Abstract
Aim: To develop a reproducible biofilm model consisting of Enterococcus faecalis (E. faecalis) and Porphyromonas gingivalis (P. gingivalis) and to evaluate the interaction between the two bacterial species. Methodology: E. faecalis and P. gingivalis were grown in mono-culture, sequential, and co-culture models for 96 h in a 96-well polystyrene microtiter plate under both aerobic and anaerobic conditions separately. The viability of the two bacterial species in the biofilms was quantified by polymerase chain reaction (qPCR). Biofilm thickness and protein contents were measured using confocal laser scanning microscopy (CLSM). Two-way analysis of variance (ANOVA) was performed to analyze cell viability and biofilm thickness among different culture models cultivated under either aerobic or anaerobic conditions. The level of significance was set at p < 0.05. Results: Different culture models tested did not show any significant difference between the viable cell counts of both E. faecalis and P. gingivalis cultivated under aerobic and anaerobic conditions (p > 0.05). Biofilm was significantly thicker (p < 0.05) in the co-culture models compared to the mono-culture and sequential models. Protein contents in the biofilms were more pronounced when both bacterial species were co-cultured under aerobic conditions. Conclusions: E. faecalis appeared to shield P. gingivalis and support its continued growth in oxic (aerobic) conditions. The co-culture model of E. faecalis and P. gingivalis produced a significantly thicker biofilm irrespective of the presence or absence of oxygen, while increased protein contents were only observed in the presence of oxygen.
1. Introduction
The oral cavity is a well-oxygenated environment, where the elevated oxygen tension modulates oxygen-tolerating bacterial cells to increase their enzymatic and non-enzymatic reduction of molecular oxygen to superoxide anions. This results in the formation of hydrogen peroxide and oxygen by dismutation, and the former reacts with superoxide anions to form hydroxyl ions in the presence of iron complexes [1]. These free oxygen radicals are highly reactive and able to cleave nucleic acids and oxidize essential proteins and lipids [2,3]. In contrast, obligate anaerobic microorganisms, such as P. gingivalis, do not possess the mechanism of anti-oxidation. Thus, the reactive oxygen species must be detoxified to minimize the undesirable effects on the obligate anaerobic microorganisms.
P. gingivalis, a group of obligate anaerobic bacteria, is highly associated with periodontal diseases, ranging from reversible gingivitis to irreversible periodontitis [4]. Along with Tanneralla forsythia (T. forsythia) and Treponema denticola (T. denticola), they form the “red complex” and have been implicated as the primary causative pathogens in periodontal diseases [5,6]. They progressively degrade the periodontal collagen and non-collagenous tissues, forming periodontal pockets [7]. Enterococcus faecalis (E. faecalis) is the most frequently isolated bacterial species from symptomatic root canal-treated teeth, with reported prevalence in up to 90% of cases [8,9,10]. E. faecalis is often found in secondary or persistent cases due to its ability to survive in harsh environments with nutrient deprivation and high alkalinity despite the presence of intracanal medicaments [11,12,13,14]. The pathogenicity and difficulty of their eradication have been attributed to the ability of E. faecalis to form biofilms, which can be 1000-fold more resistant to antimicrobials than their planktonic counterparts [15,16]. Both E. faecalis and P. gingivalis have been frequently isolated in infected root canals and periodontal pockets in failed cases by culturing and molecular identification techniques [17,18].
P. gingivalis can tolerate low levels of oxygen (6–10%) and reach a steady state of growth depending on the availability of hemin [19,20]. Fusobacterium nucleatum (F. nucleatum), another obligate anaerobic bacteria, supports the growth of P. gingivalis in an unfavorable oxygenated and carbon dioxide-depleted environment [19,21]. F. nucleatum creates a microenvironment with reduced oxygen tension that protects P. gingivalis within the niche [19]. A recent study demonstrated the potential of Candida albicans (C. albicans), a yeast-like fungus, in protecting anaerobic bacteria in an aerobic environment [22]. As a facultative anaerobe, C. albicans creates a hypoxic microenvironment within the fungal biofilms, which is conducive to the growth of P. gingivalis [22]. To date, there is no research conducted to study the interaction between E. faecalis and P. gingivalis when they co-exist. Therefore, this study aimed (i) to develop a simple and reproducible biofilm model consisting of E. faecalis and P. gingivalis and (ii) to study the interaction between these two bacterial species under both aerobic and anaerobic conditions separately, either using single- or dual-species culture models.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
E. faecalis (ATCC 47077) and P. gingivalis (ATCC 33277) were used in this study. The strains were stored at −70 °C as freeze-dried cultures and recovered on blood agar supplemented with 40 mL of horse blood and 8 mL of Hemin-Vitamin K solution (Sigma-Aldrich, Burlington, MA, USA) before cultivation in Brain-heart Infusion Broth (BHI) (Thermo Fisher Scientific™, Oxoid, UK). E. faecalis was cultivated under aerobic conditions at 37 °C for 24 h, while P. gingivalis was cultivated under anaerobic conditions (85% N2, 10%H2, and 5% CO2) at 37 °C for 24 h. For dual-species biofilm (sequential and co-culture) development, both bacterial species were cultivated under aerobic or anaerobic conditions separately and independently for 48 h, with media replenished every 48 h before subjecting the biofilm to further culturing under a different condition.
2.2. Development of Biofilm and Incubation
Cultures of E. faecalis and P. gingivalis were centrifuged and washed with phosphate-buffered solution (PBS) after the supernatant removal. Fresh Pg Broth (TSB, Sigma Aldrich; Yeast extract, Thermo Fisher Scientific™) was added to the pellet. Bacterial density was adjusted to 0.271–0.279 (2 × 108 CFU/mL) using a spectrophotometer (Beckman Coulter DU530 Life Science UV/Vis Spectrophotometer, Brea, CA, USA) at an optical density of 1 and a wavelength of 660 nm, according to MacFarland Standard scale no. 2. Then, 10-fold serial dilutions of the bacterial suspension were made up to 10−2 in tubes containing sterilized Pg broth. A total of 200 µL of the bacterial suspension was deposited into 96-well microtiter plates in quadruplicates, and 200 µL of sterilized Pg broth was deposited into the same well plates in triplicates to serve as the negative control. The same procedure was repeated in another two microtiter plates. For dual-species cultures, i.e., sequential or co-culture models, an equal volume of the two mono-species cultures were combined. All the specimens were incubated either aerobically or anaerobically for 96 h, and the culture medium was replenished after the first 48 h.
2.3. Experimental Groupings
Three different models of dual-species biofilm were developed in the flat-bottomed 96-well polystyrene microtiter plates in quadruplicates. The experimental culture models were divided into the following groups:
Sequential model I: E. faecalis biofilm was formed by seeding bacterial cells into the microtiter plate wells (100 µL; 1 × 106 cells/well) in Pg broth for 48 h of aerobic incubation. Then, the medium was removed and replenished. P. gingivalis (100 µL; 1 × 106 cells/well) were added and cultured for another 48 h under aerobic and anaerobic conditions separately. After the supernatant removal, each well was washed with 200 µL of PBS for further analysis.
Sequential model II: P. gingivalis biofilm was formed by seeding bacterial cells into the microtiter plate wells (100 µL; 1 × 106 cells/well) in Pg broth for 48 h of anaerobic incubation. The medium was then removed and replenished. E. faecalis (100 µL; 1 × 106 cells/well) were added and cultured for another 48 h under aerobic and anaerobic conditions separately. Afterward, the supernatants were removed. Each well was washed with 200 µL of PBS for further analysis.
Co-culture model: Both E. faecalis and P. gingivalis were grown simultaneously for 96 h under aerobic and anaerobic conditions separately. E. faecalis (100 µL; 1 × 106 cells/well) and P.gingivalis (100 µL; 1 × 106 cells/well) cells were seeded into the microtiter plate at the same time and then incubated for 96 h under aerobic and anaerobic conditions separately. The medium was refreshed after the first 48 h. After 96 h, each well was washed with 200 µL of PBS for further analysis.
Positive control: In parallel with each of these dual-species biofilm models, E. faecalis (200 µL; 1 × 106 cells/well) and P. gingivalis (200 µL; 1 × 106 cells/well) were cultured as mono-species under aerobic and anaerobic conditions separately to serve as positive controls.
All experiments were repeated on three independent occasions.
2.4. Quantitative Analysis of Bacterial Cell Viability
After removing the supernatant, biofilms were washed with 200 µL of PBS, detached using the pipette tips, and transferred to the 1.5 mL Eppendorf tubes (SARSTEDT AG & Co. KG, Sarstedtstraß, Nümbrecht, Germany). The biofilm was re-suspended in 1 mL PBS and divided into two groups. One group was used for Propidium monoazide (PMA) staining (PMAxx™, Biotium, Fremont, CA, USA), and another group was used for DNA extraction.
For PMA staining, 1.25 µL of PMA was added to the biofilm suspension and left on ice for 10 min. Subsequently, samples were exposed to PMA-Lite LED Photolysis Device (Biotium Inc., Fremont, CA, USA) for 5 min and subjected to further centrifugation to remove the supernatant. The DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Compositional analyses were enumerated using viability PCR (v-PCR). In brief, 1 µL of extracted DNA was added to 10 µL TaqMan Mix (Life Technologies, Thermo Fisher Scientific™, Waltham, MA, USA), MilliQ water (Merck Millipore, Burlington, MA, USA), and 1 µL of 10 µM forward/reverse primers (Life Technologies, Thermo Fisher Scientific™) and Taqman probes (Life Technologies, Thermo Fisher Scientific™) that were bacterial species-specific. The probes and primers are listed in Table 1. The thermal parameters used were as follows: (i) denaturation at 95 °C for 2 min; (ii) 40 cycles at 95 °C for 10 s; (iii) 58 °C for 30 s, using the StepOne plus Real-Time PCR System (Applied Biosystems™, Thermo Fisher Scientific™); and (iv) StepOnePlus software (Applied Biosystems™, Thermo Fisher Scientific™) for data compilation. Samples were quantified by calculating the colony-forming equivalent (CFE) based on an established standard curve of microbial colony-forming units ranging from 1 × 103 to 108 CFU/mL of E. faecalis and P. gingivalis, which DNA was extracted and processed with RT-PCR’s procedures. All samples were processed in duplicates in the v-PCR, with negative control samples containing water, primers, and master-mix only to rule out possible DNA contamination.

Table 1.
(a) Species-specific primer sequences used in this study; and (b) Species-specific DNA probe sequences used in this study.
2.5. Biofilm Protein and Thickness Analysis with CLSM
The biofilm samples were stained with SyPRO biofilm matrix stain (Thermo Fisher Scientific™, Invitrogen™) and counterstained with Syto9 (Thermo Fisher Scientific™, Invitrogen™). In brief, planktonic cells were removed and washed gently with 200 µL of PBS. Biofilms were then stained, and the z-stacks were obtained from 5 different spots using CLSM (FV1000, Olympus, Shinjuku, Tokyo, Japan). The images were processed and viewed using FV10-ASW 4.2 (FV10, Olympus). Biofilm thicknesses were measured using the z-dimension of the CLSM images.
2.6. Data Analysis
All assays were carried out in three independent experiments, and the results were expressed as mean ± SD. Statistical analysis of the data was performed by two-way analysis of variance (ANOVA) using IBM SPSS Statistics version 26 (SPSS, Chicago, IL, USA). The level of significance was set at p < 0.05.
3. Results
3.1. Sequential Model I
The viability of E. faecalis in Sequential model I under aerobic or anaerobic conditions is similar to that of E. faecalis in monospecies culture (p > 0.05) (Figure 1a). The log10/mL values for E. faecalis under aerobic and anaerobic conditions were 8.30 and 8.22, respectively; in the positive control group, the values were 8.25 and 8.23, respectively (Figure 1a). The dual-species culture model and oxygen tension did not significantly alter the viability of E. faecalis (p > 0.05). The viability of P. gingivalis in Sequential model I under aerobic or anaerobic conditions is similar to that of P. gingivalis in monospecies culture (p > 0.05) (Figure 1b). The log10/mL values for P. gingivalis under the aerobic and anaerobic conditions were 3.99 and 5.59, respectively, whereas the values were 4.10 and 4.72 in the positive control group, respectively (Figure 1b).
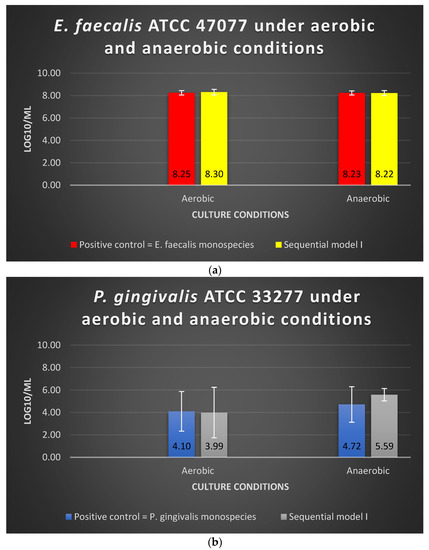
Figure 1.
The viability of (a) E. faecalis and (b) P. gingivalis, cultured under aerobic and anaerobic conditions based on the Sequential model I and mono-culture models. Data represent the mean and standard deviation.
3.2. Sequential Model II
Under aerobic or anaerobic conditions, the amount of viable E. faecalis in Sequential model II was similar to that of the E. faecalis as monospecies in the positive control group (p > 0.05) (Figure 2a). The log10/mL values for E. faecalis monospecies culture under aerobic and anaerobic conditions were 8.10 and 8.13, respectively, whereas the values for Sequential model II were 8.01 and 8.08, respectively (Figure 2a). The dual-species culture model and oxygen tension did not significantly alter the viable cell counts of E. faecalis (p > 0.05).
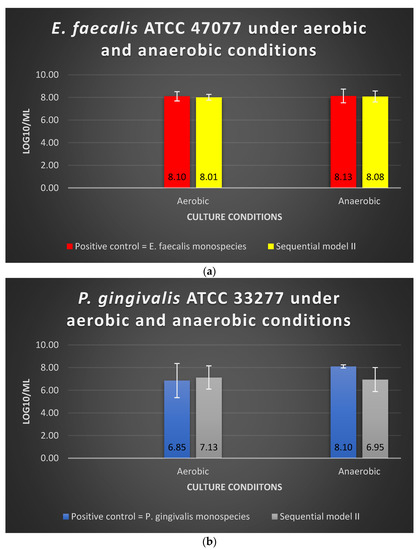
Figure 2.
The viability of (a) E. faecalis and (b) P. gingivalis, cultured independently under aerobic and anaerobic conditions based on Sequential model II and mono-culture models. Data represent the mean and standard deviation.
Under aerobic conditions, the amount of viable P. gingivalis in the Sequential model II was similar to that of the P. gingivalis as mono-culture in the positive control group (p > 0.05) (Figure 2a). Under anaerobic conditions, there was a 1-log reduction observed when P. gingivalis was grown in the Sequential model II without significant difference (p > 0.05) (Figure 2b). The log10/mL values for P. gingivalis as mono-culture under the aerobic and anaerobic conditions were 6.85 and 8.10, respectively, whereas the values for Sequential model II were 7.13 and 6.95, respectively.
3.3. Co-Culture Model
There was no significant alteration (p > 0.05) in the viable cell counts of E. faecalis and P. gingivalis in the co-culture models compared with their mono-culture counterparts. The log10/mL values for E. faecalis in mono-culture under aerobic and anaerobic conditions were 8.25 and 8.23, respectively, whereas the values in the co-culture model were 8.16 and 8.19, respectively (Figure 3). The log10/mL values for P. gingivalis in mono-culture under aerobic and anaerobic conditions were 4.10 and 4.72, respectively, whereas the values for the co-culture model were 4.44 and 3.74, respectively (Figure 3). Oxygen tension did not significantly impact the viable cell counts of these two bacterial species when they were cultured simultaneously (p > 0.05).
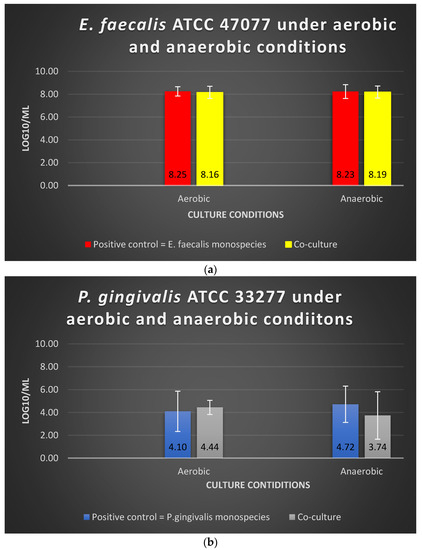
Figure 3.
The viability of (a) E. faecalis and (b) P. gingivalis, cultured independently under aerobic and anaerobic conditions based on the co-culture and mono-culture models. Data represent the mean and standard deviation.
3.4. Biofilm Thickness
In Sequential model I, no significant difference was found in biofilm thickness (p > 0.05) between the mono-cultures and dual-species sequential models. The presence or absence of oxygen did not affect the biofilm formation. However, E. faecalis formed thicker biofilms than P. gingivalis in monospecies culture, i.e., 12.7 μm and 9.1 μm, respectively, under aerobic conditions (p < 0.05). For Sequential model I, a similar biofilm thickness in the dual-species sequential model was observed under aerobic and anaerobic conditions (Figure 4a,b).
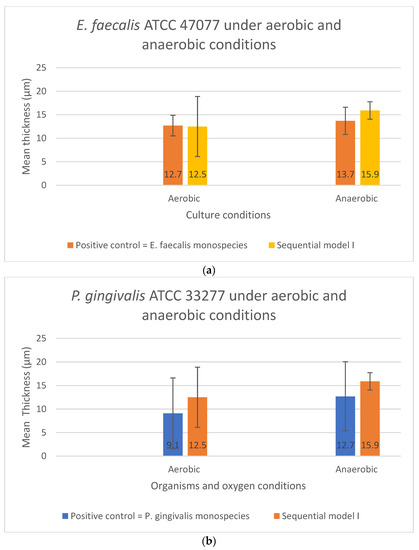
Figure 4.
The biofilm thickness of (a) E. faecalis and (b) P. gingivalis cultured independently under aerobic and anaerobic conditions based on Sequential model I and mono-culture models. Data represent the mean and standard deviation.
As shown in Figure 5a,b, no significant difference in the biofilm thickness could be observed when the two bacterial species were grown sequentially in Sequential model II compared with their monospecies culture counterparts (p > 0.05). The presence or absence of oxygen did not affect the biofilm formation and thickness. In contrast, significant differences in the biofilm thickness were found in both the E. faecalis (p = 0.029) and P. gingivalis (p = 0.043) between their co-culture and mono-culture biofilm models under both aerobic and anaerobic conditions (Figure 6a,b). In fact, a significant increase in biofilm thickness was observed in co-cultures (p < 0.05) compared to the positive controls. When co-cultured aerobically, the biofilm formed was approximately one-fold thicker than their monospecies culture counterparts (Figure 6a,b).
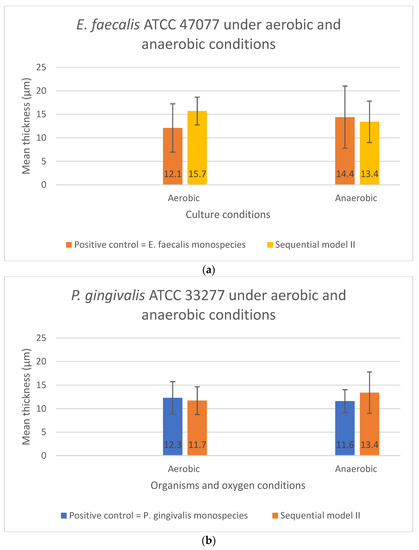
Figure 5.
The biofilm thickness of (a) E. faecalis and (b) P. gingivalis cultured independently under aerobic and anaerobic conditions based on the Sequential model II and mono-culture models. Data represent the mean and standard deviation.
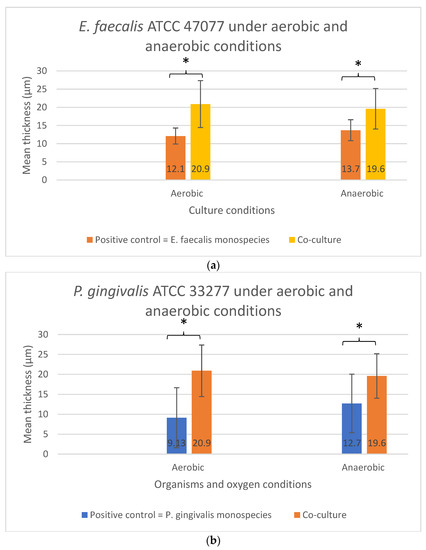
Figure 6.
The biofilm thickness of (a) E. faecalis and (b) P. gingivalis cultured independently under aerobic and anaerobic conditions based on the co-culture and mono-culture models. Asterisks (*) denote a statistically significant difference (p < 0.05). Data represent the mean and standard deviation.
3.5. Biofilm Protein Visualization
Under the aerobic condition, no difference in the protein contents was observed from the biofilms in the Sequential models I and II. The protein contents in the biofilms for both E. faecalis (Figure 7b) and P. gingivalis (Figure 7f) were more pronounced in the co-culture models compared with their mono-culture counterparts under aerobic conditions (Figure 7a,e). In anaerobic conditions, no visible difference was observed in the biofilm’s protein contents for both bacterial species in mono-culture and co-culture models (Figure 7c,d,g,h).
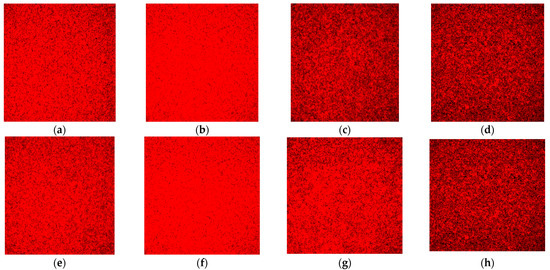
Figure 7.
Representative CLSM images of protein content in the biofilm stained with the SyPRO biofilm matrix stain: (a) E. faecalis as a monospecies culture under aerobic conditions; (b) E. faecalis as co-culture under aerobic conditions; (c) E. faecalis as a monospecies culture under anaerobic conditions; (d) E. faecalis as co-culture under anaerobic conditions; (e) P. gingivalis as a monospecies culture under aerobic conditions; (f) P. gingivalis as co-culture under aerobic conditions; (g) P. gingivalis as a monospecies culture under anaerobic conditions; and (h) P. gingivalis as co-culture under anaerobic conditions. Note: Protein contents are stained red. Images with a greater degree of redness denote a greater amount of protein contents, as seen in (b,f).
4. Discussion
A large body of evidence has suggested that bacterial behaviors in multi-species biofilm differ from the monospecies biofilm. Hence, research into the interactions between microbial species in biofilms can reveal the crucial factors that drive stability and changes leading to dysbiosis and the development of preventive and therapeutic approaches [23,24]. In vitro studies on dual- or multi-species biofilms have been carried out under controlled conditions to address the microbial community lifestyle in the root canal and periodontal pocket, assess their interactions and effects of stresses, as well as investigate and modify current therapeutic and preventive protocols [25,26,27]. This in vitro laboratory study employed dual-species bacteria, namely E. faecalis and P. gingivalis, to develop a simple and reproducible biofilm model. This dual-species biofilm model produced laboratory biofilm samples with minimal variation, which are readily assessable by microscopic and molecular analyses. A combination of techniques, including RT-PCR using viable cell staining and confocal laser scanning microscopy, were applied to study the interactions of this in vitro biofilm community. Three different models of dual-species infections were established in this study to identify biofilm-favoring conditions. In the first two sequential models, a single species of either E. faecalis or P. gingivalis was seeded prior to its colonization by the second microorganism. The co-culture model, as postulated to be more likely to occur in combined endodontic-periodontal lesions, involved the common contact of both species that shared and cross-infected two habitat niches, i.e., the root canal system and periodontal pockets. All models considered both anaerobic (anoxic) and aerobic (normoxic) conditions.
It was found by next-generation sequencing methods that the key genera in primary and persistent infected root canals included Prevotella, Fusobacterium, Porphyromonas, Parvimonas, and Streptococcus and the most abundant phyla were Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria [28,29]. Using the checkerboard DNA-DNA hybridization method, root canal and periodontal niches were found to share similar microbial profiles [18], and Gram-positive E. faecalis and Gram-negative P. gingivalis were detected in both root canals with secondary endodontic involvements and periodontal pockets in primary periodontal lesions [18]. A recent scientific report highlighted that the obligate anaerobic P. gingivalis could survive in the presence of oxygen when grown along with C. albicans, a facultative yeast-like anaerobe [22]. It was shown that C. albicans grew in their filamentous form, i.e., hyphae, and this could create a microenvironment with reduced oxygen tension and support the growth of P. gingivalis [22]. Obligate anaerobes usually do not possess any anti-oxidation mechanisms like superoxide dismutase to detoxify the oxygen challenge [19]. C. albicans is able to create a reduced redox potential environment to support the growth of some anaerobic bacteria, and a gradient of oxygen concentration down to ca. 4% has been observed in C. albicans-formed biofilms [30,31]. On the other hand, P. gingivalis cells were shown to survive and tolerate low oxygen levels (6–10%), depending on the haemin availability [32]. Haemin dimers binding to the surface of P. gingivalis would serve as a catalase-like buffer system to overcome the oxygen stress and reach a steady-state growth [19,20,33]. Whether E. faecalis possesses the same ability to form a protective “biofilm” to shield anaerobic P. gingivalis in the aerobic environment has never been investigated.
PCR is usually used to qualitatively detect the presence or absence of a particular bacterial DNA, while it does not provide quantification of the bacterial species of interest and is unable to differentiate live from dead cells [34]. To counteract these issues, quantitative PCR was used in this study to quantify the relative amount of individual living bacterial cells and propidium monoazide (PMA) stain (PMAxx™, Biotium, CA, USA) was used to detect the viable DNA. PMAxx™ dye is a photoreactive dye that preferentially binds to the dsDNA [35]. Visible blue light by PMA-Lite LED Photolysis Device will induce a photoreaction of the chemical, leading to a covalent bond with PMA and the dsDNA. PMAxx™ dye is designed to be a cell membrane-impermeant [36]; thus, only dead cells are susceptible to DNA modification due to compromised cell membranes. Thus, this unique feature of PMAxx™ is highly useful in selectively detecting live bacteria by subsequent qPCR.
CLSM, a non-destructive method often used to investigate the biofilm ecosystem and the hydrated spatial arrangement at the cellular scale, was employed in this study to assess biofilm thickness and relative protein contents [15]. This technique allows for three-dimensional reconstruction of the biomass to study the biofilm architecture [15]. SyPRO biofilm matrix stain was used in this study to allow the visualization of various protein molecules produced by the bacterial species, including glycoprotein, lipoprotein, phosphoprotein, calcium-binding protein, and fibrillar protein [37].
It was reported that C. albicans formed filamentous hyphae that facilitate the growth of P. gingivalis under the oxic challenge [22]. Similarly, another Gram-negative obligate anaerobe, F. nucleatum, could support the growth of P. gingivalis in the oxygenated environment by its ability to metabolize oxygen [19]. F. nucleatum expresses nicotinamide adenine dinucleotide phosphate (NADPH) oxidase to break down the oxygen molecule [38]. In this study, P. gingivalis was found to maintain its viability in the oxic condition in the presence of E. faecalis. Hence, the authors proposed that E faecalis could probably provide some barrier to protect the obligate anaerobe P. gingivalis from the oxygen challenges so that P. gingivalis was able to grow and proliferate in the presence of high oxygen tension. The possible mechanism that E. faecalis could act as an “oxygen reducing bacteria” is that it is a facultative anaerobe that can grow and utilizes the oxygen present in the biofilm micro-environment [39,40]. Additionally, E. faecalis can form a biofilm, and the extracellular polysaccharide (EPS) formed in the biofilm can protect the resident P. gingivalis. The EPS also offers protection against various environmental stresses such as pH shifts, osmotic shock, UV radiation, and desiccation [15]. However, a 15-species polymicrobial biofilm model showed that when E. faecalis was added to the basic biofilm that consisted of P. gingivalis and others, the total cell number was reduced [41]. This could be due to the antagonistic effect of the remaining bacteria present in the biofilm models.
Biofilm thickness was significantly greater when both species were co-cultured simultaneously, regardless of the oxygen challenge. This was further supported by the findings of increased protein contents in the co-cultured biofilms. Biofilm formation is associated with quorum sensing, which regulates bacterial gene expression in response to large cell population densities. A molecule called “autoinducers” was involved in biofilm formation [42]. E. faecalis biofilm formation is regulated by the fecal streptococci regulator (fsr) locus [43]. Enterococcal cells do not only communicate through Fsr quorum sensing but are also capable of communicating by peptide pheromones, including Cpd, Cob, and Ccf [44,45]. On the other hand, P. gingivalis’s capacity to form biofilms is related to the caseionlytic proteases (Clp), which upregulate the expression levels of fimA, mfa1, and luxS [46]. E. faecalis’ biofilm development was influenced by growth medium and nutrient availability [16]. In this study, TSB broth in Pg broth contained 3 g of glucose which helped to promote E. faecalis’ biofilm formation. It was reported that E. faecalis showed highly aggregated biofilms with higher bacteria counts and EPS bio-volume when the medium contained sucrose compared to the medium containing glucose or without sugar substrate [16].
E. faecalis possesses many virulence factors, such as enterococcal surface protein (esp), gelatinase (gelE), aggregation substance (asa1), cytolysin B (cylB), and endocarditis-specific antigen A (efaA) gene, ArgR family transcription factor (ahrC), endocarditis and biofilm-associated pili (ebpA), enterococcal polysaccharide antigen (epal), epal and OG1RF_11715 (epaOX), and (p)ppGpp-synthetase/hydrolase (relA) genes [47,48,49,50,51,52,53,54,55]. Meanwhile, P. gingivalis also possesses various virulence factors, such as lipopolysaccharide (LPS), gingipains, fimbriae/pili, collagenase, (erythrocyte) lectins, capsule, protease, and superoxide dismutase [56,57]. These virulence factors are predominantly proteins of different types, which might have attributed to the higher protein contents observed and thicker biofilm formation in this study when both microorganisms were grown together. A previous dual-species biofilm proteomic study consisting of F. nucleatum and P. gingivalis showed that more proteins were found when they were co-cultured together as compared with P. gingivalis as mono-culture but to a lesser extent than F. nucleatum mono-culture biofilm [58]. The reasons were not fully elucidated. It might be due to the antagonist effect of P. gingivalis upon F. nucleatum. To date, no studies have characterized the proteome of dual-species biofilm composed of E. faecalis and P. gingivalis. However, some facultative anaerobes such as C. albicans and Streptococcus gordonii (S. gordonii) have been shown to co-adhere to P. gingivalis in co-culture [22,59]. The co-adhesion between P. gingivalis and S. gordonii might have been mediated by the SspB protein of S. gordonii, whereas Mp65 adhesin and enolase might have played a role in the interaction between C. albicans and P. gingivalis (Chung et al., 2000, Bartnicka et al., 2019). It was postulated that the cross-talk between these proteins might have rendered the P. gingivalis in the dual-species biofilm the protective mechanism to thrive in an oxic environment. E. faecalis, a facultative anaerobe like S. gordonii and C. albicans, might have also cross-talked with P. gingivalis when they were co-cultured, resulting in an increased protein production. Chung and co-workers reported that 100-kDa protein of P. gingivalis interacted with E. faecalis to induce the cellular surface’s expression of SspB [59]. Furthermore, E. faecalis had been shown to take up heme and synthesize heme proteins, called hemoproteins [60]. The hemin content in the culture medium used in this study could have potentiated the hemoprotein synthesis, contributing to the increased protein contents when E. faecalis cells were present in the co-cultured biofilms.
In future studies, scanning electron microscopy or fluorescence in situ hybridization (FISH) combined with CLSM could be used to determine the spatial distribution of the bacteria in the biofilm [61], which may reveal the actual architecture of the dual-species biofilm. Furthermore, biofilm protein quantification assay, such as Lowry protein assay or fluorescein isothiocyanate (FITC), can be considered to quantify the protein component in the biofilms [62,63].
5. Conclusions
Within the limitations of this in vitro study, it may be concluded that E. faecalis appeared to shield P. gingivalis from oxygen challenges and support its continued growth in an oxic environment. When both E. faecalis and P. gingivalis were co-cultured simultaneously, thicker biofilms were formed under both aerobic and anaerobic conditions. A greater amount of protein was also found in the dual-species biofilm model when they were co-cultured aerobically, compared to the mono-culture and sequential culture models.
Author Contributions
Data curation, H.C.T.; Formal analysis, H.C.T. and A.H.C.L.; Funding acquisition, C.Z.; Investigation, H.C.T.; Project administration, C.Z. and A.H.C.L.; Supervision, G.S.P.C., J.W.W.C., C.Z. and A.H.C.L.; Validation, G.S.P.C. and J.W.W.C.; Writing—original draft, H.C.T.; Writing—review & editing, C.Z. and A.H.C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data may be obtained from the first and corresponding authors—H.C.T., C.Z. and A.H.C.L.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rosen, H.; Klebanoff, S.J. Bactericidal activity of a superoxide anion-generating system. A model for the polymorphonuclear leukocyte. J. Exp. Med. 1979, 149, 27–39. [Google Scholar] [PubMed]
- Brawn, K.; Fridovich, I. DNA strand scission by enzymically generated oxygen radicals. Arch. Biochem. Biophys. 1981, 206, 414–419. [Google Scholar] [PubMed]
- Harley, J.B.; Flaks, J.G.; Goldfine, H.; Bayer, M.E.; Rasmussen, H. Hyperbaric oxygen toxicity and ribosome destruction in Escherichia coli K12. Can. J. Microbiol. 1981, 27, 44–51. [Google Scholar] [CrossRef]
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [PubMed]
- Paster, B.J.; Olsen, I.; Aas, J.A.; Dewhirst, F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 2006, 42, 80–87. [Google Scholar]
- Kriebel, K.; Hieke, C.; Müller-Hilke, B.; Nakata, M.; Kreikemeyer, B. Oral Biofilms from Symbiotic to Pathogenic Interactions and Associated Disease—Connection of Periodontitis and Rheumatic Arthritis by Peptidylarginine Deiminase. Front. Microbiol. 2018, 9, 53. [Google Scholar]
- Molander, A.; Reit, C.; Dahlén, G.; Kvist, T. Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 1998, 31, 1–7. [Google Scholar]
- Siqueira, J.F., Jr.; Rôças, I.N. Polymerase chain reaction–based analysis of microorganisms associated with failed endodontic treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2004, 97, 85–94. [Google Scholar]
- Endo, M.; Ferraz, C.; Zaia, A.A.; Almeida, J.F.A.; Gomes, B.P.F.A. Quantitative and qualitative analysis of microorganisms in root-filled teeth with persistent infection: Monitoring of the endodontic retreatment. Eur. J. Dent. 2013, 7, 302–309. [Google Scholar]
- Bystrom, A.; Claesson, R.; Sundqvist, G. The antibacterial effect of camphorated paramonochlorophenol, camphorated phenol and calcium hydroxide in the treatment of infected root canals. Dent. Traumatol. 1985, 1, 170–175. [Google Scholar]
- Love, R.M. Enterococcus faecalis—A mechanism for its role in endodontic failure. Int. Endod. J. 2001, 34, 399–405. [Google Scholar] [PubMed]
- Evans, M.; Davies, J.K.; Sundqvist, G.; Figdor, D. Mechanisms involved in the resistance of Enterococcus faecalis to calcium hydroxide. Int. Endod. J. 2002, 35, 221–228. [Google Scholar] [PubMed]
- Sedgley, C.M.; Lennan, S.L.; Appelbe, O.K. Survival of Enterococcus faecalis in root canals ex vivo. Int. Endod. J. 2005, 38, 735–742. [Google Scholar] [CrossRef]
- Neelakantan, P.; Romero, M.; Vera, J.; Daood, U.; Khan, A.U.; Yan, A.; Cheung, G.S.P. Biofilms in Endodontics—Current Status and Future Directions. Int. J. Mol. Sci. 2017, 18, 1748. [Google Scholar]
- Kim, M.-A.; Rosa, V.; Min, K.-S. Characterization of Enterococcus faecalis in different culture conditions. Sci. Rep. 2020, 10, 21867. [Google Scholar]
- Gomes, B.P.; Berber, V.B.; Kokaras, A.S.; Chen, T.; Paster, B.J. Microbiomes of endodontic-periodontal lesions before and after chemomechanical preparation. J. Endod. 2015, 41, 1975–1984. [Google Scholar]
- Rovai, E.D.S.; Matos, F.D.S.; Kerbauy, W.D.; Cardoso, F.G.D.R.; Martinho, F.C.; Oliveira, L.D.D.; Valera, M.C.; Carvalho, C.A.T. Microbial profile and endotoxin levels in primary periodontal lesions with secondary endodontic involvement. Braz. Dent. J. 2019, 30, 356–362. [Google Scholar]
- Diaz, P.I.; Zilm, P.S.; Rogers, A.H. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 2002, 148, 467–472. [Google Scholar]
- Smalley, J.W.; Birss, A.J.; Silver, J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the μ-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol. Lett. 2000, 183, 159–164. [Google Scholar]
- Diaz, P.; Valm, A. Microbial Interactions in Oral Communities Mediate Emergent Biofilm Properties. J. Dent. Res. 2020, 99, 18–25. [Google Scholar] [PubMed]
- Bartnicka, D.; Karkowska-Kuleta, J.; Zawrotniak, M.; Satała, D.; Michalik, K.; Zielinska, G.; Bochenska, O.; Kozik, A.; Ciaston, I.; Koziel, J.; et al. Adhesive protein-mediated cross-talk between Candida albicans and Porphyromonas gingivalis in dual species biofilm protects the anaerobic bacterium in unfavorable oxic environment. Sci. Rep. 2019, 9, 4376. [Google Scholar] [PubMed]
- de Oliveira, R.V.D.; Bonafé, F.S.S.; Spolidorio, D.M.P.; Koga-Ito, C.Y.; de Farias, A.L.; Kirker, K.R.; James, G.A.; Brighenti, F.L. Streptococcus mutans and actinomyces naeslundii interaction in dual-species biofilm. Microorganisms 2020, 8, 194. [Google Scholar]
- Thurnheer, T.; Belibasakis, G. Streptococcus oralis maintains homeostasis in oral biofilms by antagonizing the cariogenic pathogen Streptococcus mutans. Mol. Oral Microbiol. 2018, 33, 234–239. [Google Scholar] [PubMed]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [PubMed]
- Bai, F.; Cai, Z.; Yang, L. Recent progress in experimental and human disease-associated multi-species biofilms. Comput. Struct. Biotechnol. J. 2019, 17, 1234–1244. [Google Scholar]
- Joshi, R.V.; Gunawan, C.; Mann, R. We Are One: Multispecies Metabolism of a Biofilm Consortium and Their Treatment Strategies. Front. Microbiol. 2021, 12, 635432. [Google Scholar]
- Shin, J.M.; Luo, T.; Lee, K.H.; Guerreiro, D.; Botero, T.M.; McDonald, N.J.; Rickard, A.H. Deciphering Endodontic Microbial Communities by Next-generation Sequencing. J. Endod. 2018, 44, 1080–1087. [Google Scholar]
- Manoil, D.; Al-Manei, K.; Belibasakis, G.N. A Systematic Review of the Root Canal Microbiota Associated with Apical Periodontitis: Lessons from Next-Generation Sequencing. Proteom. Clin. Appl. 2020, 14, 1900060. [Google Scholar]
- Fox, E.; Cowley, E.; Nobile, C.; Hartooni, N.; Newman, D.; Johnson, A. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar]
- Kowalski, C.H.; Morelli, K.A.; Schultz, D.; Nadell, C.D.; Cramer, R.A. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 22473–22483. [Google Scholar] [CrossRef] [PubMed]
- Diaz, P.I.; Rogers, A.H. The effect of oxygen on the growth and physiology of Porphyromonas gingivalis. Oral Microbiol. Immunol. 2004, 19, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Smalley, J.W.; Olczak, T. Heme acquisition mechanisms of Porphyromonas gingivalis—strategies used in a polymicrobial community in a heme-limited host environment. Mol. Oral Microbiol. 2017, 32, 1–23. [Google Scholar]
- Siqueira, J.F., Jr.; Rôças, I.N. Exploiting molecular methods to explore endodontic infections: Part 1—Current molecular technologies for microbiological diagnosis. J. Endod. 2005, 31, 411–423. [Google Scholar] [CrossRef]
- Rousseau, A.; Villena, I.; Dumètre, A.; Escotte-Binet, S.; Favennec, L.; Dubey, J.P.; Aubert, D.; La Carbona, S. Evaluation of propidium monoazide–based qPCR to detect viable oocysts of Toxoplasma gondii. Parasitol. Res. 2019, 118, 999–1010. [Google Scholar] [PubMed]
- Sicard, A.; Merfa, M.V.; Voeltz, M.; Zeilinger, A.R.; De La Fuente, L.; Almeida, R.P.P. Discriminating between viable and membrane-damaged cells of the plant pathogen Xylella fastidiosa. PLoS ONE 2019, 14, e0221119. [Google Scholar]
- Lopez, M.F.; Berggren, K.; Chernokalskaya, E.; Lazarev, A.; Robinson, M.; Patton, W.F. A comparison of silver stain and SYPRO Ruby Protein Gel Stain with respect to protein detection in two-dimensional gels and identification by peptide mass profiling. Electrophor. Int. J. 2000, 21, 3673–3683. [Google Scholar] [CrossRef]
- Bradshaw, D.J.; Marsh, P.D.; Watson, G.K.; Allison, C. Role of Fusobacterium nucleatum and Coaggregation in Anaerobe Survival in Planktonic and Biofilm Oral Microbial Communities during Aeration. Infect. Immun. 1998, 66, 4729–4732. [Google Scholar] [CrossRef]
- Portenier, I.; Waltimo, T.M.; Haapasalo, M. Enterococcus faecalis–the root canal survivor and ‘star’ in post-treatment disease. Endod. Top. 2003, 6, 135–159. [Google Scholar] [CrossRef]
- Rodríguez-Niklitschek, C.; Oporto, G.H. Clinical implications of Enterococcus faecalis microbial contamination in root canals of devitalized teeth: Literature review. Rev. Odontol. Mex. 2015, 19, 181–186. [Google Scholar] [CrossRef][Green Version]
- Lukic, D.; Karygianni, L.; Flury, M.; Attin, T.; Thurnheer, T. Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases. Microorganisms 2020, 8, 674. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Hancock, L.E.; Perego, M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 2004, 186, 5629–5639. [Google Scholar] [CrossRef] [PubMed]
- Dunny, G.M.; Berntsson, R.P.-A. Enterococcal Sex Pheromones: Evolutionary Pathways to Complex, Two-Signal Systems. J. Bacteriol. 2016, 198, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Clewell, D.B.; Francia, M.V.; Flannagan, S.E.; An, F.Y. Enterococcal plasmid transfer: Sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 2002, 48, 193–201. [Google Scholar] [CrossRef]
- He, L.; Wang, H.; Zhang, R.; Li, H. The regulation of Porphyromonas gingivalis biofilm formation by ClpP. Biochem. Biophys. Res. Commun. 2019, 509, 335–340. [Google Scholar] [CrossRef]
- Strateva, T.; Atanasova, D.; Savov, E.; Petrova, G.; Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016, 20, 127–133. [Google Scholar] [CrossRef]
- Lins, R.X.; Andrade, A.D.O.; Junior, R.H.; Wilson, M.J.; Lewis, M.A.; Williams, D.W.; Fidel, R.A.S. Antimicrobial resistance and virulence traits of Enterococcus faecalis from primary endodontic infections. J. Dent. 2013, 41, 779–786. [Google Scholar] [CrossRef]
- Saffari, F.; Sobhanipoor, M.H.; Shahravan, A.; Ahmadrajabi, R. Virulence genes, antibiotic resistance and capsule locus polymorphisms in Enterococcus faecalis isolated from canals of root-filled teeth with periapical lesions. Infect. Chemother. 2018, 50, 340–345. [Google Scholar] [CrossRef]
- Frank, K.L.; Barnes, A.M.; Grindle, S.M.; Manias, D.A.; Schlievert, P.M.; Dunny, G.M. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect. Immun. 2012, 80, 539–549. [Google Scholar] [CrossRef]
- Chávez de Paz, L.E.; Lemos, J.A.; Wickström, C.; Sedgley, C.M. Role of (p) ppGpp in biofilm formation by Enterococcus faecalis. Appl. Environ. Microbiol. 2012, 78, 1627–1630. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.T.; Dunny, G.M. Multiple Roles for Enterococcus faecalis Glycosyltransferases in Biofilm-Associated Antibiotic Resistance, Cell Envelope Integrity, and Conjugative Transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105. [Google Scholar] [CrossRef] [PubMed]
- Zoletti, G.O.; Pereira, E.M.; Schuenck, R.P.; Teixeira, L.M.; Siqueira, J.F.; dos Santos, K.R.N. Characterization of virulence factors and clonal diversity of Enterococcus faecalis isolates from treated dental root canals. Res. Microbiol. 2011, 162, 151–158. [Google Scholar] [CrossRef]
- Kayaoglu, G.; Ørstavik, D. Virulence factors of Enterococcus faecalis: Relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 2004, 15, 308–320. [Google Scholar] [CrossRef]
- Tendolkar, P.M.; Baghdayan, A.S.; Gilmore, M.S.; Shankar, N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 6032–6039. [Google Scholar] [CrossRef]
- How, K.Y.; Song, K.P.; Chan, K.G. Porphyromonas gingivalis: An overview of periodontopathic pathogen below the gum line. Front. Microbiol. 2016, 7, 53. [Google Scholar] [CrossRef]
- Jia, L.; Han, N.; Du, J.; Guo, L.; Luo, Z.; Liu, Y. Pathogenesis of Important Virulence Factors of Porphyromonas gingivalis via Toll-Like Receptors. Front. Cell. Infect. Microbiol. 2019, 9, 262. [Google Scholar] [CrossRef]
- Mohammed, M.M.A.; Pettersen, V.K.; Nerland, A.H.; Wiker, H.G.; Bakken, V. Quantitative proteomic analysis of extracellular matrix extracted from mono- and dual-species biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis. Anaerobe 2017, 44, 133–142. [Google Scholar] [CrossRef]
- Chung, W.O.; Demuth, D.R.; Lamont, R.J. Identification of a Porphyromonas gingivalis Receptor for the Streptococcus gordonii SspB Protein. Infect. Immun. 2000, 68, 6758–6762. [Google Scholar] [CrossRef]
- Frankenberg, L.; Brugna, M.; Hederstedt, L. Enterococcus faecalis Heme-Dependent Catalase. J. Bacteriol. 2002, 184, 6351–6356. [Google Scholar] [CrossRef]
- Almstrand, R.; Daims, H.; Persson, F.; Sörensson, F.; Hermansson, M. New methods for analysis of spatial distribution and coaggregation of microbial populations in complex biofilms. Appl. Environ. Microbiol. 2013, 79, 5978–5987. [Google Scholar] [CrossRef] [PubMed]
- Breen, C.J.; Raverdeau, M.; Voorheis, H.P. Development of a quantitative fluorescence-based ligand-binding assay. Sci. Rep. 2016, 6, 25769. [Google Scholar] [CrossRef] [PubMed]
- Polasko, A.L.; Ramos, P.; Kaner, R.B.; Mahendra, S. A multipronged approach for systematic in vitro quantification of catheter-associated biofilms. J. Hazard. Mater. Lett. 2021, 2, 100032. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).