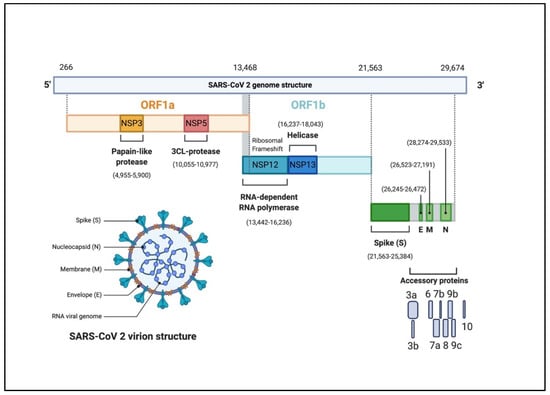
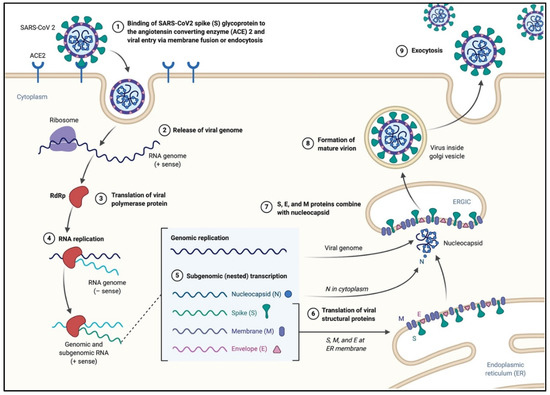
Abstract
The emerging coronavirus disease (COVID-19) swept across the world, affecting more than 200 countries and territories. Genomic analysis suggests that the COVID-19 virus originated in bats and transmitted to humans through unknown intermediate hosts in the Wuhan seafood market, China, in December of 2019. This virus belongs to the Betacoronavirus group, the same group of the 2003 severe acute respiratory syndrome coronavirus (SARS-CoV), and for the similarity, it was named SARS-CoV-2. Given the lack of registered clinical therapies or vaccines, many physicians and scientists are investigating previously used clinical drugs for COVID-19 treatment. In this review, we aim to provide an overview of the CoVs origin, pathogenicity, and genomic structure, with a focus on SARS-CoV-2. Besides, we summarize the recently investigated drugs that constitute an option for COVID-19 treatment.
1. Introduction
The coronavirus disease 2019 (COVID-19) was first identified in patients with severe respiratory disease in Wuhan, China. The causative agent was a novel coronavirus scientifically named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,2]. Since its discovery, more than 2,850,000 cases have been infected, including nearly 200,000 who have died. The worrisome features of COVID-19 are its apparent ability to spread readily and its propensity to cause severe disease in older adults and patients with existing health conditions [3,4]. Moreover, coronaviruses are well-known to mutate and recombine [5]. The genomic sequence of SARS-CoV-2 has changed since it was first reported. Some scientists believe that these changes have enhanced the virulence of the virus and there are two circulating strains, deadly strain “L”, and less virulent one “S” [6]. Currently, there are no specific antiviral treatments or vaccines available for COVID-19. Treatments are mainly focusing on symptomatic and respiratory support according to protocols issued by the health authority in each country, where many countries are following WHO protocol [3,4]. Nearly all cases with severe symptoms accept oxygen. In critical cases and life-threatening situations, passive immunization through the convalescent plasma and immunoglobulin G transfusion can be used as a rescue treatment [7]. Given the current situation, finding a treatment has become a global public health priority. Many companies are working on finding a medication or a viable vaccine for SARS-CoV-2. However, this is not a short process; scientists estimate the period to have one, about 12–18 months.
The COVID-19 is the third novel coronavirus to cause a large-scale epidemic in the twenty-first century after the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) in 2003 [8,9,10] and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012 [11,12,13]. Our knowledge and experience in fighting the previous two epidemics can be used for creating treatment strategies against this pandemic. Recently, different medications and a combination of drugs have shown promising results. For example, the combination of chloroquine and azithromycin [14]. Furthermore, the antiviral drug remdesivir and the malaria drug chloroquine have gained much attention globally as potential medications for COVID-19 treatment. On the other hand, different companies are working on finding a treatment in a creative method, antiviral RNA, for example [15].
Given the lack of specific treatment for COVID-19, many groups are working urgently to find an alternative strategy to control the replication and spreading of the virus. We have explored different scientific databases searching for the most promising options for the treatment of COVID-19 and provide an overview of our findings. We are going to review the characteristics of coronaviruses, its origin, genome structure, and replication to explain how each treatment strategy could act on preventing or slowing the viral infection.
3. Potential COVID-19 Treatment
Antiviral drugs usually interfere with the viral replication machinery in the target cells. Given that viruses have similar actions of infection, different studies suggest the use of successful antiviral drugs for the treatment of COVID-19. Oseltamivir, peramivir, and zanamivir are neuraminidase inhibitors that have been used for influenza treatment. A recent report suggests the potential activity of these drugs against SAS-CoV-2 by interfering with the virions budding and releasing from the cell [35]. However, this virus uses different mechanisms other than neuraminidase to facilitate its release out of the infected cell. Another recent study suggests that ribavirin if combined with interferon-β, might be a promising drug for inhibition of SARS-CoV-2 replication [7]. Although this claim was based on previous in vitro findings [36], no other studies have shown the same results. Moreover, two reports did not recommend the use of this drug. On one hand, because it has no significant activity against SAS-CoV, and on the other hand, ribavirin is toxic with hemolysis activity [37,38]. Even though, there are many drugs with general antiviral activity; that can interfere with vial entry or blocking the virus receptors. Some examples are listed in Table 1. These drugs are either under investigation, showed antiviral activity against SARS-CoV-2, or constituted a treatment option of COVID-19.

Table 1.
Common and potent antiviral drugs.
3.1. Chloroquine and Hydroxychloroquine
Although many drugs have been reported to show promising results against SARS-CoV-2, none of these drugs have gained much attention like the antimalarial drugs Chloroquine (CQ) and Hydroxychloroquine (HCQ) [14,49,50]. HCQ is a CQ analog that has the same mechanism of action with a safer profile making it the prioritized drug to treat malaria, viruses, and autoimmune conditions [51]. There are several potential molecular mechanisms of HCQ action against SARS-CoV-2 that have been postulated. Generally, HCQ is a weak base drug that accumulates in the cell acidic compartments such as lysosomes and endosomes. This accumulation increases the pH and inhibits the maturation of these endosomal compartments. Therefore, HCQ might be interfering with the endocytosis of SARS-CoV-2 and subsequently affecting the virus entry and exit from the host cell [7,50]. Moreover, HCQ might reduce glycosylation of ACE2, interfering with SARS-CoV-2 to bind effectively to the host cell [51]. On the other hand, a recent publication hypothesizes that HCQ reduces the production of pro-inflammatory cytokines (such as interleukin-6, IFN-alpha, and TNF), thereby inhibiting various immune pathways that might lead to acute respiratory distress syndrome (ARDS) [52]. Currently, the use of hydroxychloroquine is approved by many of COVID-19 treatment protocols, especially in combination with other antibiotics and antiviral drugs, where others seriously considering it [1,4,40,50].
3.2. Remdesivir
Remdesivir is an adenosine nucleoside analog drug showing antiviral activity against Ebola and Marburg viruses, and other RNA viruses [53]. It acts through interfering with the viral RNA polymerase and evades proofreading by viral exoribonuclease, leading to a decrease in viral RNA production [54,55]. A recent report found that a combination of remdesivir and chloroquine inhibited SARS-CoV-2 in vitro [56]. Very recently, the drug gained more attention after a recent clinical trial found that remdesivir is highly effective in stopping the replication mechanism of SARS-CoV-2.
3.3. Losartan and Telmisartan
Losartan and telmisartan are drugs that belong to the angiotensin II receptor antagonist group. They block the substances that cause blood vessels narrowing, which lowers blood pressure and improves blood flow by relaxing the blood vessels. It has been suggested that Losartan can be used to reduce the aggressiveness and mortality of SARS-CoV-2 [57,58,59]. The fact that these drugs could block the ACE2 receptor should mean the ability to interfere with the entry of SARS-CoV-2 to the target cell. Using this drug group to treat COVID-19 seems vital at this point, particularly in combination with other antiviral drugs.
3.4. Baricitinib
Baricitinib is a drug used basically for the treatment of rheumatoid arthritis. It acts as an inhibitor for Janus kinase (JAK, U.S. Food and Drug Administration, 2018). Inhibition of the JAK pathway might interrupt the virus entry and the virions intracellular assembly by interfering with the endocytosis process [42].
3.5. Lopinavir/Ritonavir
It is a combination of both lopinavir and ritonavir drugs that have been used for HIV treatment and prevention. These drugs act by inhibiting the HIV protease enzyme through forming an inhibitor-enzyme complex [60]. A recent study showed that combination of these drugs reduced SARS-CoV-2 viral load and improved the clinical symptoms during the treatment of COVID-19 patients [43].
3.6. Darunavir
Darunavir is a second generation of HIV-1 protease inhibitors. It is a non-peptidic inhibitor of protease that enters itself in the active site of protease through several hydrogen bonds [61]. A recent report of an in vitro experiment indicated that using darunavir at a concentration of 300 μM, significantly inhibits SARS-CoV-2 replication. Its inhibition efficiency increased by 280-fold in comparison to the control group [44].
3.7. Camostat Mesylate
Camostat Mesylate is a type II transmembrane serine protease (TMSPSS2) inhibitor [62]. Since that SARS-CoV-2 entry mechanism involving the use of cellular protease TMPRSS2, a protease inhibitor would block entry and thus could be a treatment option [45].
3.8. Cepharanthie, Selamectin, and Mefloquine Hydrochloride
These are medications used as anti-malarial, antiviral, and anti-inflammatory activities. Those drugs showed significantly decreased viral RNA yield in treated cells, making them potential drugs for treating COVID-19 infections [47].
3.9. SARS-CoV-Specific Human Monoclonal Antibody (CR3022)
Passive immunization (PI) is a method to obtain instant, short-term fortification against infectious agents by introducing pathogen-specific antibodies to patients. These specific antibodies can bind to the pathogen antigens and block its interaction with a cellular receptor. It is extremely applicable in the case of viral antigens that facilitate the attachment to the target receptors [63]. After exposure to a viral infection, the body of the patient creates antibodies to fight off the virus. These antibodies in the blood of a recovered patient can be collected as convalescent plasma (CP) and transferred to the blood of a newly infected patient. Consequently, it can neutralize the pathogen, boost the immunity of that patient [64]. CR3022 is a human monoclonal antibody previously isolated from a convalescent SARS patient, used to target a highly conserved epitope that enables cross-reactive binding between SARS-CoV-2 and SARS-CoV [65]. CR3022 might have the potential to be candidate therapeutics for the prevention and treatment of COVID-19 patients, especially in life-threatening situations [66].
3.10. CRISPR/Cas13d System
Clustered regularly interspaced short palindromic repeats (CRISPR) is a family of repeated DNA sequences that can be found in the genomes of bacteria and archaea. These fragments are molecular remaining of bacteriophages infections and act as a defense mechanism to the prokaryotic cell. They can detect and destroy specific foreign DNA fragments, most probably from similar bacteriophages subsequent infections. CRISPR with chaperoning Cas proteins is a powerful technique that has been derived from the bacterial defense mechanism and implemented in gene editing of our interest [67]. Currently, there is an ambitious study suggesting that CRISPR/Cas13d system can be used to accurately digest the SARS-CoV-2 RNA genome, hence limiting its ability to reproduce [15]. Theoretically, this approach is excellent not only against COVID-19 but for the treatment and prevention of different RNA viruses’ infections. However, we have no idea if it is practical or not.
4. Conclusions
The COVID-19 pandemic poses a significant threat to the global public health systems. The increasing number of cases and deaths have put the international community on alert that the worst scenarios are possible. Despite our knowledge of the SARS-CoV-2 infectious cycle, there is no clear strategy for COVID-19 patients’ treatment. Based on recent experimental findings and recommendations, physicians are investigating different potential drugs that showed antiviral activity against SARS-CoV-2. Only time to tell which one of these drugs is going to work. In the meantime, scientists around the globe are working aggressively to find clinical therapies or vaccines against the virus.
Author Contributions
The authors contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, R.H.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical characteristics of coronavirus disease 2019 in China. New Engl. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med Res. 2020, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y. Potential interventions for novel coronavirus in China: A systematic review. J. Med. Virol. 2020, 92, 479–490. [Google Scholar] [CrossRef]
- Lee, N.; Hui, D.S.; Wu, A.; Chan, P.K.S.; Cameron, P.; Joynt, G.; Ahuja, A.T.; Yung, M.Y.; Leung, C.; To, K.; et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. New Engl. J. Med. 2003, 348, 1986–1994. [Google Scholar] [CrossRef]
- Peiris, J.; Lai, S.; Poon, L.L.; Guan, Y.; Yam, L.; Lim, W.; Nicholls, J.M.; Yee, W.; Yan, W.; Cheung, M.; et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef]
- De Wit, E.; Van Doremalen, N.; Falzarano, D.; Munster, V. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Genet. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Alagaili, A.N.; Briese, T.; Mishra, N.; Kapoor, V.; Sameroff, S.C.; De Wit, E.; Munster, V.; Hensley, L.E.; Zalmout, I.S.; Kapoor, A.; et al. Middle east respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 2014, 5, e00884-14. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.; Kapoor, V.; Epstein, J.H.; AlHakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.; Osterhaus, A.D.; Fouchier, R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.-C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 105949. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-F.; Shi, Z.; Zhang, S.; Field, H.; Daszak, P.; Eaton, B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006, 12, 1834–1840. [Google Scholar] [CrossRef]
- Ge, X.-Y.; Li, J.; Yang, X.; Chmura, A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, D. Molecular mechanisms of coronavirus RNA capping and methylation. Virol. Sin. 2016, 31, 3–11. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014, 101, 45–56. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Daszak, P. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Genet. 2018, 17, 181–192. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Ithete, N.; Stoffberg, S.; Corman, V.M.; Cottontail, V.M.; Richards, L.R.; Schoeman, C.; Drosten, C.; Drexler, J.F.; Preiser, W. Close relative of human middle east respiratory syndrome coronavirus in Bat, South Africa. Emerg. Infect. Dis. 2013, 19, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Butt, K.M.; et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The Molecular Biology of Coronaviruses; Elsevier: Amsterdam, The Netherlands, 2006; Volume 66, pp. 193–292. [Google Scholar]
- Demogines, A.; Farzan, M.; Sawyer, S. Evidence for ACE2-Utilizing Coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in Bats. J. Virol. 2012, 86, 6350–6353. [Google Scholar] [CrossRef] [PubMed]
- Neuman, B.; Kiss, G.; Kunding, A.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Boil. 2010, 174, 11–22. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Álvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A severe acute respiratory syndrome coronavirus that lacks the E Gene is attenuated in vitro and in vivo. J. Virol. 2006, 81, 1701–1713. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Therapeutic Antibodies; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1282, pp. 1–23. [Google Scholar]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Qian, Z.; Travanty, E.A.; Oko, L.; Edeen, K.; Berglund, A.; Wang, J.; Ito, Y.; Holmes, K.V.; Mason, R.J. Innate immune response of human alveolar Type II cells infected with severe acute respiratory syndrome–coronavirus. Am. J. Respir. Cell Mol. Boil. 2013, 48, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Lu, H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci. Trends 2020, 14, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, B.; Michaelis, M.; Baer, P.; Doerr, H.W.; Cinatl, J., Jr. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005, 326, 905–908. [Google Scholar] [CrossRef]
- Booth, C.; Matukas, L.M.; Tomlinson, G.; Rachlis, A.R.; Rose, D.B.; Dwosh, H.A.; Walmsley, S.; Mazzulli, T.; Avendano, M.; Derkach, P.; et al. Clinical features and short-term outcomes of 144 patients with SARS in the Greater Toronto Area. JAMA 2003, 289, 2801. [Google Scholar] [CrossRef]
- Tan, E.L.; Ooi, E.E.; Lin, C.-Y.; Tan, H.C.; Ling, A.E.; Lim, B.; Stanton, L.W. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg. Infect. Dis. 2004, 10, 581–586. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C.; et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Martinez, M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob. Agents Chemother. 2020. [Google Scholar] [CrossRef]
- Richardson, P.; Griffin, I.; Tucker, C.; Smith, D.; Oechsle, O.; Phelan, A.; Stebbing, J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020, 395, e30–e31. [Google Scholar] [CrossRef]
- Lim, J.; Jeon, S.; Shin, H.-Y.; Kim, M.J.; Seong, Y.M.; Lee, W.J.; Choe, K.-W.; Kang, Y.M.; Lee, B.; Park, S.-J. Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. J. Korean Med Sci. 2020, 35, e79. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Hu, S.; Gao, J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov. Ther. 2020, 14, 58–60. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Krüger, N.; Müller, M.; Drosten, C.; Pöhlmann, S.; A Muller, M. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y.; et al. Temporary Removal: Experimental Treatment with Favipiravir for COVID-19: An open-label control study. Eng. 2020. [Google Scholar] [CrossRef]
- Fan, H.-H.; Wang, L.-Q.; Liu, W.-L.; An, X.-P.; Liu, Z.-D.; He, X.-Q.; Song, L.-H.; Tong, Y.-G. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus (2019-nCoV) related coronavirus model. Chin. Med. J. 2020. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 104787. [Google Scholar] [CrossRef]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef]
- Colson, P.; Rolain, J.-M.; Lagier, J.-C.; Brouqui, P.; Raoult, D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105932. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.-M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int. J. Antimicrob. Agents 2020, 105938. [Google Scholar] [CrossRef]
- Schrezenmeier, E.; Dorner, T. Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 2020, 16, 155–166. [Google Scholar] [CrossRef]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Gotte, M. Mechanism of inhibition of Ebola Virus RNA-Dependent RNA polymerase by remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Sparks, M.A.; South, A.; Welling, P.; Luther, J.M.; Cohen, J.; Byrd, J.B.; Burrell, L.M.; Batlle, D.; Tomlinson, L.; Bhalla, V.; et al. Sound science before quick judgement regarding RAS Blockade in COVID-19. Clin. J. Am. Soc. Nephrol. 2020. [Google Scholar] [CrossRef]
- Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Development and Approval Process (Drugs). Available online: www.fda.gov/drugs/development-approval-process-drugs (accessed on 1 March 2020).
- Sham, H.L.; Kempf, D.J.; Molla, A.; Marsh, K.C.; Kumar, G.N.; Chen, C.-M.; Kati, W.; Stewart, K.; Lal, R.; Hsu, A.; et al. ABT-378, a Highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 1998, 42, 3218–3224. [Google Scholar] [CrossRef]
- Leonis, G.; Czyżnikowska, Ż.; Megariotis, G.; Reis, H.; Papadopoulos, M.G. Computational studies of darunavir into HIV-1 protease and DMPC bilayer: Necessary conditions for effective binding and the role of the flaps. J. Chem. Inf. Model. 2012, 52, 1542–1558. [Google Scholar] [CrossRef]
- Kawase, M.; Shirato, K.; Van Der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef]
- Rajam, G.; Sampson, J.; Carlone, G.M.; Ades, E.W. an augmented passive immune therapy to treat fulminant bacterial infections. In Recent Patents Anti-Infective Drug Discovery; Bentham Science Publishers: Sharjah, UAE, 2010; pp. 157–167. [Google Scholar] [CrossRef]
- Marano, G.; Vaglio, S.; Pupella, S.; Facco, G.; Catalano, L.; Liumbruno, G.M.; Grazzini, G. Convalescent plasma: New evidence for an old therapeutic tool? Blood Transfus. 2015, 14, 152–157. [Google Scholar]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.-C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, eabb7269. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Daszak, P.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Sun, J.; Yan, Y. The functional aspects of bacterial CRISPR-cas systems and interactions between phages and its bacterial hosts–a review. Acta Microbiol. Sin. 2015, 55, 251–257. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).