Trust Your Gut: The Association of Gut Microbiota and Liver Disease
Abstract
1. Introduction
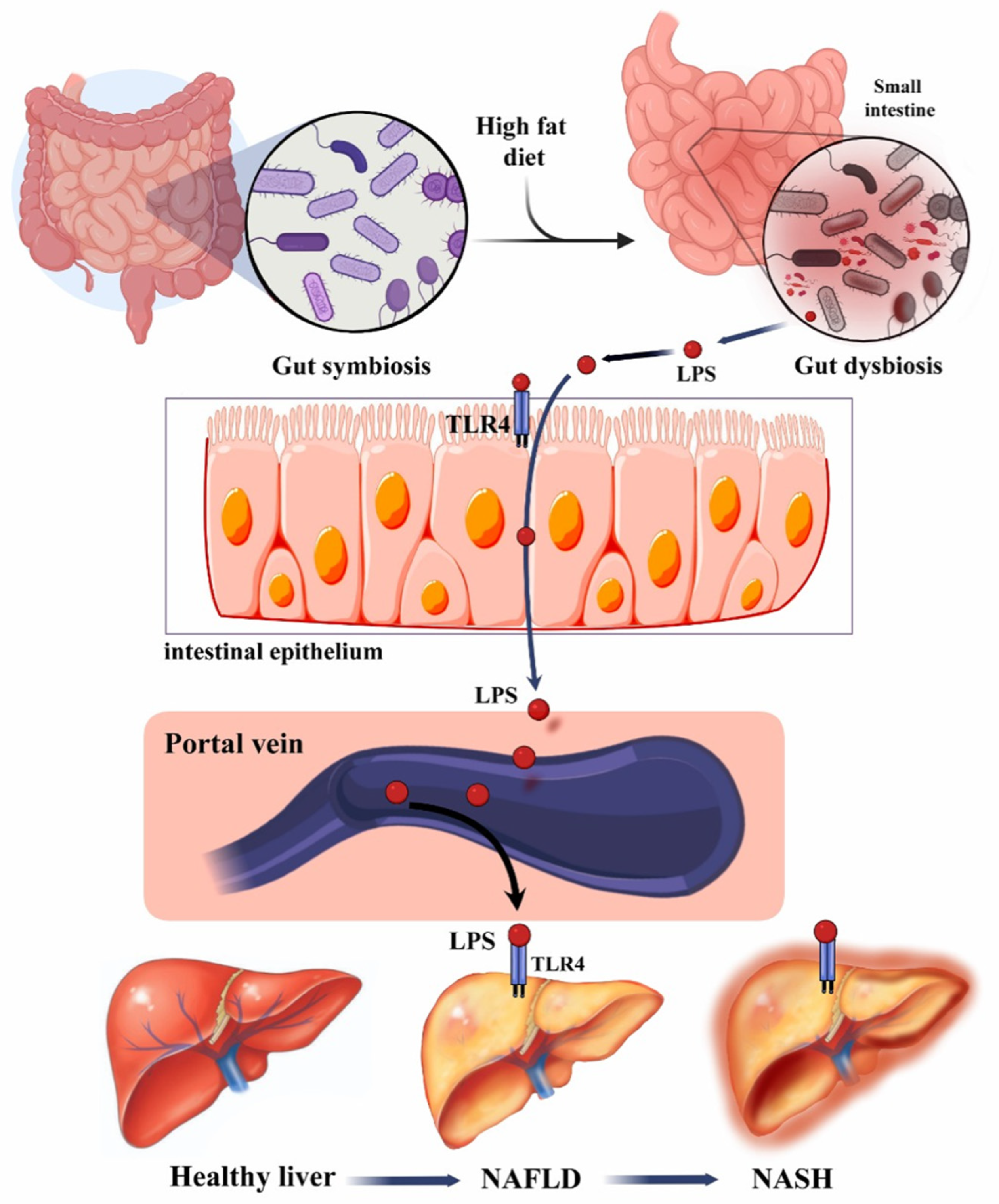
1.1. Role of Gut–Liver Axis in Liver Disease
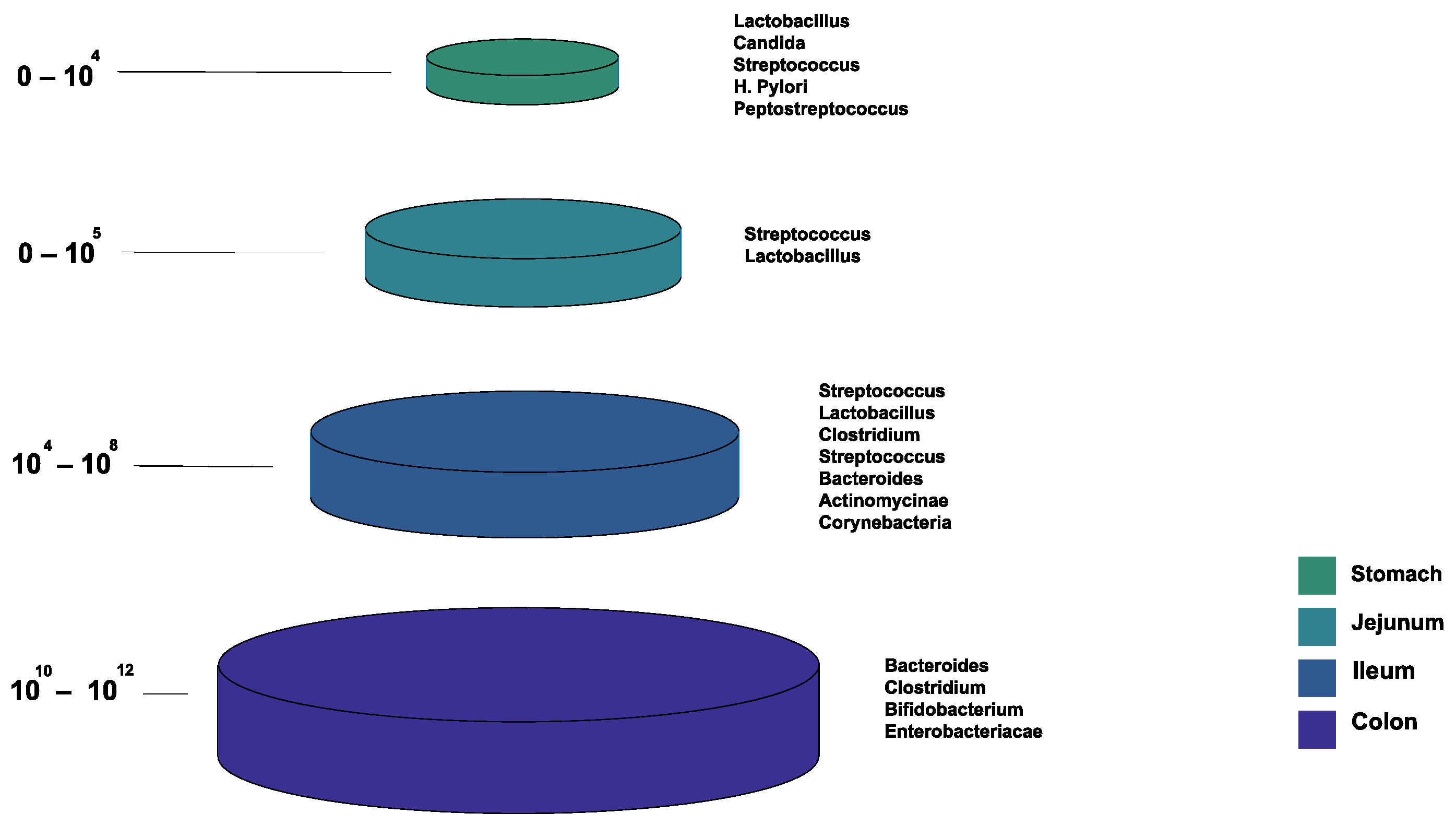
1.2. Normal Gut Microbiota Composition
2. Gut Microbiota: Link with Non-Alcoholic Liver Disease
2.1. Epidemiology, Clinical Manifestations, and Pathophysiology
2.2. Gut Microbiome Profile in NAFLD
| Author Reference | Country | Study Design | Participants | Changes in the Composition of Gut Microbiota in NAFLD | Key Findings |
|---|---|---|---|---|---|
| [38] | France | Cross-sectional study | 57 NAFLD 35 NASH | ↑ Actinobacteria ↑ Bacteroides ↑ Ruminococcus ↓ Prevotella No change in Firmicutes | ↑ Ruminococcus was significant in NASH Gut microbiota can be one of the prognostic tools to evaluate NAFLD progression and severity |
| [40] | Canada | Prospective cross-sectional study | 33 NAFLD: 11 simple steatosis 22 NASH 17 healthy controls | ↑ C. Coccoides in NASH ↓ Bacteroidetes in NASH compared to the SS and HCC | The relationship between Bacteroidetes and liver disease state was not dependent on the increase in BMI or diet |
| [41] | United States | Cross-sectional study | 44 NAFLD 29 healthy controls | ↓ Bacteroidetes ↓ Prevotella ↓ Gemmiger ↓ Oscillospira | ↓ Bacterial diversity in patients with NAFLD compared to controls contributed to an increase in the rate of inflammation in NAFLD |
| [35] | United States | Prospective, observational, cross-sectional study | 87 NAFLD 37 healthy controls | ↑ Bacteroidetes ↑ Proteobacteria ↓ Firmicutes | ↓ α-diversity in NAFLD was attributed to the differences in bacterial abundance rather than an increase in specific phyla or genus ↑ Pro-inflammatory bacterial products (LPS) in patients with NAFLD |
| [36] | United States | Case-control | 22 NASH 25 obese 16 healthy controls | ↑ Bacteroides (Prevotella) ↑ Proteobacteria (Escherichia) ↓ Firmicutes ↓ Actinobacteria | ↑ Abundance of ethanol producing bacteria (Escherichia) in patients with NASH contributed to disease progression ↑ Ethanol-producing bacteria (Escherichia) was attributed to the use of antibiotics |
| [39] | Italy | Case-control | 61 NASH/NAFL 54 healthy controls | ↑ Actinobacteria ↑ Bradyrhizobium ↑ Anaerococcus ↑ Peptoniphilus ↑ Propionibacterium acnes ↑ Enterobacteriaceae (Escherichia coli) ↑ Dorea ↑ Ruminococcus ↓ Bacteroidetes ↓ Oscillospira ↓ Rikenellaceae | ↓ Microbial diversity in NASH/NAFL ↓ Bacteroidaceae and Bacteroides were in NAFL and NASH, while it ↑ in obese patients compared to controls ↑ Ethanol-producing bacteria (Enterobacteriaceae) in NAFL/NASH compared to controls |
| [37] | Canada | Case-control | 30 NAFLD 30 healthy controls | ↑ Proteobacteria ↑ Firmicutes ↓ Bacteroidetes | Fecal ester volatile organic compounds could influence the microbiome composition of patients with NAFLD in an unfavorable way |
3. Gut Microbiota: Link with Alcoholic Liver Disease
4. Gut Microbiota: Link with Liver Cirrhosis
4.1. Epidemiology, Clinical Manifestations, and Pathophysiology
4.2. Gut Microbiome Profile in Liver Cirrhosis
| Author Reference | Country | Study Design | Participants | Changes in the Composition of the Gut Microbiota in Liver Cirrhosis | Key Findings |
|---|---|---|---|---|---|
| [54] | China | Case-control | 36 cirrhosis 24 healthy controls | ↑ Proteobacteria ↑ Fusobacteria ↑ Enterobacteriacea ↑ Veillonellacea ↑ Streptococcaceae ↓ Bacteroidetes ↓ Lachnospiraceae | Fecal microbiome composition was altered in patients with cirrhosis compared to healthy individuals, indicating there is dysbiosis ↑ Enterobacteriaceae and Streptococcaceae may affect cirrhosis prognosis |
| [55] | United States | Prospective cohort study | 25 cirrhosis: 17 with HE 8 without HE 10 controls | ↑ Bacteroidetes ↑ Veillonellaceae in HE ↑ Enterobacteriacea ↑ Alcaligeneceae ↑ Porphyromonadacea ↑ Fusobacteriaceae ↓ Ruminococcaceae ↓ Lachnospiraceae | Dysbiosis was found in patients with HE compared to healthy individuals Certain bacterial families were associated with endotoxemia, impaired cognition, and inflammation in liver cirrhosis patients in HE |
| [57] | China | Case-control | 26 cirrhosis patients with MHE 25 cirrhosis patients without MHE 26 healthy controls | ↑ Streptococcus salivarius in HE ↑ Streptococcaceae ↑ Veillonellaceae | Streptococcus salivarius was positively correlated with ammonia accumulation in MHE patients |
| [61] | United States and Japan | Cross-sectional study | 47 cirrhosis 14 healthy controls | ↑ Enterobacteriaceae ↓ Lachnospiraceae ↓ Ruminococcaceae ↓ Blautia | ↑ Pathogenic bacteria due to gut dysbiosis in cirrhotic patients altered bile acid composition |
| [58] | China | Case-control | 98 cirrhosis 83 controls | ↑ Proteobacteria ↑ Veillonella ↑ Streptococcus ↓ Bacteroidetes | In liver cirrhosis, there was an invasion of the gut by oral bacterial species |
| [64] | Unites States | Case-control | 87 with HE 40 healthy controls | ↑ Enterobacteriaceae ↓ Lachnospiraceae ↓ Ruminococcaceae | Specific bacterial families were associated with astrocytic and neuronal MRI changes Gut dysbiosis in cirrhosis was linked with systemic inflammation, elevated ammonia levels, and neuronal dysfunction |
| [62] | China | Case-control | 30 cirrhosis 28 healthy controls | ↑ Veillonella ↑ Megasphaera ↑ Dialister ↑ Atopobium ↑ Prevotella ↑ Firmicutes | ↑ Oral bacteria in duodenal mucosal microbiota in cirrhotic patients |
| [65] | China | Cross-sectional study | 36 cirrhosis 20 healthy controls | ↑ Firmicutes ↓ Bacteroidetes | ↑ Microbial dysbiosis in cirrhotic patients with Child-Pugh scores > 5 led to slower small bowel transit |
| [66] | Austria | Case-control | 90 cirrhosis: 50 on PPI therapy 40 not on PPI therapy | ↑ Streptococcus salivarius ↑ Veillonella parvula | ↑ Gut dysbiosis in cirrhotic patients with long-term PPI therapy |
| [67] | Spain | Prospective cohort study | 182 cirrhosis | ↑ Enterococcus ↑ Streptococcus in ACLF ↑ Faecalibacterium ↑ Ruminococcus ↑ Eubacterium in decompensated patients | As cirrhosis progressed from compensated to uncompensated to ACLF, there was a linear progression in reduction in gene and metagenomic richness |
| [56] | Russia | Case-control | 48 cirrhosis 21 healthy controls | ↑ Enterobacteriaceae ↑ Proteobacteria ↑ Lactobacillaceae ↓ Firmicutes ↓ Clostridia | Severe dysbiosis was an independent risk factor for death Levels of Clostridia and Bacilli determined death within a year Levels of Proteobacteria and Enterobacteriaceae determined the long-term prognosis (death over the subsequent three years) |
5. Gut Microbiota: Link with Hepatocellular Carcinoma
5.1. Epidemiology, Clinical Manifestations, and Pathophysiology
5.2. Gut Microbiome Profile in HCC
| Author Reference | Country | Study Design | Participants | Changes in the Composition of Gut Microbiota in HCC | Key Findings |
|---|---|---|---|---|---|
| Human Studies | |||||
| [79] | Poland | Cross-sectional | 15 HCC 5 without HCC All participants had cirrhosis and underwent liver transplantation. | ↑ Escherichia coli ↑ Enterobacteriaceae ↑ Enterococcus ↑ Lactobacillus ↑ H2O2-producing Lactobacillus species | ↑ Fecal counts of E coli were noted in the cirrhotic-HCC group, demonstrating its role in HCC development |
| [88] | Australia | Cohort study; metagenomics and metabolomics analysis | 32 NAFLD-HCC 28 NAFLD-cirrhosis 30 non-NAFLD controls | ↑ Proteobacteria ↑ Enterobacteriaceae ↑ Bacteroides xylanisolvens ↑ B. caecimuris ↑ Ruminococcus gnavus ↑ Clostridium bolteae ↑ Veillonella parvula ↑ Bacteroides caecimuris ↑ Veillonella parvula ↑ Clostridium bolteae ↑ Ruminococcus gnavus ↓ Oscillospiraceae ↓ Erysipelotrichaceae ↓ Eubacteriaceae | ↑ B. caecimuris and Veillonella parvula distinguished NAFLD-HCC from NAFLD-cirrhosis and non-NAFLD controls ↓ Gut microbial α-diversity ↑ SCFAs serum levels in NAFLD-HCC compared to NAFLD-cirrhosis and non-NAFLD control Gut microbiota in NAFLD-HCC microbiota contribute to immunosuppression |
| [89] | China | Cohort | 75 with early HCC 40 liver cirrhosis 75 healthy controls | ↑ Actinobacteria ↑ Gemmiger ↑ Parabacteroides ↑ Paraprevotella ↑ Klebsiella ↑ Haemophilus ↓ Verrucomicrobia ↓ Alistipes ↓ Phascolarctobacterium ↓ Ruminococcus ↓ Oscillibacter ↓ Faecalibacterium ↓ Clostridium IV ↓ Coprococcus | ↓ Butyrate-producing bacteria ↑ LPS-producing bacteria in early HCC versus healthy controls |
| [82] | China | Case-control | 57 HCC (35 with HBV related HCC, 22 with non-HBV non-HCV related HCC) 33 healthy controls | ↑ Bifidobacterium ↑ Lactobacillus ↓ Proteobacteria ↓ Firmicutes | ↓ Anti-inflammatory and ↑ pro-inflammatory bacteria in non-HBC non-HCV related HCC patients which correlated with their increased alcohol consumption |
| [81] | China | Case-control | 68 with primary HCC: (23 Stage I, 13 Stage II, 30 Stage III, 2 Stage IV) 18 healthy controls | ↑ Dysbiosis index Proteobacteria (Enterobacter, Haemophilus) ↑ Desulfococcus ↑ Prevotella ↑ Veillonella ↓ Cetobacterium | ↑ Dysbiosis index in patients with primary HCC compared with healthy controls |
| [83] | Italy | Cohort | 21 with NAFLD-related cirrhosis with HCC 20 NAFLD-related cirrhosis without HCC 20 healthy controls | ↑ Bacteroides ↑ Ruminococcaceae ↓ Bifidobacterium | ↑ Fecal calprotectin in HCC patients, which explains increased inflammation |
| [90] | Argentina | Case-control | 407 Cirrhosis: 25 with HCC, 25 w/o HCC 25 healthy controls | ↑ Erysipelotrichaceae ↑ Odoribacter ↑ Butyricimonas ↓ Leuconostocaceae ↓ Fusobacterium ↓ Lachnospiraceae | ↓ Prevotella in cirrhotic patients with HCC, which is associated with the activation of several inflammatory pathways such as the NLR signalling pathways |
| [91] | China | Case-control | 24 PLC 24 cirrhosis 23 healthy controls | ↑ Enterobacter ludwigii ↑ Enterococcaceae ↑ Lactobacillales ↑ Bacilli ↑ Gammaproteobacteria ↑ Veillonella ↓ diversity of Firmicutes ↓ Clostridia ↓ Subdoligranulum | Veillonella positively correlated with AFP Subdoligranulum negatively correlated with AFP Subdoligranulum contains SCFA-producing lineages |
| [84] | China | Case-control | 24 hepatitis 24 cirrhosis 75 HCC (35 with HBV, 25 with HCV, 15 with ALD) 20 healthy controls | ↑ Neisseria ↑ Enterobacteriaceae ↑ Veillonella ↑ Limnobacter ↓ Enterococcus ↓ Phyllobacterium ↓ Clostridium ↓ Ruminococcus ↓ Coprococcus | ↑ LPS by harmful bacteria generated liver inflammatory reactions through TLR4 |
| Animal experimental model studies | |||||
| [78] | Japan | Mice | 24 STZ-HFD (streptozocin-high-fat diet)-induced 24 controls | ↑ Bacteroides ↑ Bacteroides vulgatus ↑ Bacteroides uniformis ↑ Clostridium ↑ Clostridium xylanolyticum ↑ Clostridium fusiformis ↑ Roseburia ↑ Allobaculum sp. id4 ↑ Subdoligranulum ↑ Anaerotruncus ↑ Oscillibacter ↑ Xylanibacter ↑ Mucispirillum schaedleri ↑ Pseudobutyrivibrio ↑ Desulfovibrio ↑ Dehalobacterium ↑ Oscillospira ↑ Sarcina ↑ Atopobium ↑ Peptococcus ↓ Parasutterella ↓ Bacteroides acidofaciens ↓ Odoribacter ↓ Barnesiella ↓ Moryella ↓ Paraprevotella ↓ Lactobacillus intestinalis ↓ Akkermansia | Clostridium, Bacteroides, and Desulfovibrio were involved in bile acid dysregulation; their increased levels resulted in the preservation of high concentrations of bile acids, further contributing to hepatocarcinogenesis |
6. Gut Microbiota: Link with Autoimmune Hepatitis
6.1. Epidemiology, Clinical Manifestations, and Pathophysiology
6.2. Gut Microbiome Profile in Autoimmune Hepatitis
| Author Reference | Country | Study Design | Participants | Changes in the Composition of Gut Microbiota in AIH | Key Findings |
| [107] | China | Case-control | 24 AIH 8 healthy controls | ↓ Bifidobacterium ↓ Lactobacillus Escherichia coli and Enterococcus were unchanged | ↑ Intestinal permeability and gut dysbiosis ↑ Bacterial translocation, indicated by increased LPS, was correlated with AIH disease severity |
| [104] | Egypt | Case-control | 5 AIH 10 healthy controls | ↑ Faecalibacterium ↑ Blautia ↑ Streptococcus ↑ Veillonella ↑ Eubacterium ↑ Lachnospiraceae ↑ Butyricicoccus ↑ Haemophilus ↑ Bacteroides ↑ Clostridium ↑ Ruminococcaceae ↓ Prevotella ↓ Parabacteroides ↓ Dilaster | ↓ Bacterial diversity in AIH ↑ Butyrate forming bacteria (e.g., Butyricicoccus and Ruminococcaceae) |
| [106] | Germany | Case-control | 72 AIH 95 healthy controls 99 primary biliary cholangitis 81 ulcerative colitis | ↑ Proteobacteria ↑ Veillonella ↑ Streptococcus ↑ Lactobacillus ↓ Firmicutes in all groups ↓ Faecalibacterium ↓ Bifidobacterium | ↓ Bifidobacterium in AIH was associated with increased disease activity and failure to achieve remission ↓ α-diversity in AIH patients vs. healthy controls |
| [105] | China | Cross-sectional | 119 steroid-naïve AIH 132 healthy controls | ↑ Veillonella ↑ Streptococcus ↑ Klebsiella ↑ Lactobacillus ↓ Clostridiales ↓ Ruminococcaceae ↓ Rikenellaceae ↓ Oscillospira ↓ Parabacteroides ↓ Coprococcus | ↑ LPS biosynthesis ↓ α-diversity Veillonella showed a strong association with AIH and was link with ↑ AST and progression of liver inflammation Veillonella, Lactobacillus, Oscillospira, and Clostridiales have high diagnostic value in AIH |
| [111] | China | Case-control | 37 AIH 78 healthy controls | ↑ Veillonella ↑ Faecalibacterium ↑ Akkermansia ↑ Klebsiella ↑ Enterobacteriaceae_unclassified ↑ Megasphaera ↓ Pseudobutyrivibrio ↓ Lachnospira ↓ Ruminococcaceae ↓ Blautia ↓ Erysipelotrichaceae_incertae_sedis ↓ Phascolarctobacterium | A combination of Bacteroides, Ruminococcaceae, Lachnospiraceae, Veillonella, Roseburia, and Ruminococcaceae could distinguish AIH patients from healthy controls |
| [108] | China | Case-control | 32 AIH 20 NAFLD 20 healthy controls | ↑ Escherichia coli ↓ Bifidobacterium ↓ Lactobacillus ↓ Bacteroides ↓ C. leptum | ↑ Serum LPS in comparison to NAFLD and healthy controls |
7. Gut Microbiota: Link with Other Liver Diseases
8. Gut Microbiota: Fungal and Viral Changes
9. Therapeutic Gut–Microbiome Interaction
9.1. Probiotics and Prebiotics
9.2. Antibiotics
9.3. Fecal Microbiota Transplantation (FMT)
9.4. Other Therapies
9.5. Liver Transplant
| Author Reference | Country | Study Design | Intervention | Participants | Changes in the Composition of the Gut Microbiota | Key Findings |
|---|---|---|---|---|---|---|
| NAFLD | ||||||
| [158] | Japan | Prospective cohort | Weight reduction | 26 Pediatric NAFLD patients | Not mentioned in the study | ↓ In liver stiffness and fat deposition |
| [138] | China | Animal experimental model (rats) | Probiotics (cholesterol-lowering probiotics and anthraquinone from Cassia obtusifolia L.) | 30 male rats: 6 NAFLD 18 NAFLD rats received treatment 6 normal diet | ↑ Bacteroides ↑ Lactobacillus P ↑ Arabacteroides ↓ Oscillospira | Probiotic use ameliorated intestinal mucosal barrier ↓ Endotoxemia and inflammatory cytokines |
| [139] | China | Animal experimental model (mice) | Probiotics (Lactobacillus reuteri GMNL-263) | 12 male mice: 6 HS received treatment 6 controls | ↑ Bifidobacteria ↑ Lactobacilli ↓ Clostridia | ↓ BG levels, TNF-α and IL-6 production by adipose tissue in those taking probiotics Probiotics also modulate insulin level and can prevent type 2 diabetes |
| [140] | China | Animal experimental model (mice) | Probiotics | 24 male mice: 8 NAFLD no treatment 8 NAFLD with treatment 8 controls | ↑ Ruminococcu ↑ Saccharibacteria (TM7 phylum) ↓ Verrucomicrobia ↓ Veillonella | ↓ TC, TG, lipid deposition and inflammation in the probiotic groups |
| [141] | Netherlands | Double-blind, randomized controlled | FMT (allogenic vs autologous) | 21 NAFLD patients: 10 allogenic 11 autologous | Allogenic FMT: ↑ Ruminococcus ↑ Eubacterium hallii ↑ Faecalibacterium ↑ Prevotella copri Autologous FMT: ↑ Lachnospiraceae | Improved liver endothelial function ↓ Liver necro-inflammation and steatosis There was no change in duodenal microbial diversity in both groups |
| [42] | China | Randomized control trial | Probiotics | 16 NASH: 7 received treatment 9 no treatment 22 healthy controls | ↑ Parabacteroide ↑ Allisonella ↓ Faecalibacterium ↓ Anaerosporobacter | Bacterial biodiversity did not differ between NASH patients and controls and did not differ with probiotic treatment ↑ Bacteroidetes and ↓ Firmicutes was noted in the probiotic group |
| Liver Cirrhosis | ||||||
| [159] | Czech Republic | Double-blind randomized clinical trial | Probiotics (E. coli Nissle strain) | 39 cirrhosis patients: 17 placebo 22 treatment group | ↑ Lactobacillus species ↑ Bifidobacterium species ↓ Proteus hauseri ↓ Citrobacter species ↓ Morganella species | Statistically significant improvement in gut microbiome in those taking the probiotic for 42 days ↓ Endotoxemia, bilirubin, and ascites |
| [136] | India | Double-blind, randomized, placebo-controlled clinical trial | Probiotics (VSL #3) | 130 cirrhosis patients: 66 probiotic group 64 placebo group | ↑ Lactobacillus species | ↓ Hospitalization due to HE with daily intake of the probiotic for 6 months |
| [160] | United States | Double-blind, randomized, placebo-controlled clinical trial (phase I) | Probiotics (Lactobacillus GG) | 30 cirrhosis patients: 14 probiotic group 16 placebo group | ↑ Firmicutes species ↓ Enterobacteriaceae ↓ Porphyromonadacea | ↓ Endotoxemia and TNF-α in patients taking probiotic for 8 weeks ↓ Dysbiosis due to decreased Enterobacteriaceae and increased Firmicutes species |
| [151] | United States | Randomized clinical trial | FMT | 20 HE patients: 10 FMT 10 placebo | ↑ Lactobacillaceae ↑ Bifidobacteriaceae ↑ Bacteroidetes ↑ Firmicutes | Reduction in hospitalizations, improved cognition, improved dysbiosis, and SCFAs in FMT group |
| [157] | United States | Case-control | Liver transplant | 45 liver transplant patients 45 healthy controls | ↑ Ruminococcaceae ↑ Lachnospiraceae ↓ Enterobacteriaceae | Post LT: ↓ pathogenic bacteria ↑ gut diversity and ↑ autochthonous bacteria Compared to controls, there was still residual dysbiosis |
| [161] | United States | Case-control | Periodontal therapy | 24 cirrhosis patients, no therapy 26 cirrhosis patients, periodontal therapy 20 healthy controls, periodontal therapy | ↑ Ruminococcaceae ↑ Lachnospiraceae ↓ Enterobacteriaceae ↓ Porphyromonadaceae ↓ Streptococcaceae (oral origin) | ↓ dysbiosis and endotoxemia with periodontal therapy for 30 days, especially in those who had HE |
| [137] | Austria | Randomized clinical trial | Probiotics (multispecies strain) | 26 cirrhosis patients on probiotic therapy 32 cirrhosis patients on placebo | ↑ Lactobacillus (brevis, salivarius, lactis) ↑ Faecalibacterium prausnitzii ↑ Syntrophococcus sucromutans ↑ Alistipes shahii ↑ Bacteroides vulgatus ↑ Prevotella | Probiotic therapy for 6 months enriched the gut microbiome in compensated cirrhosis patients and improved gut barrier function Changes seen were transient |
| HCC | ||||||
| [80] | China | Animal experimental model (rats) | Probiotics (VSL #3) Antibiotics (penicillin) | 13 DEN-induced HCC mice: 7 probiotics 6 controls Penicillin group Dextran Sulfate sodium (DSS) group DEN + DSS + Penicillin group | ↓ Escherichia coli ↓ Atopobium cluster ↓ B. fragilis ↓ Prevotella ↑ Escherichia coli ↑ Atopobium ↓ Bifidobacterium ↓ Lactobacillus | High-dose probiotic administration into DEN-induced HCC mice showed a restoration of gut homeostasis and inhibition of DEN-induced hepatocarcinogenesis There was an association between increased gut dysbiosis, inflammation, intestinal mucosa damage in the penicillin groups and the increased cell proliferation, hence demonstrating the contribution of antibiotics to hepatocarcinogensis |
| [145] | China | Animal experimental model (mice) | Probiotics (Prohep: Lactobacillus rhamnosus GG (LGG), viable Escherichia coli Nissle 1917 (EcN), and heat-inactivated VSL#3) | 8 probiotics 8 cisplatin 8 control | ↑ Alistipes ↑ Butyricimonas ↑ Mucispirillum ↑ Oscillibacter ↑ Parabacteroides ↑ Paraprevotella ↑ Prevotella ↑ Bacteroidetes ↓ Firmicutes ↓ Proteobacteria | In the probiotics group: ↑ anti-inflammatory bacteria ↓ Th17-inducing bacteria and segmented filamentous bacteria which are pro-inflammatory This stayed the same in control group |
| AIH | ||||||
| [109] | China | Animal experimental model (mice) | Probiotics (Bifidobacterium and Lactobacillus) | 16 experimental AIH mice, no treatment 13 experimental AIH mice, probiotics 13 experimental AIH mice, dexamethasone 16 controls | ↑ Bacteroidetes ↑ Bifidobacterium ↑ Bacteroides ↑ Clostridium ↑ Ruminococcus ↑ Anaerostipes ↑ Blautia ↓ Firmicutes ↓ Faecalibacterium ↓ Helicobacter ↓ Staphylococcus | Probiotics group: ↑ Treg differentiation ↑ SCFAs ↓ infiltration of inflammatory cells in the liver ↓ ALT, AST ↓ Th1, Th17 cells (-) LPS translocation to the liver (-) activation of the TLR/NF-kB pathway |
| [110] | China | Animal experimental model (mice) | Probiotics (Bifidobacterium animalis spp. Lactis 420) | 6 experimental AIH mice, no treatment 6 experimental AIH mice, probiotic 6 controls | ↑ Lactobacillus ↑ Alistipes ↑ Rikenella ↑ Clostridia ↓ Bacteroides ↓ Ruminococcus | Probiotics reduced liver injury and improved immune homeostasis via: Upregulation of tight junction proteins ↓ Serum endotoxin levels ↑ Fecal SCFAs ↑ α-diversity Regulation of pro-inflammatory cytokines (-) RIP3-MLKL signalling pathway of liver macrophages |
| [108] | China | Animal experimental model (mice) | FMT | Antibiotic-induced gut dysbiosis AIH group, FMT therapy AIH group, FMT therapy Control group | ↑ Bifidobacterium ↑ Lactobacillus ↓ Escherichia coli | ↓ AST, ALT and serum IgG, regulation of TFR/TFH immune imbalance and restoration of microbiome in both treatment groups, thus slowing AIH progression in mice |
10. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AIH | Autoimmune hepatitis |
| ALD | Alcohol-associated liver disease |
| CHB | Chronic hepatitis B |
| E. coli | Escherichia coli |
| FMT | Fecal microbiota transplant |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| HE | Hepatic encephalopathy |
| IBD | Inflammatory bowel disease |
| LPS | Lipopolysaccharide |
| LT | Liver transplant |
| NAFLD | Nonalcoholic fatty liver disease |
| NASH | Nonalcoholic steatohepatitis |
| PBC | Primary biliary cholangitis |
| PSC | Primary sclerosing cholangitis |
| PSC-IBD | Primary sclerosing cholangitis-inflammatory bowel disease |
| shortSCFAs | Short-chain fatty acids |
| TNF-α | Tumor necrosis factor alpha |
References
- Schwenger, K.J.; Clermont-Dejean, N.; Allard, J.P. The role of the gut microbiome in chronic liver disease: The clinical evidence revised. JHEP Rep. 2019, 1, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ohnishi, H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 7381–7391. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, L.; Nemeş, S.-A.; Szabo, K.; Teleky, B.-E.; Vodnar, D.-C. Guts imbalance imbalances the brain: A review of gut microbiota association with neurological and psychiatric disorders. Front. Med. 2022, 9, 813204. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Fukui, H. Role of gut dysbiosis in liver diseases: What have we learned so far? Diseases 2019, 7, 58. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R.; Szabo, G. Interleukin-1 and inflammasomes in alcoholic liver disease/acute alcoholic hepatitis and nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology 2016, 64, 955–965. [Google Scholar] [CrossRef]
- Miele, L.; Marrone, G.; Lauritano, C.; Cefalo, C.; Gasbarrini, A.; Day, C.; Grieco, A. Gut-liver axis and microbiota in NAFLD: Insight pathophysiology for novel therapeutic target. Curr. Pharm. Des. 2013, 19, 5314–5324. [Google Scholar] [CrossRef]
- Bruneau, A.; Hundertmark, J.; Guillot, A.; Tacke, F. Molecular and Cellular Mediators of the Gut-Liver Axis in the Progression of Liver Diseases. Front. Med. 2021, 8, 725390. [Google Scholar] [CrossRef] [PubMed]
- Odenwald, M.A.; Turner, J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Suk, K.T.; Kim, D.J. Gut microbiota: Novel therapeutic target for nonalcoholic fatty liver disease. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 193–204. [Google Scholar] [CrossRef]
- Martín-Mateos, R.; Albillos, A. The Role of the Gut-Liver Axis in Metabolic Dysfunction-Associated Fatty Liver Disease. Front. Immunol. 2021, 12, 660179. [Google Scholar] [CrossRef]
- Wiest, R.; Albillos, A.; Trauner, M.; Bajaj, J.S.; Jalan, R. Corrigendum to “Targeting the gut-liver axis in liver disease” [J Hepatol 67 (2017) 1084-1103]. J. Hepatol. 2018, 68, 1336. [Google Scholar] [CrossRef] [PubMed]
- Hugon, P.; Dufour, J.-C.; Colson, P.; Fournier, P.-E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef]
- Albhaisi, S.A.M.; Bajaj, J.S.; Sanyal, A.J. Role of gut microbiota in liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G84–G98. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Deschasaux, M.; Bouter, K.E.; Prodan, A.; Levin, E.; Groen, A.K.; Herrema, H.; Tremaroli, V.; Bakker, G.J.; Attaye, I.; Pinto-Sietsma, S.-J.; et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018, 24, 1526–1531. [Google Scholar] [CrossRef]
- Rogler, G.; Biedermann, L.; Scharl, M. New insights into the pathophysiology of inflammatory bowel disease: Microbiota, epigenetics and common signalling pathways. Swiss Med. Wkly 2018, 148, w14599. [Google Scholar] [CrossRef]
- Lee, N.Y.; Suk, K.T. The role of the gut microbiome in liver cirrhosis treatment. Int. J. Mol. Sci. 2020, 22, 199. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives. Biomolecules 2021, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver (EASL); European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO) EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1388–1402. [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012, 55, 2005–2023. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef]
- Arshad, T.; Paik, J.M.; Biswas, R.; Alqahtani, S.A.; Henry, L.; Younossi, Z.M. Nonalcoholic Fatty Liver Disease Prevalence Trends Among Adolescents and Young Adults in the United States, 2007–2016. Hepatol. Commun. 2021, 5, 1676–1688. [Google Scholar] [CrossRef]
- Sheka, A.C.; Adeyi, O.; Thompson, J.; Hameed, B.; Crawford, P.A.; Ikramuddin, S. Nonalcoholic steatohepatitis: A review. JAMA 2020, 323, 1175–1183. [Google Scholar] [CrossRef]
- Jasirwan, C.O.M.; Lesmana, C.R.A.; Hasan, I.; Sulaiman, A.S.; Gani, R.A. The role of gut microbiota in non-alcoholic fatty liver disease: Pathways of mechanisms. Biosci. Microbiota Food Health 2019, 38, 81–88. [Google Scholar] [CrossRef]
- Khan, A.; Ding, Z.; Ishaq, M.; Bacha, A.S.; Khan, I.; Hanif, A.; Li, W.; Guo, X. Understanding the effects of gut microbiota dysbiosis on nonalcoholic fatty liver disease and the possible probiotics role: Recent updates. Int. J. Biol. Sci. 2021, 17, 818–833. [Google Scholar] [CrossRef]
- Ge, H.; Li, X.; Weiszmann, J.; Wang, P.; Baribault, H.; Chen, J.-L.; Tian, H.; Li, Y. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology 2008, 149, 4519–4526. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ye, J.; Shao, C.; Zhong, B. Compositional alterations of gut microbiota in nonalcoholic fatty liver disease patients: A systematic review and Meta-analysis. Lipids Health Dis. 2021, 20, 22. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.R.; Hoyles, L.; Flint, H.J.; Dumas, M.-E. Colonic bacterial metabolites and human health. Curr. Opin. Microbiol. 2013, 16, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Tokuhara, D. Role of the Gut Microbiota in Regulating Non-alcoholic Fatty Liver Disease in Children and Adolescents. Front. Nutr. 2021, 8, 700058. [Google Scholar] [CrossRef]
- Schwimmer, J.B.; Johnson, J.S.; Angeles, J.E.; Behling, C.; Belt, P.H.; Borecki, I.; Bross, C.; Durelle, J.; Goyal, N.P.; Hamilton, G.; et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology 2019, 157, 1109–1122. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875.e1. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. [Google Scholar] [CrossRef]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandonà, A.; Paci, P.; Capuani, G.; et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; McGilvray, I.D.; Allard, J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef]
- Monga Kravetz, A.; Testerman, T.; Galuppo, B.; Graf, J.; Pierpont, B.; Siebel, S.; Feinn, R.; Santoro, N. Effect of Gut Microbiota and PNPLA3 rs738409 Variant on Nonalcoholic Fatty Liver Disease (NAFLD) in Obese Youth. J. Clin. Endocrinol. Metab. 2020, 105, e3575–e3585. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.-S.; Tse, C.-H.; Lam, T.T.-Y.; Wong, G.L.-H.; Chim, A.M.-L.; Chu, W.C.-W.; Yeung, D.K.-W.; Law, P.T.-W.; Kwan, H.-S.; Yu, J.; et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis—A longitudinal study. PLoS ONE 2013, 8, e62885. [Google Scholar] [CrossRef] [PubMed]
- DuPont, A.W.; DuPont, H.L. The intestinal microbiota and chronic disorders of the gut. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Dubinkina, V.B.; Tyakht, A.V.; Odintsova, V.Y.; Yarygin, K.S.; Kovarsky, B.A.; Pavlenko, A.V.; Ischenko, D.S.; Popenko, A.S.; Alexeev, D.G.; Taraskina, A.Y.; et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome 2017, 5, 141. [Google Scholar] [CrossRef]
- Mutlu, E.A.; Gillevet, P.M.; Rangwala, H.; Sikaroodi, M.; Naqvi, A.; Engen, P.A.; Kwasny, M.; Lau, C.K.; Keshavarzian, A. Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G966–G978. [Google Scholar] [CrossRef]
- Chen, P.; Stärkel, P.; Turner, J.R.; Ho, S.B.; Schnabl, B. Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology 2015, 61, 883–894. [Google Scholar] [CrossRef]
- Wang, L.; Fouts, D.E.; Stärkel, P.; Hartmann, P.; Chen, P.; Llorente, C.; DePew, J.; Moncera, K.; Ho, S.B.; Brenner, D.A.; et al. Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe 2016, 19, 227–239. [Google Scholar] [CrossRef]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- GBD 2017 Cirrhosis Collaborators. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar] [CrossRef]
- Ginès, P.; Krag, A.; Abraldes, J.G.; Solà, E.; Fabrellas, N.; Kamath, P.S. Liver cirrhosis. Lancet 2021, 398, 1359–1376. [Google Scholar] [CrossRef]
- Malhi, H.; Gores, G.J. Cellular and molecular mechanisms of liver injury. Gastroenterology 2008, 134, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, G.; Garcia-Tsao, G.; Pagliaro, L. Natural history and prognostic indicators of survival in cirrhosis: A systematic review of 118 studies. J. Hepatol. 2006, 44, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Ridlon, J.M.; Hylemon, P.B.; Thacker, L.R.; Heuman, D.M.; Smith, S.; Sikaroodi, M.; Gillevet, P.M. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G168–G175. [Google Scholar] [CrossRef] [PubMed]
- Maslennikov, R.; Ivashkin, V.; Efremova, I.; Alieva, A.; Kashuh, E.; Tsvetaeva, E.; Poluektova, E.; Shirokova, E.; Ivashkin, K. Gut dysbiosis is associated with poorer long-term prognosis in cirrhosis. World J. Hepatol. 2021, 13, 557–570. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhai, H.; Geng, J.; Yu, R.; Ren, H.; Fan, H.; Shi, P. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am. J. Gastroenterol. 2013, 108, 1601–1611. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Betrapally, N.S.; Gillevet, P.M.; Bajaj, J.S. Gut microbiome and liver disease. Transl. Res. 2017, 179, 49–59. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Alves, J.M.; Hylemon, P.B.; Bajaj, J.S. Cirrhosis, bile acids and gut microbiota: Unraveling a complex relationship. Gut Microbes 2013, 4, 382–387. [Google Scholar] [CrossRef]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ji, F.; Guo, J.; Shi, D.; Fang, D.; Li, L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci. Rep. 2016, 6, 34055. [Google Scholar] [CrossRef] [PubMed]
- Patidar, K.R.; Bajaj, J.S. Covert and overt hepatic encephalopathy: Diagnosis and management. Clin. Gastroenterol. Hepatol. 2015, 13, 2048–2061. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, V.; Betrapally, N.S.; Hylemon, P.B.; White, M.B.; Gillevet, P.M.; Unser, A.B.; Fagan, A.; Daita, K.; Heuman, D.M.; Zhou, H.; et al. Impaired Gut-Liver-Brain Axis in Patients with Cirrhosis. Sci. Rep. 2016, 6, 26800. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, Y.; Li, J.; Zhao, L.; Li, Z.; Xu, J.; Zhao, F.; Feng, J.; Chen, H.; Fang, C.; et al. Small bowel transit and altered gut microbiota in patients with liver cirrhosis. Front. Physiol. 2018, 9, 470. [Google Scholar] [CrossRef]
- Horvath, A.; Rainer, F.; Bashir, M.; Leber, B.; Schmerboeck, B.; Klymiuk, I.; Groselj-Strele, A.; Durdevic, M.; Freedberg, D.E.; Abrams, J.A.; et al. Biomarkers for oralization during long-term proton pump inhibitor therapy predict survival in cirrhosis. Sci. Rep. 2019, 9, 12000. [Google Scholar] [CrossRef]
- Solé, C.; Guilly, S.; Da Silva, K.; Llopis, M.; Le-Chatelier, E.; Huelin, P.; Carol, M.; Moreira, R.; Fabrellas, N.; De Prada, G.; et al. Alterations in Gut Microbiome in Cirrhosis as Assessed by Quantitative Metagenomics: Relationship With Acute-on-Chronic Liver Failure and Prognosis. Gastroenterology 2021, 160, 206–218.e13. [Google Scholar] [CrossRef]
- Akkız, H. The gut microbiome and hepatocellular carcinoma. J. Gastrointest. Cancer 2021, 52, 1314–1319. [Google Scholar] [CrossRef]
- Balogh, J.; Victor, D.; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, R.M.; Monsour, H.P. Hepatocellular carcinoma: A review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef]
- Mittal, S.; El-Serag, H.B. Epidemiology of hepatocellular carcinoma: Consider the population. J. Clin. Gastroenterol. 2013, 47 (Suppl. S2–S6). [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Bandoh, S.; Roberts, L.R. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. [version 1; peer review: 4 approved]. F1000Res. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, D.; Srivastava, U.; Thaler, M.; Kleinhans, K.N.; N’kontchou, G.; Scheffold, A.; Bauer, K.; Kratzer, R.F.; Kloos, N.; Katz, S.-F.; et al. Telomerase gene mutations are associated with cirrhosis formation. Hepatology 2011, 53, 1608–1617. [Google Scholar] [CrossRef] [PubMed]
- Calado, R.T.; Brudno, J.; Mehta, P.; Kovacs, J.J.; Wu, C.; Zago, M.A.; Chanock, S.J.; Boyer, T.D.; Young, N.S. Constitutional telomerase mutations are genetic risk factors for cirrhosis. Hepatology 2011, 53, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M. p53 mutation in hepatocellular carcinoma after aflatoxin exposure. Lancet 1991, 338, 1356–1359. [Google Scholar] [CrossRef]
- Cleary, S.P.; Jeck, W.R.; Zhao, X.; Chen, K.; Selitsky, S.R.; Savich, G.L.; Tan, T.-X.; Wu, M.C.; Getz, G.; Lawrence, M.S.; et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013, 58, 1693–1702. [Google Scholar] [CrossRef]
- Deng, Y.-B.; Nagae, G.; Midorikawa, Y.; Yagi, K.; Tsutsumi, S.; Yamamoto, S.; Hasegawa, K.; Kokudo, N.; Aburatani, H.; Kaneda, A. Identification of genes preferentially methylated in hepatitis C virus-related hepatocellular carcinoma. Cancer Sci. 2010, 101, 1501–1510. [Google Scholar] [CrossRef]
- Su, P.-F.; Lee, T.-C.; Lin, P.-J.; Lee, P.-H.; Jeng, Y.-M.; Chen, C.-H.; Liang, J.-D.; Chiou, L.-L.; Huang, G.-T.; Lee, H.-S. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int. J. Cancer 2007, 121, 1257–1264. [Google Scholar] [CrossRef]
- Xie, G.; Wang, X.; Liu, P.; Wei, R.; Chen, W.; Rajani, C.; Hernandez, B.Y.; Alegado, R.; Dong, B.; Li, D.; et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 2016, 7, 19355–19366. [Google Scholar] [CrossRef]
- Grąt, M.; Wronka, K.M.; Krasnodębski, M.; Masior, Ł.; Lewandowski, Z.; Kosińska, I.; Grąt, K.; Stypułkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of gut microbiota associated with the presence of hepatocellular cancer in patients with liver cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef]
- Zhang, H.-L.; Yu, L.-X.; Yang, W.; Tang, L.; Lin, Y.; Wu, H.; Zhai, B.; Tan, Y.-X.; Shan, L.; Liu, Q.; et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J. Hepatol. 2012, 57, 803–812. [Google Scholar] [CrossRef]
- Ni, J.; Huang, R.; Zhou, H.; Xu, X.; Li, Y.; Cao, P.; Zhong, K.; Ge, M.; Chen, X.; Hou, B.; et al. Analysis of the relationship between the degree of dysbiosis in gut microbiota and prognosis at different stages of primary hepatocellular carcinoma. Front. Microbiol. 2019, 10, 1458. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, F.; Zhuang, Y.; Xu, J.; Wang, J.; Mao, X.; Zhang, Y.; Liu, X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef]
- Yamada, S.; Takashina, Y.; Watanabe, M.; Nagamine, R.; Saito, Y.; Kamada, N.; Saito, H. Bile acid metabolism regulated by the gut microbiota promotes non-alcoholic steatohepatitis-associated hepatocellular carcinoma in mice. Oncotarget 2018, 9, 9925–9939. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Kawamoto, K.; Horibe, I.; Uchida, K. Purification and characterization of a new hydrolase for conjugated bile acids, chenodeoxycholyltaurine hydrolase, from Bacteroides vulgatus. J. Biochem. 1989, 106, 1049–1053. [Google Scholar] [CrossRef]
- Behary, J.; Amorim, N.; Jiang, X.-T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 2021, 12, 187. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef]
- Piñero, F.; Vazquez, M.; Baré, P.; Rohr, C.; Mendizabal, M.; Sciara, M.; Alonso, C.; Fay, F.; Silva, M. A different gut microbiome linked to inflammation found in cirrhotic patients with and without hepatocellular carcinoma. Ann. Hepatol. 2019, 18, 480–487. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, Y.-N.; Chen, T.; Ren, C.-H.; Li, X.; Liu, G.-X. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Czaja, A.J. Global disparities and their implications in the occurrence and outcome of autoimmune hepatitis. Dig. Dis. Sci. 2017, 62, 2277–2292. [Google Scholar] [CrossRef] [PubMed]
- Tunio, N.A.; Mansoor, E.; Sheriff, M.Z.; Cooper, G.S.; Sclair, S.N.; Cohen, S.M. Epidemiology of Autoimmune Hepatitis (AIH) in the United States Between 2014 and 2019: A Population-based National Study. J. Clin. Gastroenterol. 2021, 55, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Lata, J. Diagnosis and treatment of autoimmune hepatitis. Dig. Dis. 2012, 30, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Baven-Pronk, M.A.M.C.; Biewenga, M.; van Silfhout, J.J.; van den Berg, A.P.; van Buuren, H.R.; Verwer, B.J.; van Nieuwkerk, C.M.J.; Bouma, G.; van Hoek, B. Role of age in presentation, response to therapy and outcome of autoimmune hepatitis. Clin. Transl. Gastroenterol. 2018, 9, 165. [Google Scholar] [CrossRef]
- de Boer, Y.S.; van Gerven, N.M.F.; Zwiers, A.; Verwer, B.J.; van Hoek, B.; van Erpecum, K.J.; Beuers, U.; van Buuren, H.R.; Drenth, J.P.H.; den Ouden, J.W.; et al. Study of Health in Pomerania Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology 2014, 147, 443–452.e5. [Google Scholar] [CrossRef]
- Rigopoulou, E.I.; Smyk, D.S.; Matthews, C.E.; Billinis, C.; Burroughs, A.K.; Lenzi, M.; Bogdanos, D.P. Epstein-barr virus as a trigger of autoimmune liver diseases. Adv. Virol. 2012, 2012, 987471. [Google Scholar] [CrossRef]
- Björnsson, E.; Talwalkar, J.; Treeprasertsuk, S.; Kamath, P.S.; Takahashi, N.; Sanderson, S.; Neuhauser, M.; Lindor, K. Drug-induced autoimmune hepatitis: Clinical characteristics and prognosis. Hepatology 2010, 51, 2040–2048. [Google Scholar] [CrossRef]
- Lammert, C. Genetic and environmental risk factors for autoimmune hepatitis. Clin. Liver Dis. 2019, 14, 29–32. [Google Scholar] [CrossRef]
- Floreani, A.; Restrepo-Jiménez, P.; Secchi, M.F.; De Martin, S.; Leung, P.S.C.; Krawitt, E.; Bowlus, C.L.; Gershwin, M.E.; Anaya, J.-M. Etiopathogenesis of autoimmune hepatitis. J. Autoimmun. 2018, 95, 133–143. [Google Scholar] [CrossRef]
- Mieli-Vergani, G.; Vergani, D.; Czaja, A.J.; Manns, M.P.; Krawitt, E.L.; Vierling, J.M.; Lohse, A.W.; Montano-Loza, A.J. Autoimmune hepatitis. Nat. Rev. Dis. Primers 2018, 4, 18017. [Google Scholar] [CrossRef] [PubMed]
- Ehser, J.; Holdener, M.; Christen, S.; Bayer, M.; Pfeilschifter, J.M.; Hintermann, E.; Bogdanos, D.; Christen, U. Molecular mimicry rather than identity breaks T-cell tolerance in the CYP2D6 mouse model for human autoimmune hepatitis. J. Autoimmun. 2013, 42, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Holdener, M.; Hintermann, E.; Bayer, M.; Rhode, A.; Rodrigo, E.; Hintereder, G.; Johnson, E.F.; Gonzalez, F.J.; Pfeilschifter, J.; Manns, M.P.; et al. Breaking tolerance to the natural human liver autoantigen cytochrome P450 2D6 by virus infection. J. Exp. Med. 2008, 205, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Elsherbiny, N.M.; Rammadan, M.; Hassan, E.A.; Ali, M.E.; El-Rehim, A.S.A.; Abbas, W.A.; Abozaid, M.A.A.; Hassanin, E.; Hetta, H.F. Autoimmune Hepatitis: Shifts in Gut Microbiota and Metabolic Pathways among Egyptian Patients. Microorganisms 2020, 8, 1011. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Li, Y.; Yan, L.; Sun, C.; Miao, Q.; Wang, Q.; Xiao, X.; Lian, M.; Li, B.; Chen, Y.; et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020, 69, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Casar, C.; Ruehlemann, M.C.; Bang, C.; Sebode, M.; Hohenester, S.; Denk, G.; Lieb, W.; Lohse, A.W.; Franke, A.; et al. A disease-specific decline of the relative abundance of Bifidobacterium in patients with autoimmune hepatitis. Aliment. Pharmacol. Ther. 2020, 51, 1417–1428. [Google Scholar] [CrossRef]
- Lin, R.; Zhou, L.; Zhang, J.; Wang, B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int. J. Clin. Exp. Pathol. 2015, 8, 5153–5160. [Google Scholar]
- Liang, M.; Liwen, Z.; Jianguo, S.; Juan, D.; Fei, D.; Yin, Z.; Changping, W.; Jianping, C. Fecal microbiota transplantation controls progression of experimental autoimmune hepatitis in mice by modulating the TFR/TFH immune imbalance and intestinal microbiota composition. Front. Immunol. 2021, 12, 728723. [Google Scholar] [CrossRef]
- Liu, Q.; Tian, H.; Kang, Y.; Tian, Y.; Li, L.; Kang, X.; Yang, H.; Wang, Y.; Tian, J.; Zhang, F.; et al. Probiotics alleviate autoimmune hepatitis in mice through modulation of gut microbiota and intestinal permeability. J. Nutr. Biochem. 2021, 98, 108863. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, M.; Liu, X.; Zhong, W.; Li, Y.; Ran, Y.; Guo, L.; Chen, X.; Zhao, J.; Wang, B.; et al. Bifidobacterium animalis ssp. Lactis 420 Mitigates Autoimmune Hepatitis Through Regulating Intestinal Barrier and Liver Immune Cells. Front. Immunol. 2020, 11, 569104. [Google Scholar] [CrossRef]
- Lou, J.; Jiang, Y.; Rao, B.; Li, A.; Ding, S.; Yan, H.; Zhou, H.; Liu, Z.; Shi, Q.; Cui, G.; et al. Fecal microbiomes distinguish patients with autoimmune hepatitis from healthy individuals. Front. Cell. Infect. Microbiol. 2020, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhang, X.; Liu, J.; Zhang, Q.; Zhao, Y.; Peng, J.; Feng, Q.; Dai, J.; Sun, S.; et al. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front. Microbiol. 2017, 8, 2222. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wu, Z.; Xu, W.; Yang, J.; Chen, Y.; Li, L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb. Ecol. 2011, 61, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.M.; Adel, A.; El-Gendy, A.O.; Essam, T.M.; Aziz, R.K. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016, 8, 42. [Google Scholar] [CrossRef]
- Heidrich, B.; Vital, M.; Plumeier, I.; Döscher, N.; Kahl, S.; Kirschner, J.; Ziegert, S.; Solbach, P.; Lenzen, H.; Potthoff, A.; et al. Intestinal microbiota in patients with chronic hepatitis C with and without cirrhosis compared with healthy controls. Liver Int. 2018, 38, 50–58. [Google Scholar] [CrossRef]
- Dyson, J.K.; Beuers, U.; Jones, D.E.J.; Lohse, A.W.; Hudson, M. Primary sclerosing cholangitis. Lancet 2018, 391, 2547–2559. [Google Scholar] [CrossRef]
- Torres, J.; Palmela, C.; Brito, H.; Bao, X.; Ruiqi, H.; Moura-Santos, P.; Pereira da Silva, J.; Oliveira, A.; Vieira, C.; Perez, K.; et al. The gut microbiota, bile acids and their correlation in primary sclerosing cholangitis associated with inflammatory bowel disease. United Eur. Gastroenterol. J. 2018, 6, 112–122. [Google Scholar] [CrossRef]
- Bajer, L.; Kverka, M.; Kostovcik, M.; Macinga, P.; Dvorak, J.; Stehlikova, Z.; Brezina, J.; Wohl, P.; Spicak, J.; Drastich, P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 2017, 23, 4548–4558. [Google Scholar] [CrossRef]
- Lichtman, S.N.; Keku, J.; Clark, R.L.; Schwab, J.H.; Sartor, R.B. Biliary tract disease in rats with experimental small bowel bacterial overgrowth. Hepatology 1991, 13, 766–772. [Google Scholar] [CrossRef]
- Vaughn, B.P.; Kaiser, T.; Staley, C.; Hamilton, M.J.; Reich, J.; Graiziger, C.; Singroy, S.; Kabage, A.J.; Sadowsky, M.J.; Khoruts, A. A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin. Exp. Gastroenterol. 2019, 12, 9–19. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Shaheen, W.; Oo, Y.H.; Iqbal, T.H. Immunological mechanisms underpinning faecal microbiota transplantation for the treatment of inflammatory bowel disease. Clin. Exp. Immunol. 2020, 199, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Orlandini, B.; Tenori, L.; Biagini, M.R.; Milani, S.; Renzi, D.; Luchinat, C.; Calabrò, A.S. Metabolic Signature of Primary Biliary Cholangitis and Its Comparison with Celiac Disease. J. Proteome Res. 2019, 18, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.-X.; Fang, D.-Q.; Shi, D.; Chen, D.-Y.; Yan, R.; Zhu, Y.-X.; Chen, Y.-F.; Shao, L.; Guo, F.-F.; Wu, W.-R.; et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ. Microbiol. 2016, 18, 2272–2286. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Wei, Y.; Li, Y.; Chen, W.; Chen, H.; Wang, Q.; Yang, F.; Miao, Q.; Xiao, X.; Zhang, H.; et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 2018, 67, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Stärkel, P.; Fan, J.-G.; Fouts, D.E.; Bacher, P.; Schnabl, B. The gut mycobiome: A novel player in chronic liver diseases. J. Gastroenterol. 2021, 56, 1–11. [Google Scholar] [CrossRef]
- Gao, W.; Zhu, Y.; Ye, J.; Chu, H. Gut non-bacterial microbiota contributing to alcohol-associated liver disease. Gut Microbes 2021, 13, 1984122. [Google Scholar] [CrossRef]
- Hsu, C.L.; Duan, Y.; Fouts, D.E.; Schnabl, B. Intestinal virome and therapeutic potential of bacteriophages in liver disease. J. Hepatol. 2021, 75, 1465–1475. [Google Scholar] [CrossRef]
- Stasiewicz, M.; Kwaśniewski, M.; Karpiński, T.M. Microbial Associations with Pancreatic Cancer: A New Frontier in Biomarkers. Cancers 2021, 13, 3784. [Google Scholar] [CrossRef]
- Chu, H.; Duan, Y.; Lang, S.; Jiang, L.; Wang, Y.; Llorente, C.; Liu, J.; Mogavero, S.; Bosques-Padilla, F.; Abraldes, J.G.; et al. The Candida albicans exotoxin candidalysin promotes alcohol-associated liver disease. J. Hepatol. 2020, 72, 391–400. [Google Scholar] [CrossRef]
- Spatz, M.; Richard, M.L. Overview of the potential role of malassezia in gut health and disease. Front. Cell. Infect. Microbiol. 2020, 10, 201. [Google Scholar] [CrossRef]
- Nd, A.M. Non-Alcoholic Fatty Liver Disease, an Overview. Integr. Med. 2019, 18, 42–49. [Google Scholar]
- You, N.; Zhuo, L.; Zhou, J.; Song, Y.; Shi, J. The role of intestinal fungi and its metabolites in chronic liver diseases. Gut Liver 2020, 14, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Sikaroodi, M.; Shamsaddini, A.; Henseler, Z.; Santiago-Rodriguez, T.; Acharya, C.; Fagan, A.; Hylemon, P.B.; Fuchs, M.; Gavis, E.; et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 2021, 70, 1162–1173. [Google Scholar] [CrossRef] [PubMed]
- Castillo, V.; Figueroa, F.; González-Pizarro, K.; Jopia, P.; Ibacache-Quiroga, C. Probiotics and Prebiotics as a Strategy for Non-Alcoholic Fatty Liver Disease, a Narrative Review. Foods 2021, 10, 1719. [Google Scholar] [CrossRef] [PubMed]
- Pineiro, M.; Asp, N.-G.; Reid, G.; Macfarlane, S.; Morelli, L.; Brunser, O.; Tuohy, K. FAO Technical meeting on prebiotics. J. Clin. Gastroenterol. 2008, 42 Pt 2 (Suppl. S3), S156–S159. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, R.K.; Rana, B.; Agrawal, S.; Garg, A.; Chopra, M.; Thumburu, K.K.; Khattri, A.; Malhotra, S.; Duseja, A.; Chawla, Y.K. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: A randomized, controlled trial. Gastroenterology 2014, 147, 1327–1337.e3. [Google Scholar] [CrossRef]
- Horvath, A.; Durdevic, M.; Leber, B.; di Vora, K.; Rainer, F.; Krones, E.; Douschan, P.; Spindelboeck, W.; Durchschein, F.; Zollner, G.; et al. Changes in the Intestinal Microbiome during a Multispecies Probiotic Intervention in Compensated Cirrhosis. Nutrients 2020, 12, 1874. [Google Scholar] [CrossRef]
- Mei, L.; Tang, Y.; Li, M.; Yang, P.; Liu, Z.; Yuan, J.; Zheng, P. Co-Administration of Cholesterol-Lowering Probiotics and Anthraquinone from Cassia obtusifolia L. Ameliorate Non-Alcoholic Fatty Liver. PLoS ONE 2015, 10, e0138078. [Google Scholar] [CrossRef]
- Hsieh, F.-C.; Lee, C.-L.; Chai, C.-Y.; Chen, W.-T.; Lu, Y.-C.; Wu, C.-S. Oral administration of Lactobacillus reuteri GMNL-263 improves insulin resistance and ameliorates hepatic steatosis in high fructose-fed rats. Nutr. Metab. 2013, 10, 35. [Google Scholar] [CrossRef]
- Liang, Y.; Liang, S.; Zhang, Y.; Deng, Y.; He, Y.; Chen, Y.; Liu, C.; Lin, C.; Yang, Q. Oral Administration of Compound Probiotics Ameliorates HFD-Induced Gut Microbe Dysbiosis and Chronic Metabolic Inflammation via the G Protein-Coupled Receptor 43 in Non-alcoholic Fatty Liver Disease Rats. Probiotics Antimicrob. Proteins 2019, 11, 175–185. [Google Scholar] [CrossRef]
- Witjes, J.J.; Smits, L.P.; Pekmez, C.T.; Prodan, A.; Meijnikman, A.S.; Troelstra, M.A.; Bouter, K.E.C.; Herrema, H.; Levin, E.; Holleboom, A.G.; et al. Donor fecal microbiota transplantation alters gut microbiota and metabolites in obese individuals with steatohepatitis. Hepatol. Commun. 2020, 4, 1578–1590. [Google Scholar] [CrossRef] [PubMed]
- Bomhof, M.R.; Parnell, J.A.; Ramay, H.R.; Crotty, P.; Rioux, K.P.; Probert, C.S.; Jayakumar, S.; Raman, M.; Reimer, R.A. Histological improvement of non-alcoholic steatohepatitis with a prebiotic: A pilot clinical trial. Eur. J. Nutr. 2019, 58, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Ferolla, S.M.; Couto, C.A.; Costa-Silva, L.; Armiliato, G.N.A.; Pereira, C.A.S.; Martins, F.S.; Ferrari, M.d.L.A.; Vilela, E.G.; Torres, H.O.G.; Cunha, A.S.; et al. Beneficial Effect of Synbiotic Supplementation on Hepatic Steatosis and Anthropometric Parameters, But Not on Gut Permeability in a Population with Nonalcoholic Steatohepatitis. Nutrients 2016, 8, 397. [Google Scholar] [CrossRef] [PubMed]
- Asgharian, A.; Askari, G.; Esmailzade, A.; Feizi, A.; Mohammadi, V. The Effect of Symbiotic Supplementation on Liver Enzymes, C-reactive Protein and Ultrasound Findings in Patients with Non-alcoholic Fatty Liver Disease: A Clinical Trial. Int. J. Prev. Med. 2016, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sung, C.Y.J.; Lee, N.; Ni, Y.; Pihlajamäki, J.; Panagiotou, G.; El-Nezami, H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E1306–E1315. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Song, J.; Li, X.; Li, N.; Dai, J. Immunomodulation and antitumor activity of kappa-carrageenan oligosaccharides. Cancer Lett. 2006, 243, 228–234. [Google Scholar] [CrossRef]
- Kang, D.J.; Kakiyama, G.; Betrapally, N.S.; Herzog, J.; Nittono, H.; Hylemon, P.B.; Zhou, H.; Carroll, I.; Yang, J.; Gillevet, P.M.; et al. Rifaximin exerts beneficial effects independent of its ability to alter microbiota composition. Clin. Transl. Gastroenterol. 2016, 7, e187. [Google Scholar] [CrossRef]
- Amarapurkar, D.N. Prescribing medications in patients with decompensated liver cirrhosis. Int. J. Hepatol. 2011, 2011, 519526. [Google Scholar] [CrossRef][Green Version]
- Zoratti, C.; Moretti, R.; Rebuzzi, L.; Albergati, I.V.; Di Somma, A.; Decorti, G.; Di Bella, S.; Crocè, L.S.; Giuffrè, M. Antibiotics and liver cirrhosis: What the physicians need to know. Antibiotics 2021, 11, 31. [Google Scholar] [CrossRef]
- Hassouneh, R.; Bajaj, J.S. Gut microbiota modulation and fecal transplantation: An overview on innovative strategies for hepatic encephalopathy treatment. J. Clin. Med. 2021, 10, 330. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Kassam, Z.; Fagan, A.; Gavis, E.A.; Liu, E.; Cox, I.J.; Kheradman, R.; Heuman, D.; Wang, J.; Gurry, T.; et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology 2017, 66, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Gavis, E.A.; Fagan, A.; Wade, J.B.; Thacker, L.R.; Fuchs, M.; Patel, S.; Davis, B.; Meador, J.; Puri, P.; et al. A randomized clinical trial of fecal microbiota transplant for alcohol use disorder. Hepatology 2021, 73, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Sinakos, E.; Lindor, K. Treatment options for primary sclerosing cholangitis. Expert Rev. Gastroenterol. Hepatol. 2010, 4, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Michaels, A.; Levy, C. The medical management of primary sclerosing cholangitis. Medscape J. Med. 2008, 10, 61. [Google Scholar] [PubMed]
- Chascsa, D.M.H.; Lindor, K.D. Emerging therapies for PBC. J. Gastroenterol. 2020, 55, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Peter, J.A.; Nelson, D.R.; Keach, J.; Petz, J.; Cabrera, R.; Clark, V.; Firpi, R.J.; Morelli, G.; Soldevila-Pico, C.; et al. Pilot study: Fenofibrate for patients with primary biliary cirrhosis and an incomplete response to ursodeoxycholic acid. Aliment. Pharmacol. Ther. 2011, 33, 235–242. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Fagan, A.; Sikaroodi, M.; White, M.B.; Sterling, R.K.; Gilles, H.; Heuman, D.; Stravitz, R.T.; Matherly, S.C.; Siddiqui, M.S.; et al. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl. 2017, 23, 907–914. [Google Scholar] [CrossRef]
- Isoura, Y.; Cho, Y.; Fujimoto, H.; Hamazaki, T.; Tokuhara, D. Effects of obesity reduction on transient elastography-based parameters in pediatric non-alcoholic fatty liver disease. Obes. Res. Clin. Pract. 2020, 14, 473–478. [Google Scholar] [CrossRef]
- Lata, J.; Novotný, I.; Príbramská, V.; Juránková, J.; Fric, P.; Kroupa, R.; Stibůrek, O. The effect of probiotics on gut flora, level of endotoxin and Child-Pugh score in cirrhotic patients: Results of a double-blind randomized study. Eur. J. Gastroenterol. Hepatol. 2007, 19, 1111–1113. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; Puri, P.; Sterling, R.K.; Luketic, V.; Stravitz, R.T.; Siddiqui, M.S.; Fuchs, M.; et al. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment. Pharmacol. Ther. 2014, 39, 1113–1125. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Matin, P.; White, M.B.; Fagan, A.; Deeb, J.G.; Acharya, C.; Dalmet, S.S.; Sikaroodi, M.; Gillevet, P.M.; Sahingur, S.E. Periodontal therapy favorably modulates the oral-gut-hepatic axis in cirrhosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G824–G837. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manzoor, R.; Ahmed, W.; Afify, N.; Memon, M.; Yasin, M.; Memon, H.; Rustom, M.; Al Akeel, M.; Alhajri, N. Trust Your Gut: The Association of Gut Microbiota and Liver Disease. Microorganisms 2022, 10, 1045. https://doi.org/10.3390/microorganisms10051045
Manzoor R, Ahmed W, Afify N, Memon M, Yasin M, Memon H, Rustom M, Al Akeel M, Alhajri N. Trust Your Gut: The Association of Gut Microbiota and Liver Disease. Microorganisms. 2022; 10(5):1045. https://doi.org/10.3390/microorganisms10051045
Chicago/Turabian StyleManzoor, Ridda, Weshah Ahmed, Nariman Afify, Mashal Memon, Maryam Yasin, Hamda Memon, Mohammad Rustom, Mohannad Al Akeel, and Noora Alhajri. 2022. "Trust Your Gut: The Association of Gut Microbiota and Liver Disease" Microorganisms 10, no. 5: 1045. https://doi.org/10.3390/microorganisms10051045
APA StyleManzoor, R., Ahmed, W., Afify, N., Memon, M., Yasin, M., Memon, H., Rustom, M., Al Akeel, M., & Alhajri, N. (2022). Trust Your Gut: The Association of Gut Microbiota and Liver Disease. Microorganisms, 10(5), 1045. https://doi.org/10.3390/microorganisms10051045






