpolar bonds (i.e., hydrophobic interactions) makes keratin an extremely recalcitrant protein [14,15]. Keratin is insoluble in water and resistant to weakly acidic or alkaline conditions. It is also resistant to common proteolytic enzymes such as pepsin and trypsin [16]. Different types of keratins exist in nature, and they are characterized by their secondary structure and sulfur content [15,17]. According to their secondary structure, keratins are categorized into alpha and beta keratins. While alpha keratins are present in the mammalian epidermis (e.g., hair, wool, bristles, etc.), beta keratins are found in reptile scales and feathers [18,19]. The most abundant amino acids in feather keratin are serine, cysteine, glutamine, and proline [20]. The high cysteine content in feather keratin contributes to the abundance of disulfide bonds, which stabilize the main structure of the feather and impart robustness and resistance to proteolysis and chemical destruction [19,21]. The main sources of keratin waste are feathers produced during poultry farming and wool production. It has been estimated that more than 2 million tons of wool and 20,000 tons of feathers are produced annually worldwide, which raises environmental concerns [9]. Keratin-laden tissues represent a major challenge in the animal rendering industry. Two-thirds of animal products are discarded because of undigested keratin-containing compounds. Keratin-laden biowaste is generally hydrolyzed by mechanical or chemical treatments to obtain feedstock, fertilizers, glues, or foils [9,16]; however, keratin is only partially degraded by these processes, which are inefficient and expensive. Furthermore, most of the essential amino acids that could be recovered (e.g., serine, cysteine, and proline) are lost and wasted [22]. The current solution to process the huge amount of keratin-based biowaste is to bury it in landfills or incinerate it. In this context, the metabolic versatility of the Fervidobacterium group, their thermophilic features, and the increasing availability of thermostable proteases for the degradation of proteinaceous biowaste from agriculture and fisheries have immense potential for biotechnological applications [23,24]. In this study, we describe a new Fervidobacterium pennivorans subspecies recovered from a hot spring in Tajikistan, which effectively breaks down native chicken feathers under anaerobic conditions at temperatures up to 75 °C.
2. Materials and Methods
2.1. Sampling and Cultivation
The sample was collected from the Khodja-Obi-Garm geothermal field, located on the Bank of the Varzob River, 50 km North of Dushanbe, Tajikistan, at 38°53′41.45″ N, 68°47′15.64″ E, at an altitude of 1800 m. The temperature and pH of the spring water were 93 °C and 8.5, respectively, with a conductivity of 4378.3 µS/cm [25]. Water samples were placed in 50 mL tightly sealed anaerobic serum flasks and transported to the laboratory at ambient temperature.
Anaerobic enrichment cultures were prepared according to a modified Hungate technique [26]. The mineral medium (MMF) used for the enrichment and cultivation was composed of the following ingredients (per liter): NaCl, 1 g; MgSO4·7H2O, 0.3 g; KCl, 0.3 g; NH4Cl, 0.5 g; CaCl2·2H2O, 0.1 g; and KH2PO4, 0.3 g. A trace element solution (1 mL) was also added, containing (per liter): HCl (25%), 10 mL; FeCl2·4H2O, 1.5 g; CoCl2·6H2O, 190 mg; MnCl2·H2O, 100 mg; ZnCl2, 70 mg; Na2MoO4·2H2O, 36 mg; NiCl2·6H2O, 24 mg; H3BO3, 6 mg; and CuCl2·H2O, 2 mg. Finally, 0.5 mL of resazurin (0.2%) was added to the medium as a redox indicator. After sterilization at 121 °C for 20 min and cooling to approximately 60 °C under continuous flushing with nitrogen gas, 10 mL of the vitamin solution was added, which contained the following ingredients: 4-aminobenzoic acid, 8 mg/L; D(+) biotin, 2 mg/L; nicotinic acid, 20 mg/L; Ca-D(+) pantothenic acid, 10 mg/L; pyridoxamine·2HCl, 30 mg/L; thiamine dichloride, 20 mg/L; and vitamin B12, 10 mg/L. Finally, 25% HCl-cysteine (2 mL) was added, and the pH was adjusted to 7.0 with 1 M HCl. The medium was transferred to 50 or 100 mL serum flasks using the Hungate technique, closed with butyl rubber stoppers, crimped with metal seals [26], and subsequently supplemented with 0.5% peptone and 0.05% yeast extract. The headspace contained nitrogen gas. Following inoculation, the cultures were incubated at 65 °C.
To obtain pure cultures, ten-fold serial dilutions for extinction experiments were carried out, yielding a bacterial isolate termed strain T. A battery of eight flasks filled with 9 mL of MMF medium supplemented with 0.5% peptone and 0.05% yeast extract was prepared. One mL of the original culture was transferred to one of these new flasks to make a 10−1 dilution; from this one, a ten-fold dilution series up to 10−9 was prepared. All the flasks were incubated at 65 °C for 48 h and the process was repeated using inoculum from the highest dilution that yielded growth. The most diluted growth-yielding culture was examined by phase contrast microscopy and appeared to be pure, and this was used as a stock culture for all the following experiments. The strain was stored at 4 °C, and fresh cultures were prepared by inoculating 1 mL of inoculum from the stock culture into 30 mL of an anoxic MMF-based medium as described above. The standard incubation temperature was 65 °C. Additional yeast extract (0.1%) was used for determining the temperature and pH-dependency and when denser cultures were needed.
2.2. Microscopy
A phase-contrast microscope (Nikon Eclipse E400, Guangzhou, China) equipped with an oil immersion lens (100×) and a Nikon DS-Fi3 camera was used to observe the bacterial cells and obtain photomicrographs. Scanning electron microscopy (SEM) was performed at the Molecular Imaging Center (MIC) (https://www.uib.no/ en/rg/mic) (accessed on 11 October 2022) and the Electron Microscopy Laboratory (ELMILab) of the University of Bergen (https://www.uib.no/en/geo/111662/scanning-electron-microscope) (accessed on 11 October 2022) using a JEOL JSM-7400F (Tokyo, Japan) and Zeiss Supra 55VP scanning electron microscope (Oberkochen, Germany), respectively. The samples were fixed in 2.5% glutaraldehyde (diluted in the culture media) and stored at 4 °C for 24 h until further processed, as described previously [27].
2.3. Physiological Characterization
The strain was tested for growth under strict anoxic conditions on the following substrates: sucrose, lactose, arabinose, galactose, mannose, sorbitol, mannitol, starch, CM-cellulose, glucose, and peptone using an MMF medium supplemented with 0.05% yeast extract. After 24 and 48 h of incubation at 65 °C, the growth was determined using phase-contrast microscopy. Doubling of the cell density after 24 or 48 h was scored as positive or slow growth, respectively. To determine the temperature range and optimal temperature for growth, the bacterial strain was subjected to 38, 55, 65, 70, 75 and 80 °C and tested in duplicates using the MMF medium supplemented with peptone (0.5%) and yeast extract (0.1%). A growth curve was prepared by measuring the optical density of the strain at 600 nm using a UV MIN 1240 Shimadzu spectrophotometer. The cells were grown in batch cultures in 50 or 100 mL serum flasks containing 30 mL of MMF supplemented with 0.5% peptone and 0.05% yeast extract. One milliliter aliquots from a freshly inoculated culture incubated at 65 °C were taken every two hours and centrifuged at 12,000× g for 7 min to get rid of the resazurin. The supernatant was discarded, and the cell pellet was resuspended in 1 mL of phosphate buffered saline (PBS) containing (per liter) Na2HPO4 (2.5 g), NaCl (8 g), KCl (0.2 g), and KH2PO4 (0.2 g) prior to an OD measurement. PBS was used as the blank. All the measurements were performed in triplicates. The generation time (g) was calculated from the exponential part of the growth curve using the equation:
where r is the growth rate of the organism, calculated using the equation:
where OD2 and OD1 are the optical densities measured at times t2 and t1, respectively.
2.4. 16S rRNA Gene Sequence Analysis
Genomic DNA was extracted and purified from cell pellets using the GenEluteTM Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s guidelines. The 16S rRNA gene was amplified using the primers 27F (5′-GAGTTTGATCCTGGCTCAG) and 1525R (5′-GAAAGGAGGTGATCCAGCC) [28]. The PCR program recommended for Taq DNA Polymerase (BioLabs, Durham, NC, USA) was used, and the PCR products were sequenced using Sanger sequencing [29]. The 16S rRNA gene sequence of strain T was aligned to the 16S rRNA gene sequences of eight members of the genus Fervidobacterium including the type strains of the six described species using the ClustalW tool [30]. A phylogenetic tree was constructed using the Neighbor-Joining [31] algorithm of the Mega11 software suite [32,33]. The nucleotide sequence distance was measured using the maximum composite likelihood method [34]. The tree was tested by bootstrapping using 1000 replicates [35]. The 16S rRNA gene sequence of Thermosipho africanus was used as the outgroup.
2.5. Genome Sequencing and Phylogenomics
Genomic DNA from strain T was extracted using the GenEluteTM Bacterial Genomic DNA Kit (Sigma-Aldrich) following the manufacturer’s guidelines. The genomic DNA was sequenced using PacBio technology at Eurofins Genomics, Constance, Baden, Germany (https://eurofinsgenomics.eu/) (accessed on 11 October 2022). Processing of the reads and de novo assembly were performed using CLC Genomic Workbench version 21 (QIAGEN Bioinformatics, Redwood City, CA, USA), which yielded a complete genome sequence of 2,002,515 bases with a coverage of 254×. The genome sequence was submitted to the NCBI database (accession No. CP050868) and annotated using an automatic NCBI annotation pipeline. A genome-based phylogenetic analysis was conducted using the Type (Strain) Genome Server (TYGS) available at the DSMZ website (https://tygs.dsmz.de) (accessed on 11 October 2022) [36] and the Ortho Average Nucleotide Identity (ANI) algorithm [37].
2.6. Keratinase Activity Test
The keratinolytic activity was assessed using native chicken breast feathers as the keratin-based substrates. The feathers were washed in a methanol: ethanol solution (1:1) before autoclaving (121 °C, 20 min). Then, 15 ± 5 mg of the feathers was added to 30 mL of MMF medium supplemented with yeast extract (0.05%), to which a 1 mL inoculum of the bacterium was added. An uninoculated flask containing only medium with feather was used as the negative control. The cultures were incubated anaerobically at 70 °C. Every 24 h, the flasks were visually inspected to check for changes in the feather integrity.
3. Results
3.1. Microscopy and Morphology
Following two dilution-to-extinction series in the MMF medium supplemented with peptone, an apparently pure culture was obtained, and termed strain T, with a cellular morphology like other members of the Thermotogales: straight to slightly curved cells (around 1.5 µm × 5 µm) with a membranous sheath-like toga. A spheroid extension of the toga was observed at the end of the cells and was visible under a phase-contrast and scanning electron microscope (Figure 1). Strain T-cells occurred mainly as single entities, although couples or short chains were also common. In addition, some cell aggregates were observed. Spores were not observed. After reaching a stationary phase, many round or irregular translucent cells were observed, indicating cell lysis.
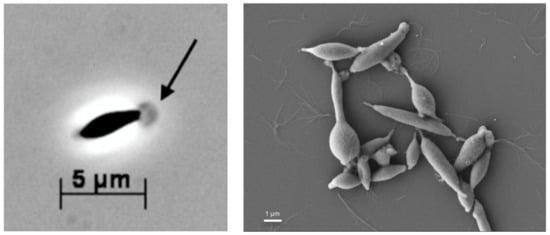
Figure 1.
Photomicrographs of strain T cells taken by a phase-contrast microscope (left) and a scanning electron microscope (right). The arrow points to the globular structure of the toga at the termini of the cells. The bars indicate the size in µm.
3.2. Phylogenetic Identification
A Neighbor-Joining phylogenetic analysis of the 16S rRNA gene sequence of strain T showed that it belongs to the genus Fervidobacterium, with a 98.88% sequence identity to the 16S rRNA gene sequence of Fervidobacterium pennivorans DSM 9078T (CP003260.1). A phylogenetic tree based on the 16S rRNA gene sequence was constructed for strain T and other members of the genus Fervidobacterium (Figure 2). According to the branching order, F. pennivorans strain T was placed between F. pennivorans type strain (DSM 9078) and F. pennivorans strain DYC. A maximum-likelihood [38,39] tree confirmed this branching order (Figure S1).
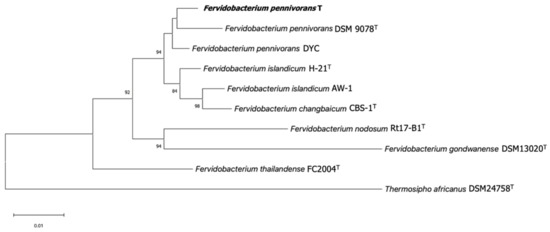
Figure 2.
Neighbor-Joining phylogenetic tree of 16S rRNA gene sequences of members of the genus Fervidobacterium showing the relationship between strain T (in bold) and other representative strains in this genus. The 16S rRNA gene sequence of Thermosipho africanus was used as an outgroup. Bootstrap values as a percentage of 1000 replications are presented at nodes. Bar: 0.01 changes per nucleotide position. Accession numbers: F. pennivorans T (ON652409), Fervidobacterium pennivorans DSM 9078 (HE582749.1), Fervidobacterium pennivorans DYC (NZ_CP011393.1), Fervidobacterium islandicum H-21 (M59176.2), Fervidobacterium islandicum AW-1 (AF434670.1), Fervidobacterium changbaicum CBS-1 (AY878719.2), Fervidobacterium nodosum Rt17-B1 (M59177.1), Fervidobacterium gondwanense DSM 13020 (Z49117.1), Fervidobacterium thailandense FC2004 (JF339226.1), and Thermosipho africanus DSM 24758 (DQ647058.1). All ambiguous positions were removed. There were a total of 1285 positions in the final dataset.
3.3. Physiology
A low concentration of yeast extract (0.05%) was essential for the growth of strain T. The strain clearly possessed a heterotrophic metabolism with a preference for carbohydrates such as lactose, galactose, glucose, and sorbitol, as well as protein-derived substrates such as peptone (Table 1). It also grew well on CM-cellulose, but slowly on sucrose, mannose, or starch. No growth was observed when arabinose or mannitol was used as the substrate. Table 1 shows a physiological comparison of all the described members of the genus Fervidobacterium. F. pennivorans strain T can degrade native chicken feathers at temperatures up to 70 °C, a characteristic that is also exhibited by other fervidobacteria, except F. nodosum and F. gondwanense.

Table 1.
Characteristics of strain T and all described Fervidobacterium type strains.
All the tested strain T cultures were capable of growing on peptone and glucose. Strain T grew within a narrow pH range (6.0 to 7.5 when tested in 0.5 pH increments from pH 5 to 8, with an optimum pH of 6.5). Growth was not observed at NaCl concentrations higher than 3%, with an optimal salt concentration of 1%. Strain T grew within a temperature range of 55 to 75 °C, with an optimal temperature of 65 °C. Growth was not observed at 50 or 80 °C.
The growth curve of F. pennivorans strain T was measured under conditions of 65 °C in MMF supplemented with 0.5% peptone and 0.1% yeast extract (Figure S2). With a fresh inoculum of cells, the lag phase lasted approximately 2 h. The logarithmic phase spanned 11 h, with an estimated minimal generation time of 150 min. After 13 h, the cultures entered a long-lasting stationary phase, never exceeding an OD of 0.2. After 24 h of incubation, the culture flasks did not lose turbidity, and no death phase was observed after a week of incubation.
3.4. Feather Degradation
Strain T degraded native chicken feathers in the MMF medium supplemented with 0.05% yeast extract. The degradation of the breast feathers was almost complete after 48 h of incubation (Figure 3). For wing feathers, which were more robust, degradation started after 7 days of incubation under the same conditions, with a complete degradation within 10 days. Trace amounts of yeast extract were essential for the feather degradation, indicating that feathers cannot serve as the sole carbon and energy source or that yeast extract triggers the keratinolytic activity of the strain.
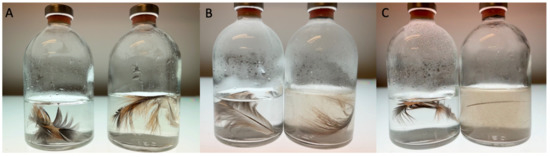
Figure 3.
Degradation of native chicken breast feathers by F. pennivorans subsp. keratinolyticus strain T after 2 days incubation at 70 °C. (A–C) show cultures at times 0, 24, and 48 h, respectively. The left flask in each panel corresponds to the negative control, containing only mineral medium with feather but without inoculation. Both flasks were held at the same temperature for the same inoculation time.
An active feather-degrading culture was examined using scanning electron microscopy. Bacterial cells were found to attach to the keratin fibers (Figure 4), indicating the involvement of cell-bound factors in keratin degradation.
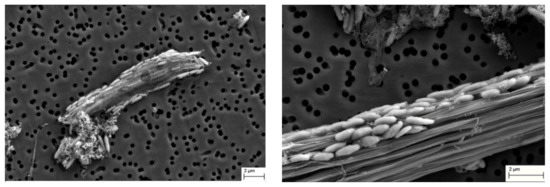
Figure 4.
Scanning electron microscope images of strain T grown in the presence of chicken feather. Images were taken after 36 (left) and 48 h (right) of incubation at 70 °C. Bar = 2 µm.
3.5. Genome Characteristics and Phylogenomics
The genome of F. pennivorans strain T was sequenced using PacBio technology. The complete genome consists of a 2,002,515 bp chromosome, with an average GC-content of 39.0%. By annotation with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP), 1875 protein-coding sequences were found, 194 of which were assigned to a subsystem according to the annotation conducted by the RAST server. Additionally, 57 RNA genes were identified (Table 2).

Table 2.
General characteristics of the members of the Fervidobacterium genus *.
Most annotated genes were involved in metabolic functions, such as protein metabolism, carbohydrate metabolism, amino acid anabolism, and the biosynthesis of cofactors or other secondary metabolites. Twenty-one genes encoding putative protein-degrading enzymes belonging to different families of serine (e.g., fervidolysin) and metallopeptidases were annotated. Enzymes involved in the reduction of disulfide bonds were also identified; thus, strain T possesses an arsenal of keratin-degrading enzymes. Moreover, this approach resulted in the identification of known genes predicted to play a role in the resistance to environmental stresses, such as oxidative stress, cold shock (the CSP family), heat shock, and detoxification. In this regard, the organism seemed to possess many defense mechanisms against heavy metals (i.e., copper, cobalt, zinc, and cadmium) and general drugs via a multidrug resistance efflux pump. Two complete 16S rRNA operons were identified and aligned, showing a 100% sequence identity. Three CRISPR arrays, including 19 genes encoding CRISPR-associated proteins, were identified. Although motility has not been reported in fervidobacteria, 56 genes involved in chemotaxis and flagellar motility were identified. The organism did not contain plasmids.
A pairwise phylogenomic comparison of F. pennivorans strain T against the complete genomes of the members of the genus Fervidobacterium was performed using in silico DNA–DNA hybridization (dDDH) analysis using the TYGS server, average nucleotide identity (ANI) calculation, and phylogenomic tree reconstructions. Based on the TYGS analysis, strain T was most closely affiliated with F. pennivorans type strain DSM9078, sharing a dDDH identity value of 76.9% (Figure S3), which was consistent with the results of the 16S rRNA-based phylogenetic tree (Figure 2). F. pennivorans strain DYC also clustered with strains T and DSM9078, but with dDDH values < 47%, which is significantly below the recommended species threshold value of 70%. This indicates that strain T belongs to the same genome species as that of F. pennivorans DSM9078, whereas strain DYC constitutes a separate Fervidobacterium species; however, the TYGS analysis indicates that strain T is sufficiently different from DSM9078 to be placed in a separate subspecies cluster, which is also supported by certain metabolic differences, for example, the utilization of cellulose and lactose for growth (Table 2). The other Fervidobacterium members formed distinct and separate species clusters, with dDDH values less than 21% compared with strain T and separated by high pseudo-bootstrap values (Figure S3). An Ortho ANI-based phylogenomic tree and heatmap confirmed the TYGS analysis, showing a clustering of strains T, DSM9078, and DYC (Figure 5), with an ANI value of 97.42% between the strains T and DSM9078. Strain DYC shared an ANI value of less than 92% with strains T and DSM9078, supporting the notion that this strain constitutes a separate genome species. Notably, the F. changbaicum strain DSM 17883 and F. islandicum AW-1 clustered closely together and shared dDDH and ANI values of 88.7% and 98.79%, respectively, strongly suggesting that strain AW-1 belongs to F. changbaicum based on the high overall genome sequence similarity. We proposed the name F. pennivorans supsp. keratinolyticus subsp. nov. for strain T as it is a particularly efficient keratin degrader.
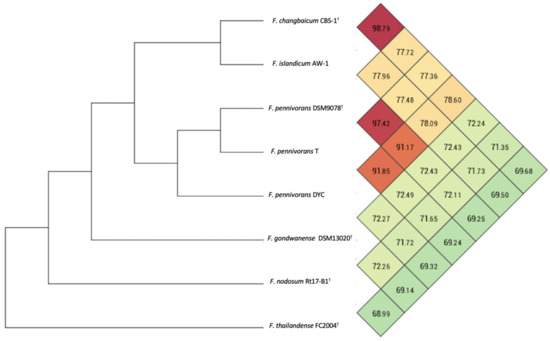
Figure 5.
Ortho ANI heatmap and tree calculated with the genome sequences of members of the genus Fervidobacterium. The values represent the percentage overall similarity among the different genome sequences. The species cut-off was set at 96%.
The genomes of representative strains of Fervidobacterium spp. were also compared using the BLAST Ring Generator (BRIG) [40], with the genome of strain T as a reference (Figure 6). In addition, the genome of strain T was used as an input for three different tools: the CRISPRCasFinder [41] program was run to detect CRISPR/Cas9 clusters, as shown in Figure 6. Moreover, regions of probable horizontal transfer origin were predicted using the IslandViewer web server [42]. These predicted genomic islands are also displayed in Figure 6, which includes, among other features, a zinc metallopeptidase (QIV78721.1) and nine transposase-related genes. Finally, a region identified as a putative prophage by the PHASTER pipeline [43,44] can also be seen in the same figure, partially overlapped by one of the predicted genomic islands.
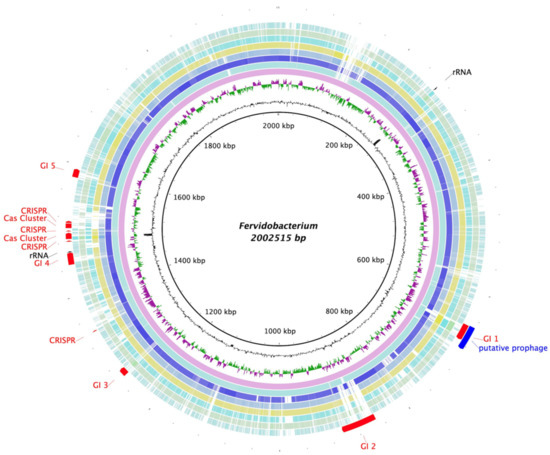
Figure 6.
Blast Ring Generator (BRIG) representation of the comparison of the Fervidobacterium species’ genomes using strain T as a reference. The innermost circles show strain T’s G + C content (black) and sections with a GC skew (purple/green). The external circles represent the different genomes of the members of Fervidobacterium genus, starting with F. pennivorans strain T (pink) and followed, from inside to outside, by F. pennivorans DSM9078 (pale blue), F. pennivorans DYC (navy blue), F changbaicum CBS-1 (blue grey), F. islandicum AW-1 (yellow), F. nodosum Rt17-B1 (turquoise), F. gondwanense (green), and F. thailandense (sky blue). Finally, the most external area of the figure shows the 16S rRNA genes (black), the predicted genomic islands, CRISPR and Cas9 clusters (red), and a putative prophage, all found in strain T’s genome.
A full genome alignment of F. pennivorans strain T and type strain DSM 9078 was performed using Mauve [45] (Figure 7). By annotation with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP), 41 transposases were predicted in DSM 9078 and 21 in strain T. Figure 6 shows this full genome alignment, with the position of the predicted transposases indicated by arrows. In addition, three large genomic inversions were detected when aligning these two genomes, all of which were flanked by transposase-encoding genes.
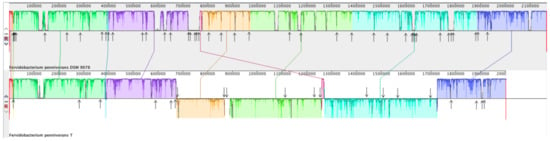
Figure 7.
Full genome alignment of Fervidobacterium pennivorans DSM9078 (species type strain) and F. pennivorans strain T using Mauve. The upper panel represents the DSM9078 genome and the bottom one corresponds to strain T. Fragments with the same color belong to homologous regions, and sequences below the horizontal division correspond to inverted regions. The arrows point to transposase genes, as predicted by genome annotation.
4. Discussion
The new isolate described in this study belongs to the F. pennivorans species, although some minor differences between this strain and the species-type strain (DSM9078) could be identified, suggesting that these two bacteria may be classified as different subspecies. The strain grew well on substrates such as glucose, peptone, and galactose; however, turbid cultures were obtained only when yeast extract was added as a supplemental nutrient, suggesting the need for a mixture of the auxiliary nutrients for growth. This behavior is not unique to this organism and has also been described for other strains belonging to the F. pennivorans species [46] as well as for many other strict anaerobes. The first batch growth curve for a member of the F. pennivorans species was established. It showed a lag phase dependent on both physical and physiological conditions: an exponential phase of 11 h with a generation time of 150 min and a long stationary phase. The total genome size of strain T was 2,002,515 base pairs, similar to those of the other members of the genus Fervidobacterium, with a similar number of coding gene sequences. Whole-genome alignment with the type strain showed three large, inverted regions flanked by transposase-encoding genes. Chromosomal rearrangements are not unusual for this group and have also been reported in other Thermotogales species. Chromosomal rearrangements have been suggested to affect species evolution [47,48].
The phylogenetic analyses showed that strain T belongs to the same species group as the type strain (DSM 9078), whereas the DYC strain forms a deeper and separate branch and shares less than a 92% ANI value with strain T and DSM9078, suggesting that strain DYC constitutes a separate Fervidobacterium species. However, F. changbaicum CB5-1 and F. islandicum AW-1 were sufficiently similar to be grouped into the same genome species, with an ANI value of 98.79% (Figure 4). According to the TYGS and OrthoANI analyses, strain T should be considered a separate subspecies.
Analysis of the genome of F. pennivorans strain T allowed the identification of several keratinase enzyme candidates. The strain showed strong keratinolytic activity and could completely degrade chicken feathers after 48 h incubation at 75 °C. Furthermore, scanning electronic microscope images revealed that this bacterium physically attaches to the feathers during the degradation process, suggesting that the cell wall or membrane-bound enzymes participate in the process. Nevertheless, the features required for keratin degradation are not fully understood, and most research in this field centers on screening novel microorganisms with keratinolytic activity, as well as their secreted protease arsenal. Furthermore, for other complex substrates, such as cellulose, it has been suggested that the degradation of keratin does not occur by the action of a single enzyme but requires a mixture of different enzymes with hydrolytic activity. F. pennivorans is an anaerobic and thermophilic bacterium, making it especially convenient for industrial processes, as neither oxygen supply nor refrigeration systems are required.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms11010022/s1: Figure S1 (Phylogenetic tree based on the 16S rRNA gene inferred by the Maximum Likelihood method), Figure S2 (Growth curve of F. pennivorans subsp. keratinolyticus strain T in anaerobic MMF medium supplemented with 0.1 % yeast extract and 0.5 % glucose) and Figure S3 (Phylogenomic tree of F. pennivorans subsp. keratinolyticus strain T and related Fervidobacterium species and strains constructed by using the TYGS genome server (https://tygs.dsmz.de, accessed on 11 October 2022)).
Author Contributions
Conceptualization, N.-K.B.; field sampling, K.B., M.D. and N.-K.B.; isolation, R.J.-L.; physiological characterization, R.J.-L. and E.M.; phylogenetic and genome analysis, R.J.-L., E.M. and N.-K.B.; electron microscopy, R.J.-L.; data curation, R.J.-L.; writing—original draft preparation, R.J.-L., E.M. and N.-K.B.; writing—review and editing, N.-K.B.; visualization, R.J.-L.; supervision, N.-K.B.; funding acquisition, N.-K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the ERA-NET Cofund on Food Systems and Climate (FOSC) under the European Union’s Horizon 2020 Research and Innovation Program (grant number 862555), the Research Council of Norway (grant number 328955), and the Norwegian Directorate for Higher Education and Skills (grant number CPEA-LT-2017/10061).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete genome sequence is available under GenBank accession number CP050868.
Acknowledgments
We are grateful to the Molecular Imaging Center (MIC) (https://www.uib.no/en/rg/mic) (accessed on 11 October 2022) and ELMILAB (https://www.uib.no/en/elmi) (accessed on 11 October 2022) at the University of Bergen for performing the electron microscopy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Frock, A.D.; Notey, J.S.; Kelly, R.M. The genus Thermotoga: Recent developments. Environ. Technol. 2010, 31, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Achenbachrichter, L.; Gupta, R.; Stetter, K.O.; Woese, C.R. Were the Original Eubacteria Thermophiles. Syst. Appl. Microbiol. 1987, 9, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, V.; Gupta, R.S. Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Anton. Leeuw. Int. J. G. 2015, 108, 1281. [Google Scholar] [CrossRef]
- Huber, R.; Langworthy, T.A.; Konig, H.; Thomm, M.; Woese, C.R.; Sleytr, U.B.; Stetter, K.O. Thermotoga-maritima Sp-Nov Represents a New Genus of Unique Extremely Thermophilic Eubacteria Growing up to 90-Degrees-C. Arch. Microbiol. 1986, 144, 324–333. [Google Scholar] [CrossRef]
- Gupta, R.S.; Bhandari, V. Phylogeny and molecular signatures for the phylum Thermotogae and its subgroups. Anton. Leeuw. Int. J. G. 2011, 100, 1–34. [Google Scholar] [CrossRef]
- Patel, B.K.C.; Morgan, H.W.; Daniel, R.M. Fervidobacterium nodosum Gen-Nov and Spec-Nov, a New Chemoorganotrophic, Caldoactive, Anaerobic Bacterium. Arch. Microbiol. 1985, 141, 63–69. [Google Scholar] [CrossRef]
- Andrews, K.T.; Patel, B.K.C. Fervidobacterium gondwanense sp. nov., a new thermophilic anaerobic bacterium isolated from nonvolcanically heated geothermal waters of the Great Artesian Basin of Australia. Int. J. Syst. Bacteriol. 1996, 46, 265–269. [Google Scholar] [CrossRef]
- Cai, J.; Wang, Y.; Liu, D.; Zeng, Y.; Xue, Y.; Ma, Y.; Feng, Y. Fervidobacterium changbaicum sp. nov., a novel thermophilic anaerobic bacterium isolated from a hot spring of the Changbai Mountains, China. Int. J. Syst. Evol. Microbiol. 2007, 57, 2333–2336. [Google Scholar] [CrossRef]
- Friedrich, A.B.; Antranikian, G. Keratin Degradation by Fervidobacterium pennavorans, a Novel Thermophilic Anaerobic Species of the Order Thermotogales. Appl. Environ. Microbiol. 1996, 62, 2875–2882. [Google Scholar] [CrossRef]
- Huber, R.; Woese, C.R.; Langworthy, T.A.; Kristjansson, J.K.; Stetter, K.O. Fervidobacterium-Islandicum sp.-nov., a New Extremely Thermophilic Eubacterium Belonging to the Thermotogales. Arch. Microbiol. 1990, 154, 105–111. [Google Scholar] [CrossRef]
- Kanoksilapatham, W.; Pasomsup, P.; Keawram, P.; Cuecas, A.; Portillo, M.C.; Gonzalez, J.M. Fervidobacterium thailandense sp. nov., an extremely thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2016, 66, 5023–5027. [Google Scholar] [CrossRef]
- Podosokorskaya, O.A.; Merkel, A.Y.; Kolganova, T.V.; Chernyh, N.A.; Miroshnichenko, M.L.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Fervidobacterium riparium sp. nov., a thermophilic anaerobic cellulolytic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2011, 61, 2697–2701. [Google Scholar] [CrossRef]
- Gerday, C.; Glansdorff, N.; American Society for Microbiology. Physiology and Biochemistry of Extremophiles; ASM Press: Washington, DC, USA, 2007; p. xvi. 429p. [Google Scholar]
- Parry, D.A.; Crewther, W.G.; Fraser, R.D.; MacRae, T.P. Structure of alpha-keratin: Structural implication of the amino acid sequences of the type I and type II chain segments. J. Mol. Biol. 1977, 113, 449–454. [Google Scholar] [CrossRef]
- Shavandi, A.; Silva, T.H.; Bekhit, A.A.; Bekhit, A.E.A. Keratin: Dissolution, extraction and biomedical application. Biomater. Sci. 2017, 5, 1699–1735. [Google Scholar] [CrossRef]
- Lee, Y.J.; Dhanasingh, I.; Ahn, J.S.; Jin, H.S.; Choi, J.M.; Lee, S.H.; Lee, D.W. Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1. Biochem. Biophys. Res. Commun. 2015, 468, 927–933. [Google Scholar] [CrossRef]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature—A new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef]
- Qiu, J.; Wilkens, C.; Barrett, K.; Meyer, A.S. Microbial enzymes catalyzing keratin degradation: Classification, structure, function. Biotechnol. Adv. 2020, 44, 107607. [Google Scholar] [CrossRef]
- Li, Q. Progress in Microbial Degradation of Feather Waste. Front. Microbiol. 2019, 10, 2717. [Google Scholar] [CrossRef]
- Laba, W.; Zarowska, B.; Chorazyk, D.; Pudlo, A.; Piegza, M.; Kancelista, A.; Kopec, W. New keratinolytic bacteria in valorization of chicken feather waste. AMB Express 2018, 8, 9. [Google Scholar] [CrossRef]
- Tesfaye, T.; Sithole, B.; Ramjugernath, D.; Chunilall, V. Valorisation of chicken feathers: Characterisation of chemical properties. Waste Manag. 2017, 68, 626–635. [Google Scholar] [CrossRef]
- Williams, C.M.; Lee, C.G.; Garlich, J.D.; Shih, J.C.H. Evaluation of a Bacterial Feather Fermentation Product, Feather-Lysate, as a Feed Protein. Poultry. Sci. 1991, 70, 85–94. [Google Scholar] [CrossRef]
- Papadopoulos, M.C. Effect of Processing on High-Protein Feedstuffs—A Review. Biol. Waste 1989, 29, 123–138. [Google Scholar] [CrossRef]
- Conners, S.B.; Mongodin, E.F.; Johnson, M.R.; Montero, C.I.; Nelson, K.E.; Kelly, R.M. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. Fems. Microbiol. Rev. 2006, 30, 872–905. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Tello, E.; Fardeau, M.L.; Thomas, P.; Ramirez, F.; Casalot, L.; Cayol, J.L.; Garcia, J.L.; Ollivier, B. Petrotoga mexicana sp nov., a novel thermophilic, anaerobic and xylanolytic bacterium isolated from an oil-producing well in the Gulf of Mexico. Int. J. Syst. Evol. Micr. 2004, 54, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Dzhuraeva, M.M.; Margaryan, A.A. (Eds.) Microbial Diversity of High-Altitude Geothermal Springs in Tajikistan; Springer: Singapore, 2021; Volume 32. [Google Scholar]
- Miller, T.L.; Wolin, M.J. Serum Bottle Modification of Hungate Technique for Cultivating Obligate Anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [CrossRef]
- Mashzhan, A.; Javier-Lopez, R.; Kistaubayeva, A.; Savitskaya, I.; Birkeland, N.K. Metagenomics and Culture-Based Diversity Analysis of the Bacterial Community in the Zharkent Geothermal Spring in Kazakhstan. Curr. Microbiol. 2021, 78, 2926–2934. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Neron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing, G.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- DeBoy, R.T.; Mongodin, E.F.; Emerson, J.B.; Nelson, K.E. Chromosome evolution in the Thermotogales: Large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 2006, 188, 2364–2374. [Google Scholar] [CrossRef]
- Xu, Z.; Puranik, R.; Hu, J.; Xu, H.; Han, D. Complete genome sequence of Thermotoga sp. strain RQ7. Stand. Genom. Sci. 2017, 12, 62. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).