Abstract
Ewingella americana is a cosmopolitan bacterial pathogen that has been isolated from many hosts. Here, we sequenced a high-quality genome of E. americana B6-1 isolated from Flammulina filiformis, an important cultivated mushroom, performed a comparative genomic analysis with four other E. americana strains from various origins, and tested the susceptibility of B6-1 to antibiotics. The genome size, predicted genes, and GC (guanine-cytosine) content of B6-1 was 4.67 Mb, 4301, and 53.80%, respectively. The origin of the strains did not significantly affect the phylogeny, but mobile genetic elements shaped the evolution of the genus Ewingella. The strains encoded a set of common genes for type secretion, virulence effectors, CAZymes, and toxins required for pathogenicity in all hosts. They also had antibiotic resistance, pigments to suppress or evade host defense responses, as well as genes for adaptation to different environmental conditions, including temperature, oxidation, and nutrients. These findings provide a better understanding of the virulence, antibiotic resistance, and host adaptation strategies of Ewingella, and they also contribute to the development of effective control strategies.
1. Introduction
Ewingella americana is a Gram-negative, oxidase-negative, catalase-positive, lactose-fermenting, non-fluorescent, rod-shaped, motile, and facultatively anaerobic bacterium [1]. The bacterium was first isolated from clinical sources and described by Grimont et al. in 1983 [2] as a new genus and species in the family Enterobacteriaceae. The genus Ewingella is currently transferred to the class Gammaproteobacteria, order Enterobacterales, and family Yersiniaceae [1]. E. americana is the only known species in the genus. The bacterium has a wide range of hosts, including human [3], mollusks [4], plants [5,6], vacuum-packed meat [7], nutria carcasses [8], and mushrooms [9,10,11]. In mushrooms, E. americana is known to cause internal stipe necrosis in Agaricus bisporus [9,12] and brown rot on cultivated Flammulina filiformis [10].
E. americana has emerged as a global problem with reports from many regions of the world, including the United States [13], Spain [14], Kingdom of Saudi Arabia [15], Egypt [16], and China [6,10]. It is generally resistant to many antibiotics. To date, six Ewingella species’ genome sequences have become publicly available at the National Centre for Biotechnology Information (NCBI) genome database. However, few studies have been done on the comprehensive analysis of E. americana genomes. Consequently, the pathogenicity, functional roles, metabolic capabilities, and the genetic adaptation to its hosts remain unknown.
Next-generation sequencing technologies have expedited sequencing and increased the number of bacterial genomes [17]. The advent of bioinformatics tools in recent years has aided comparative analysis within different taxon levels of microorganisms [18]. Genomic comparisons can address issues in taxonomy, phylogenetics, virulence, and genotype resistance profiles from different hosts [19,20,21]. It can also facilitate a better understanding of the specific mechanisms deployed by the bacteria to adapt to different hosts and environmental conditions.
In this study, we used Single-Molecule-Real-Time (SMRT) technology [22] to generate the whole genome sequence of E. americana B6-1 isolated from cultivated F. filiformis. The genome sequence was annotated and compared with the representative genomes of other E. americana strains isolated from different hosts/habitat to assess their phylogeny, pan- and core-genome, virulence, antibiotic resistance genes, mobile genetic elements, and defense system. The comparison of the five E. americana genomes will enhance our knowledge of pathogenicity and adaptability to different hosts.
2. Results
2.1. Genomic Features of E. americana B6-1
Strain B6-1 was confirmed as E. americana from the phenotypic, biochemical characters (Supplementary Table S1), and the 16S rRNA gene phylogenetic analysis from previous work [10]. The whole-genome sequence of E. americana B6-1 was assembled into one circular chromosome (4.67 Mbp in size with GC content of 53.80%) and two plasmids (330 Kbp and 104 Kbp in size with GC content of 53.26% and 50.98%, respectively) (Figure 1). The N50 and L50 were 18,509 bp and 13,069 bp, respectively. Total 4301 protein-coding sequences (CDS) and 99 RNA genes (including 77 tRNA, eight 5S rRNA, seven 16S rRNA, and seven 23S rRNA genes) were detected from the assembled genome. The protein-coding genes were assigned to 25 clusters of orthologous groups (COG) and functional categories.
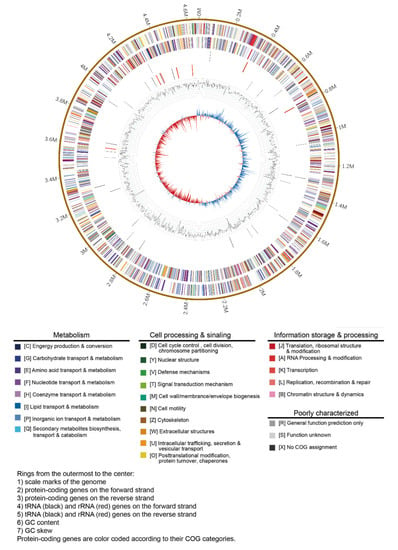
Figure 1.
Circos plot of E. americana strain B6-1 genome showing the densities of GC (guanine-cytosine) content, tRNA, rRNA, and protein-coding genes.
2.2. Average Nucleotide Identity Calculations and Phylogenetic Analyses
The 16S rRNA-based phylogeny and whole genome-based phylogeny were produced to determine the phylogenetic relationship among the E. americana strains. The strains had similar 16S rRNA gene sequences with 98.97 to 100% similarity. The maximum-likelihood (ML) phylogenetic tree using 16S rRNA gene sequences placed all the five strains in the genus Ewingella (Figure 2A).
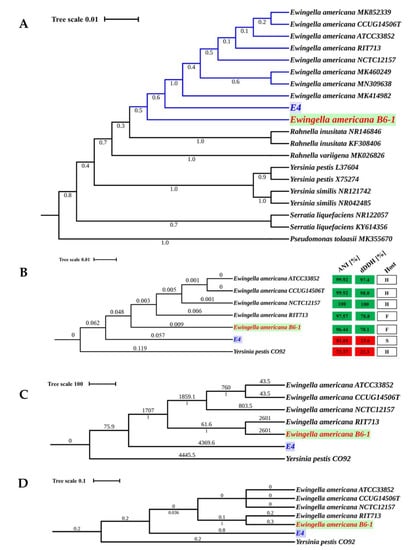
Figure 2.
Phylogenetic analyses of the five strains of E. americana and other related species. (A). Maximum-likelihood phylogenetic tree computed using 16S rRNA gene sequences of the five E. americana strains studied. (B). Maximum-likelihood phylogenetic tree using 2,095 core- proteins for the five strains of E. americana and other related species generated with 1000 bootstrap replications. (C). Whole-genome MLST (wgMLST) using 10,373 alleles for the five strains of E. americana and other related species (D). Phylogenetic tree based on the SNPs of the five E. americana strains and other related species. Yersinia pestis CO92 was used as an outgroup and root for trees (B–D). Pseudomonas tolaasii was used as an outgroup and root for trees (A).
The whole genome-based phylogeny consisted of a core set of 2095 single-copy orthologues proteins for the genus (Figure 2B), whole-genome multi-locus sequence typing (wgMLST) (Figure 2C), single-nucleotide polymorphism (SNP) (Figure 2D), and family trees (Supplementary Figure S3), respectively. The genus core-proteins, wgMLST, and SNP trees showed that the E. americana strains (ATCC, CCUG, and NCTC) isolated from humans are more closely related than the other two strains. The wgMLST and SNP trees clustered the strains according to their host, but the genus core-protein tree clustered the strains according to their evolution. All the whole-genome based trees showed that strain E4 is the ancestor of E. americana strains.
Moreover, the genetic relatedness among the five E. americana strains in the phylogenetic analyses was confirmed by results from the average nucleotide identity (ANI), and digital DNA-DNA hybridization (dDDH) values based on the genomic sequences. The pairwise ANI and dDDH values ranged from 81.01% to 100%, and 23.6 to 100%, respectively. The detailed results of ANI and dDDH values are presented in Supplementary Table S4. The ANI and dDDH values of strain E4 (81.01% and 23.6%) were well below the defined thresholds for species delineation, 95–96% for ANI, and 70% for GGDC. The Tetra Correlation Search (TCS) calculations for strain E4 was related to Serratia sp. Leaf51 (Supplementary Table S5). However, the 16S rRNA sequence showed higher similarity to E. americana compared to Serratia sp. Leaf51. Besides, there was slightly more variability in the ANI and dDDH values between E4 and the E. americana (81.01% and 23.6%) compared to Serratia sp. Leaf51 (78.92% and 22.2%). Since strain E4 is probably a new species, it was not used in the subsequent analyses.
2.3. Orthology and Pan-Genome Analysis
The orthologous gene clusters shared among the five strains of E. americana were identified. All five strains of E. americana formed a total of 4519 clusters and shared 3735 orthologous clusters (Figure 3). The singletons ranged from 5 to 790 gene clusters. The unique orthologue gene clusters within the species were 13, 3, and 0 for RIT713, B6-1, and other three strains, respectively. Strain B6-1 shared a higher number of orthologous clusters with RIT713 (41) compared to the other strains
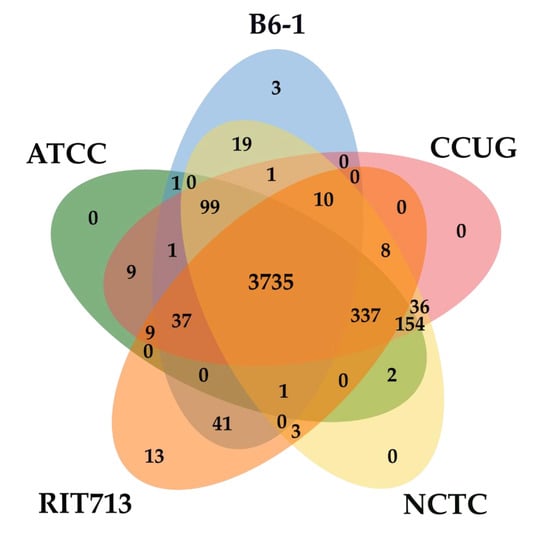
Figure 3.
Venn diagram of the unique and shared number of orthologous gene clusters among the genomes of the five E. americana strains.
Functional analysis of the gene clusters showed that biological processes were the most assigned Gene Ontology (GO) terms. A total of 10,359 shared orthologous gene clusters were assigned to biological processes GO terms within the five E. americana strains. Some of the GO annotations among the core shared orthologous proteins include glycogen catabolic process (GO:0005980), anaerobic respiration (GO:0009061), pathogenesis (GO:0009405), rhamnose catabolic, and metabolic processes (GO:0019301 and GO:0019299) and terpenoid biosynthetic process (GO:0016114). Oxidoreductase activity (GO:0016491; p-value = 9.47 × 10−5) was the only enriched GO term for the shared orthologous gene clusters among the five strains.
The GO enrichment analysis between B6-1 and RIT713 revealed enriched GO terms for the lipopolysaccharide core region biosynthetic process (GO:0009244; p-value = 4.48 × 10−5) and cellular response to DNA damage stimulus (GO:0006974; p-value = 4.48 × 10−5). Protein secretion by the type II secretion system (p-value = 1.85 × 10−12) was the enriched GO term found in the unique gene cluster of RIT713. The annotation of unique orthologous clusters in B6-1 showed classification in biological processes as the main category, with genes related to the biological process, metabolic process, toxin metabolic process, secondary metabolic process, cellular metabolic process, and heterocycle metabolic process as a subcategory. The toxin metabolic process contains two protein-coding genes involved in the aflatoxin biosynthetic process (GO:0045122).
The E. americana pan-genome (Supplementary Table S6) for the five strains contained 5103 gene families, and 43 to 421 new genes were found in four genomes (B6-1, CCUG, NCTC, and RIT713). The genes of the pan-genome increased from 4275 to 5104, and core genes decreased from 4275 to 3677 with the addition of a new genome sequence. The pan-genome curve (Supplementary Figure S2) did not reach the plateau by the addition of new genes with each additional genome. The expansion parameter ’b’ was 0.102 (>0).
2.4. Resistome and Antimicrobial Susceptibility Profile of B6-1
A total of 27 unique antibiotic resistance genes were identified in all the genomes of the E. americana strains, with four different mechanisms of resistance from the comprehensive antibiotic resistance (CARD) database. The multiple antibiotic resistance genes (ranging from 20 to 33) in the strains (Supplementary Table S7) were associated with an aminoglycoside, aminocoumarin, carbapenem, cephalosporin, diaminopyrimidine, fluoroquinolone, fosfomycin, macrolide, nitroimidazole, a peptide antibiotic, phenicol, rifamycin, and tetracycline antibiotic. Fosfomycin resistant gene (fosA) was found in all the strains. The most abundant antimicrobial resistance gene families were encoding multi-efflux pump (23 genes).
Strain B6-1 was tested for antibiotic susceptibility (Table 1) by the Kirby–Bauer test. It showed resistance to ampicillin, cefazolin, clindamycin, novobiocin, rifampicin, tetracycline, and vancomycin. However, B6-1 showed intermediate resistance to cefixime and erythromycin, but it was susceptible to aztreonam, ceftriaxone, ciprofloxacin, gentamicin, kanamycin, ofloxacin, and streptomycin.

Table 1.
Antimicrobial susceptibility profiles of strain B6-1.
2.5. Mobile Genetic Elements (MGE)
From the INTEGRALL database search, four and five putative class 1 integron (In1) genes with a variety of cassette arrays (Table 2 and Supplementary Table S9) were found in the genomes of (B6-1, ATCC, CCUG, and NCTC) and RIT713, respectively. All the five E. americana strains contained one class 1 Integron (In1) with catB8j-aacA4-aadA5 cassette arrays. Class 1 integron (In1) with cassette arrays dfrA12-gcuF-aadA2 and dfrA14b-arr-2-cmlA5-blaOXA-10-aadA1-qacED1-sul1 were only found in B6-1 and RIT713.

Table 2.
Total number of Integron, plasmids, genome islands CRISPR, R-M system and TA system in the E. americana strains.
A total of four different insertion sequence (IS) families were detected in all the five genomes of E. americana strains (Supplementary Table S10). IS3 and Tn3 were the dominant IS families. All the E. americana strains (ATCC, CCUG, and NCTC) isolated from humans contained four IS gene families, while those isolated from mushrooms (B6-1 and RIT713) had the least number of IS families (n = 2). Among the E. americana strains, only B6-1 possessed plasmids (n = 2). The plasmids pB61a and pB61b were 330 kb and 104 kb in size, respectively. Both plasmids contained genes coding for antibiotic resistance, insertion sequences, and toxin-antitoxin system (Table 2 and Supplementary Table S11).
Intact phages were found in all the strains (Table 3). All the phages were circular excerpt ATCC33852, which had linear phage. Strain B6-1 contained two sequences with two phage regions. From the IslandViewer4, the five genomes of E. americana strains contained 19 to 24 genome islands (GIs) (Table 2) of total length ranging from 8 to 10 kb. The gene annotation showed that most of the genes were hypothetical proteins with unknown function, while other genes were associated with replication, recombination, repair, integrases, transposases, and other genome mobility-related genes.

Table 3.
The distribution of phages among the E. americana strains.
2.6. CRISPR-CAS System, Restriction Modification System, and Toxin-Antitoxin System
A total of five (ranging from 2 to 5 spacers) clustered regularly interspaced short palindromic repeats (CRISPR) encoding type I CRISPR-Cas systems were identified in the genomes of the E. americana strains through CRISPRCasFinder (Table 2). ATCC33852 had the highest number of CRISPR spacers (n = 5). Strains B6-1, CCUG, and NCTC had four CRISPR spacers. None of the strains contained the Cas element.
All the strains contained putative genes for the type II restriction-modification system (R-M system). Type I R-M system was found in four strains (ATCC, B6-1, CCUG, and NCTC), and the type IV R-M system was found in only two strains (B6-1 and RIT713). Putative genes for the toxin-antitoxin system (TA) were found in all the E. americana strains (Supplementary Table S12). Complete type II and type IV TA gene modules were found in all the genomes of the five E. americana strains. Type I TA gene modules were present in only strains B6-1 and RIT713.
2.7. Pathogenicity and Virulence Factors
The genomes of the five E. americana strains were surveyed to investigate pathogenicity and virulence-associated genes. Strain B6-1 had a predicted probability score (P score) of 46.24 and the probability of being a human pathogen of 0.60. It was matched to 24 pathogenic families. The predicted probability score (P score), probability of being a human pathogen, and pathogenic families matched to all the E. americana strains ranged from 46.21–53.60, 0.60–0.63, and 24–25, respectively (Table 4). The five E. americana strains (B6-1, RIT713, ATCC, CCUG, and NCTC) were all predicted as a human pathogen.

Table 4.
The predicted pathogenicity score for the Ewingella americana strains from the PathogenFinder.
From the virulence factor database search, a total of 82 putative genes virulence-associated were found in genomes of the five E. americana strains (Supplementary Table S13). Out of the 82 genes, those found in each genome ranged from 66 to 70 genes. B6-1 contained 67 putative virulence genes. The most abundant virulence features in the genomes of all the five E. americana strains were secretion system, adherence, invasion, chemotaxis and motility, and immune evasion. Pore-forming toxins were found in all the five genomes of the E. americana strains.
Further examination of the macromolecular secretion system revealed the E. americana strains encode putative genes related to flagella, Type I, Type III, Type IV, Type V and Type VI secretion systems (Supplementary Table S14). The total number of putative genes for the macromolecular secretion system among the strains ranged from 136 to 199. The least and most abundant macromolecular secretion putative genes were found in B6-1 (136 genes) and RIT713 (199 genes), respectively. The most abundant macromolecular secretion system was type VI, followed by Type I, Type II, and Type III secretion systems. Type II secretion genes (gspD, gspE, gspF, gspG, gspM, gspH, gspI, gspJ, gspK, and gspC) were found in RIT713.
2.8. Stress Response
Genes associated with stress response were identified in all the five strains of E. americana. Strain B6-1 had the highest number of putative genes (90) for stress response compared to the other strains (86–87 genes). Genes coding for oxidative stress (35.40%) followed by stress response (21.60%) and osmotic stress (17.06%) were the most abundant in the genomes of the five strains of E. americana. A persister cell-related gene (cell division inhibitor SulA) and sporulation associated gene (peptidyl-tRNA hydrolase (EC 3.1.1.29)) were found in all five strains (Supplementary Table S15). In addition, five genes coding for sulfate and thiosulfate import ATP-binding protein CysA (EC 3.6.3.25), DedA protein, S-formylglutathione hydrolase (EC 3.1.2.12), TsgA protein and S-(hydroxymethyl) glutathione dehydrogenase (EC 1.1.1.284) for detoxification, and a cold shock protein CspA for cold stress tolerance were also found in all the genomes of the five E. americana strains.
2.9. Annotation of Carbohydrate-Active Enzymes (CAZymes)
The presence of carbohydrate-active enzymes (CAZymes), including the variety of families of glycoside hydrolases (GHs), glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate-binding molecule (CBM), carbohydrate Esterase (CE), and auxiliary activities (AA), that synthesis, metabolize, and transport carbohydrates were identified using the dbCAN2 meta server. Table 5 shows the total number of predicted CAZymes genes that were found in the genomes of the five E. americana strains. The total number of CAZymes ranged from 151 to 168. Strain B6-1 contained 160 CAZymes, while the most abundant and least CAZymes were found in NCTC and RIT713, respectively. The large average sets of genes of CAZyme families among the pathogens were GH 43.43% (68.8 genes) and GT 33.46% (53 genes). GT2 (chitin synthase (EC 2.4.1.16)) was the most abundant CAZyme module (Figure 4). Other abundant GT modules are GT4 (sucrose synthase (EC 2.4.1.13)), GT9 (lipopolysaccharide N-acetylglucosaminyltransferase (EC 2.4.1.56)) and GT51 (murein polymerase (EC 2.4.1.129)). The abundant GH modules among the genomes of the five E. americana strains were GH23, GH13, and GH1. Two to three genes encoding GH18 (chitinase (EC 3.2.1.14)) were identified in the genomes of E. americana strains. GH75 (chitosanases) were absent in all the genomes of E. americana. The genomes of E. americana strains contained a large number of CBM 50 (8-10 genes), which are found to be attached to various enzymes from families GH18, GH19, GH23, GH24, GH25, and GH73, to degrade the chitinous fungal or peptidoglycan bacterial cell walls. All the genomes encode PL17, a putative alginate lyase (EC 4.2.2.3), or oligoalginate lyase (EC 4.2.2.26) and PL7 (poly(β-mannuronate) lyase/M-specific alginate lyase (EC 4.2.2.3)). The E. americana genomes encoded putative genes for CE1, CE4, CE8, CE9, CE11, and CE12, which degraded xylans, chitin, peptidoglycan, pectin, N-acetylglucosamine (GlcNAc), and lipids.

Table 5.
Total CAZymes found in the E. americana genomes.
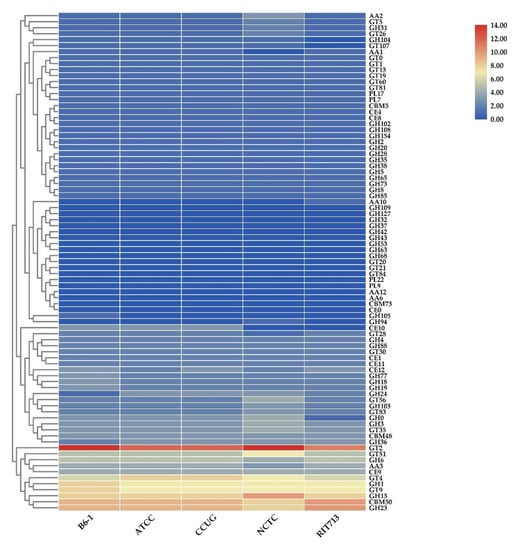
Figure 4.
Heatmap showing the abundance of each CAZyme family in each genome of the E. americana strains.
2.10. Secondary Metabolites and Bacteriocins
The genomes of the five E. americana strains were searched against the antibiotics and secondary metabolite analysis shell (antiSMASH) 5.0 (Supplementary Table S16) and BAGEL 4 web server. Five and seven putative biosynthetic gene clusters were found in four (ATCC, CCUG, NCTC, and RIT713) and B6-1, respectively. Three known secondary metabolites, aryl polyenes, O-antigen, and desferrioxamine E, which are arylpolyene, thiopeptide, and siderophore, respectively, were commonly found among all the five E. americana strains. The other two to four gene clusters have no known annotation. The E. americana genomes contained multiple genes related to bacteriocin production (bottromycin, colicin-M, and microcin). Bottromycin was found in the five genomes. Microcin was found in four genomes (ATCC, CCUG, NCTC, and RIT713) and plasmid pB61a of strain B6-1. Colicin-M was found in only two genomes (B6-1 and RIT713).
3. Discussion
Ewingella americana is known to cause disease in hosts from different kingdoms, and there is an emergence of its multi-drug resistance worldwide [13,15,23]. The study aimed to examine the phylogeny, resistome, mobilome, virulome, and defense systems of five strains of E. americana. The genome size and the number of genes of strain B6-1 were slightly small but comparable to those in the genus. The differences in the genome sizes and the number of genes may be due to the influence of mobile genetic elements [24].
The 16S rRNA gene-based phylogenetic tree showed that all the strains were related to E. americana. However, E4 showed ANI and dDDH values were below the acceptable threshold used to differentiate the closely related species [25]. The genome-based phylogenetic trees for the E. americana corroborates the ANI and dDDH values. Therefore, strain E4 should be reclassified as a different species based on morphological and physiological characteristics. The results confirm that 16S rRNA gene sequences alone cannot resolve the phylogeny of bacteria at the species or genera level [26]. The results suggest that morphological, biochemical, 16S rRNA gene sequence, as well as genome-based comparative, remain essential in delineating bacterial taxa [27].
The highly conserved orthologous genes mean all the strains evolved through speciation events from the last common ancestor [28]. Annotation of the conserved orthologous genes revealed they play an active role in essential biological processes, metabolic functions, and cellular processes. The differences in accessory and unique genes among the E. americana strains confirms the previous report that strains within a bacterial species usually have a set of conserved core genes and a variable set of accessory genes [29].
Interestingly, annotation of the unique clusters in B6-1 revealed putative genes related to the biosynthesis of toxins, namely aflatoxin. Aflatoxins are carcinogenic and mutagenic secondary metabolites produced by members of Aspergillus sp. (commonly by Aspergillus flavus and A. parasiticus) that contaminate many food crops [30]. Besides fungi, bacteria residing within the fungal cytosol could produce mycotoxins, such as rhizoxin and rhizonin [31]. We speculate that B6-1 could have acquired the putative aflatoxin genes from mushroom substrate contaminated with A. flavus and A. parasiticus [32] for self-defense against other predatory fungi. Therefore, strain B6-1 has the potential to contaminate mushrooms by producing aflatoxins. This result suggests the need for effective control of E. americana and other pathogenic fungi in mushroom production and food processing to prevent the adverse effect on human health.
The E. americana strains showed an open but soon to be closed pan-genome. The results indicate that every newly sequenced genome contributed new genes to the species, but the availability of a large number of complete genomes for this species will fail to add new genes to the E. americana pan-genome. Genome size, protein-coding genes, lifestyle, isolated niche, and natural environment of bacteria may influence the size of the pan-genome [33]. The E. americana strains from this study were isolated from different hosts, habitat, and environment; hence, they might have influenced the pan-genome. The pan-genome analysis suggests that additional high-quality reference genomes representing different eco-species may provide a better understanding of the biology of E. americana.
The putative antibiotic resistance genes found among all the strains confirm that E. americana were frequently resistant to several classes of antimicrobial agents [3,15,23]. The predominance of antibiotic resistance genes in the core and accessory genomes of the strains in this study may be associated with the evolution of multidrug resistance in the taxa [34,35,36] and the ability to successfully acquire antibiotic resistance genes encoded by mobile genetic elements such as insertion sequences, transposons, integrons, and plasmids from their host or different bacterial cells [37]. The E. americana strains may be out-competing strains with lower resistance multiplicity in their habitat [34].
In addition, increased use of antibiotics in healthcare and animal farms, as well as in the mushroom industry, could have led to the emergence, spread, and persistence of multidrug-resistant E. americana strains [36]. From the orthology analysis, rhamnose metabolism was found in all the E. americana. Therefore, new therapeutic compounds targeting rhamnose biosynthesis can be used to control the pathogen [38].
Virulence genes play an important role in pathogenicity, and several of such genes were found in the genomes of all the E. americana strains in this study. In silico prediction of the pathogenic potential revealed that about 60% of the pathogenic families were linked to the family Yersiniaceae. The pathogenicity of the E. americana strains (B6-1, ATCC, CCUG, NCTC, and RIT713) are weak pathogens of humans compared to the type species in the family Yersiniaceae, Yersinia pestis CO92 (1569 matched pathogenic families). This result confirms which E. americana is an opportunistic pathogen that infects immunosuppressed patients due to other illnesses [13,39]. However, the predictive method is not sufficient to arrive at a conclusion about the pathogenesis of the microorganism [40]. Therefore, there is a need to conduct further pathogenicity testing to confirm the pathogenesis of all the other four E. americana strains. The virulence factors found in the core genome, such as flagella, pili, and type secretions, were mostly conserved across the strains and were involved in adherence and immune system evasion. The virulence genes are ubiquitous as they likely play a role in the fitness of E. americana in different environments, whereas accessory virulence factors offer additional functions for improved environmental fitness. The acquisition of plasmids by B6-1 could increase its virulence. The E. americana strains’ putative toxins may play a significant role in its pathogenesis and survival.
The identification of only class 1 Integron and the abundance of IS3 and Tn3 family elements in E. americana strains confirms the report that they are the most widely distributed mobile DNA elements in bacteria and contribute to the dissemination of antibiotic resistance and the emergence of multi-resistant pathogens worldwide [41,42,43]. In addition, the acquisition of two plasmids by strain B6-1, absent in the other strains, may play an essential role in virulence, antibiotic resistance, detoxification, and ecological interaction [44]. The results suggest that the various MGEs influenced the size of the genome islands among the strains, hence the varied number of virulence or resistance genes between the strains.
Bacteria have developed multiple systems, including CRISPR–Cas systems and restriction-modification (R-M) systems, to defend themselves against invaders such as plasmids or phages [45]. This result corroborates that multiple CRISPR elements can often be detected in bacterial genomes, but not all elements are accompanied by Cas genes [46]. However, the intact phages in the genomes of the other five E. americana strains could be due to the development of anti-CRISPR systems by phages to avoid CRISPR regulation to enable integration into the genome [47]. It is not surprising to find genes related to R-M systems (type I, II, and IV) among the E. americana strains because R-M systems are widespread and considered as an effective immune system in bacteria and archaea [48,49]. The two to three R-M systems among the strains are consistent with other bacteria. The numerous and diverse R-M systems in the E. americana strains isolated from mushrooms (B6-1 and RIT713) may provide a selective advantage by rapid genetic adaptation to its natural environment [50].
Further, the stress related genes found in the genomes of E. americana were targeted for a particular kind of stress. The results suggest all the E. americana strains can adapt to any stress, including tolerance for oxidative, osmotic, carbon starvation, nutrients, cold, and heat stress. Also, they possessed toxin-antitoxin modules, which play significant roles in persister formation when exposed to environmental stimuli [51]. TA systems are gene modules that encode a protein toxin and an antitoxin that neutralizes either the toxin’s action or its expression [52]. The abundant and complete type II TA system found among the strains confirms the reports of its wide distribution and diversity [53]. TA system is primarily involved in biological processes such as DNA replication, mRNA synthesis, cell wall synthesis, and programmed cell death [54]. Therefore, it could serve as a potential target for novel antimicrobial agents [55] to control multi-drug resistant bacteria like E. americana.
The diverse repertoire of putative CAZyme genes found in all the E. americana strains indicates that complex enzymes are produced to digest the components of their host’s cell wall, including complex polysaccharides (cellulose, hemicellulose, and pectin) in plants [56] and chitin in fungi, insects, and mollusks. The difference in CAZyme numbers and the presence or absence of some CAZyme families may indicate different substrate utilization capabilities. There were chitinase GH18 in all the strains, indicating that they share similar strategies for degrading fungal cell walls. However, strain B6-1 had more of GH18 and GH19. This suggests that it may require more chitinase to invade its fungal host. All the E. americana strains produced abundant GT2 and CMB50, which are important for evading animal/plant and fungi/plant cells, respectively [57,58]. CMB50 acts as a chitin surface binder for the plant or fungal hosts’ invasion and colonization [59]. The secreted CAZymes in the genomes of E. americana strains are potential virulence factors, particularly for the host fungi and plants. Actual experiments are required to validate and confirm the designation [59].
All the E. americana strains possessed genes for biosynthesis of secondary metabolites, which are not essential for growth, development, or reproduction of the organism but have an important ecological function [60]. The large number of secondary metabolite gene clusters in strain B6-1 may be required for its survival in the mushroom environment. The O-antigen cluster found in the strains may be required for virulence [61] and resistance to complement-mediated killing and phagocytosis [62]. The siderophores, desferrioxamine E (found in all the strains) may play an essential role in bacterial pathogenesis by scavenging iron from the host or their surrounding environment [63,64]. The siderophores may also be involved in oxidative stress tolerance and have applications in medicine and agriculture [63]. All the E. americana strains produced aryl polyene pigments, and they were similar to carotenoids [65,66]. Some bacterial species, such as Enterococcus mundtii, are known to produce carotenoid-like pigments [40]. Aryl polyene pigments play a role in protecting the bacteria from oxidative stress when exposed to the environment [66,67]. All the strains also produced bacteriocin. The production of colicin M toxin may be relevant and unique characteristics for strains B6-1 and RIT713, which were isolated from fungi. The identified putative gene-encoded antimicrobial peptides (bottromycin, microcin, and sactipeptide) and colicin M toxin may be responsible for microbial competition [67,68,69]. The genome mining of secondary metabolites from the E. americana suggests the potential to use the bacteriocins in healthcare, animal husbandry, and the food industry as well as agriculture, to replace antibiotics and to treat multi-drug resistant pathogens [70,71]. However, further work needs to be done using other detection techniques like chromatography to ascertain the production of the secondary metabolites, bacteriocins, and assessment of their efficacy for controlling multidrug-resistant pathogens.
4. Materials and Methods
4.1. Bacterial Strains and Characterization
Ewingella americana strain B6-1 was isolated from a symptomatic mushroom, Flammulina filiformis, collected from the Gaorong Biotech Company at Changchun, Jilin, China, in 2016. The other four E. americana strains used in this study include three strains isolated from the human throat (E. americana ATCC 33852 [72], E. americana CCUG 14506T [73], and E. americana NCTC12157 [74]), and one from mushroom (Craterellus sp.) E. americana RIT713 [75]. In addition, strain E4 [76], isolated from permafrost soil, was included to ascertain its name and taxonomic position. There is no report of the pathogenicity test for the five other strains.
The isolation and identification of E. americana B6-1 were described in the previous report [10]. Molecular characterization was done by amplifying 16S rRNA and gyrB genes with the primers 27F/1492R and gyrB-UP1s/gyrB-UP2sr, respectively, using polymerase chain reaction (PCR). The colony morphology of B6-1 was observed after 72 h growing at 28 °C under the dark on nutrient agar (NA), blood agar, and Kings Medium B (KB), separately. Bacterial cell morphology was examined using a transmission electron microscope (HITACHI H-7650) after 48 h growing on NA media.
For biochemical and physiological tests, API 20E, and API 50CHE kits (BioMérieux, Marcy-l’Etoile, France) were used following the methods described by Mergaert et al. [77]. The bacterium was grown at 30 °C overnight in 5 mL Luria-Bertani (LB) liquid medium. The bacterial cells were centrifuged, collected, and washed twice with sterile distilled water. The API assays were repeated three times. The pellets of bacterial cells were tested by Qingdao Kechuang Quality Testing Co. LTD. (Qingdao, Shandong Province, China) for Fatty acid methyl esters (FAME) analysis with an Agilent Technologies 7890A Gas Chromatograph using methods described by Ivanovic et al. [78].
4.2. DNA Extraction, Genome Sequencing, and Annotation
The genomic DNA was extracted from E. americana B6-1 grown overnight in 20 mL LB broth at 28 °C using BioFlux Biospin bacterial genomic DNA extraction kit (Bioer Technology Co., Ltd., Hangzhou, China). The DNA quality was examined by gel electrophoresis and quantified using the Qubit 2.0 fluorometer (Life Technologies, Darmstadt, Germany). The DNA was used to construct a Single-Molecule Real-Time (SMRT) Bell library with an insert size of 20 kb at Tianjin Biochip Corporation (Tianjin, China). The library was sequenced using the PacBio RSII platform (Pacific Biosciences, Menlo Park, CA, USA). The low-quality reads were filtered out by the SMRT analysis software 2.3.0 [79], and the filtered reads were de novo assembled by a hierarchical genome assembly process (HGAP) [79] with the SMRT portal software.
The genome annotation was done by Prokka, prokaryotic genome annotation software [80]. Functional annotation of the genes was performed by the BLASTP search against the Cluster of Orthologous Groups of proteins database (COG, https://www.ncbi.nlm.nih.gov/COG/) [81], and the Kyoto Encyclopedia of Genes and Genomes database (KEGG, http://www.genome.jp/kegg/) [82]. Gene ontology (GO) was analyzed using InterProScan 5 [83,84]. The genome of E. americana B6-1 was deposited in the GenBank database under the accession CP048243 in the Genome and SAMN13930952 in the BioSample. In addition, four other E. americana genomes (including E. americana ATCC 33852 [72], E. americana CCUG 14506T [73], E. americana NCTC12157 [74], and E. americana RIT713 [75]), and a wrongly named species E4 [76] (Supplementary Table S3) were downloaded from the National Centre for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) genome repository and reannotated following the same pipeline as B6-1. The quality of all the genome assembly was assessed using Quast (v5.0.2) [85].
4.3. Average Nucleotide Identity and Phylogenetic Analyses
The Average Nucleotide Identity (ANI) was calculated using ANI Calculator (https://www.ezbiocloud.net/tools/ani) [86], and the digital DNA-DNA hybridization (dDDH) was calculated using Genome-to-Genome Distance Calculator 2.0 (GGDC) server (http://ggdc.dsmz.de/ggdc.php#) [87]. Additionally, Tetra Correlation Search (TCS) was used to search strain E4 genome against the entire genomes reference database (GenomesDB) of JSpecies Web Server (http://jspecies.ribohost.com/jspeciesws/) [88] to provide insights into the relationships to other organisms.
The phylogenetic relationship among the E. americana strains and its closest neighbors was determined by the whole genome-based and 16S rRNA sequences (Supplementary Table S2). The 16S rRNA sequences of each E. americana strain were obtained from the NCBI GenBank database and aligned using ClustalW application in MEGA X [89]. A phylogenetic tree of the 16S rRNA gene was constructed using the maximum likelihood method based on the JTT matrix-based model [90] with 1000 bootstraps replications in MEGA X [89]. OrthoMCL 2.0.9 [91] was used to cluster the protein sequences of each E. americana strain. Each set of orthologous proteins were individually aligned using MUSCLE [92]. The poorly aligned and divergent positions of protein sequences were trimmed with Gblocks v0.19b [93]. The final conserved blocks were concatenated to create a core-proteome alignment and used for the construction of a maximum likelihood phylogenetic tree by the LG matrix, and the Gamma model of rate heterogeneity [94] with bootstrap supporting of 1000 replicates in RAxML v8.0.29 [95].
The whole-genome multi-locus sequence typing (wgMLST) tree was constructed using the PGAdb-builder web service [96]. The PGAdb profile of five E. americana strains and two other related species genomes was compared by BLASTn, with 90% coverage and 90% identity. The SNP-based phylogenomic tree was constructed using the CSI Phylogeny-1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) web service [97]. Yersinia pestis CO9 genome sequence was used as an outgroup for the wgMLST, SNP, and core-protein genus trees, and Pseudomonas tolaasii was used as outgroup for 16S rRNA and core-protein family tree. A total of 28 genomes (Supplementary Table S3) was used to construct the phylogenetic family tree by the genome BLAST distance phylogeny method (GBDP) in the Type Strain Genome Server (TYGS) platform (https://tygs.dsmz.de/) [98]. All phylogenetic trees were visualized using an interactive tree of life (iTOL) v5 (https://itol.embl.de/) online tool [99].
4.4. Orthology and Pan-Genome Analyses
OrthoVenn2 (https://orthovenn2.bioinfotoolkits.net/home) webserver [100] was used to identify orthologous gene clusters that are unique and shared among the E. americana strains. The analysis was performed with default parameters for the protein-coding genes of the strains. The protein-coding genes were also clustered using USEARCH [101]. The pan-genome (core, accessory, and unique genes) of the E. americana strains were calculated and annotated in the COGs and KEGG databases using the Bacterial Pan Genome Analysis tool (BPGA) pipeline [102].
4.5. Resistome and Antimicrobial Susceptibility Profile of B6-1
The comprehensive antibiotic resistance database (CARD) (https://card.mcmaster.ca/home) [103] was used to detect antibiotic resistance genes. Antibiotic susceptibility test was done by Kirby–Bauer disk diffusion method [104] following the recommendations of the Clinical and Laboratory Standards Institute [105]. E. americana strain B6-1 was cultured on Mueller-Hinton Agar (MHA) (Solarbio Life sciences, Beijing, China). Escherichia coli ATCC 25922 was used as the control for these assays. The antibiotic discs (Oxoid, Wesel, Germany) used included ampicillin (AMP, 10 μg), ciprofloxacin (CIP, 5 μg), cefazolin (30 μg), cefixime (5 μg), erythromycin (E, 15 μg), streptomycin (S, 10 μg), kanamycin (K, 30 μg), gentamicin (10 μg), tetracycline (TET, 30 μg), rifampicin (5 μg), vancomycin (30 μg), ceftriaxone (30 μg), ofloxacin (5 μg), clindamycin (22 μg), aztreonam (30 μg) and novobiocin (5 μg). The diameters of the inhibition zone used for classifying the microorganism’s antibiotic susceptibilities are shown in Table S8. All antibiotic resistance determinations were conducted in triplicates.
4.6. Mobile Genetic Elements (MGE) and Bacterial Defense
Genomic islands were predicted using IslandViewer 4 [106]. Insertion sequences, plasmids, and integrons were predicted using Isfinder [107], PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) [108], and INTEGRALL database (http://integrall.bio.ua.pt/) [109], respectively. PHAge Search Tool—Enhanced Release (PHASTER) web server (www.phaster.ca.) [110] was used to predict bacteriophage sequences. Putative CRISPR loci and Cas clusters were examined using clustered regularly interspaced short palindromic repeats (CRISPR) (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index) [111]. The restriction-modification system and toxin-antitoxin system (TA system) were identified from the KEGG annotation.
4.7. Pathogenicity, and Virulence Factors
BLASTP (E-value cutoff of 1 × 10−5) was used to detect virulence genes detected by searching against the virulence factor database (VFDB; http://www.mgc.ac.cn/VFs/) [112]. Pathogenfinder (https://cge.cbs.dtu.dk/services/PathogenFinder/) was used to predict pathogenicity towards humans [113]. Type secretion systems were detected using MacSyFinder v1.0.2 [114]. Genes related to dormancy, sporulation, and stress response were inferred from the KEGG annotation.
4.8. CAZymes and Secondary Metabolites
Carbohydrate-active enzymes (CAZymes) and secondary metabolites gene clusters were predicted using the dbCAN2 meta server (http://cys.bios.niu.edu/dbCAN2) [115] and antiSMASH 5.0 (https://antismash.secondarymetabolites.org/#!/start) [116], respectively. BAGEL4 (http://bagel4.molgenrug.nl/) web server [117] was used to annotate bacteriocins found in the genomes of the five E. americana strains.
5. Conclusions
In this study, we performed a comparative genomic analysis of five E. americana. There were significant differences in the genome size and number of predicted genes between strains isolated from different host-habitats. Our phylogenetic analysis revealed that the strains formed clusters according to their host and evolution, but the habitat was involved in shaping the genomes of the strains. The pan-genome analysis revealed conserved and variable genes involved in all fundamental life processes, including growth, development, virulence, antibiotic resistance, detoxification, and adaptation to host and environment. Additionally, all the E. americana strains are weak pathogens and/but contain genes from virulence factors, macromolecular secretion system, toxins, antibiotic resistance, CAZymes, secondary metabolites, and stress response that aid the pathogen to colonize hosts across kingdoms and different environments.
Further analysis revealed that the acquisition of mobile genetic elements is a significant source of genome diversity in the genus and possesses highly conserved defense systems made up of CRISPR elements, R-M system, and TA systems. The rhamnose biosynthesis, R-M system, and TA system are potential targets for future new drugs to control the bacterium. Additionally, the E. americana strains have putative genes for the production of bacteriocin and the biodegradation of toxic compounds. This result provides the opportunity for the development and commercialization of useful products such as control of multidrug-resistant bacteria and bioremediation. However, further work is required to ascertain the production of these compounds and testing to validate their efficacy.
The findings suggest that multiple high-quality genome sequences of the pathogen from a different host and geographical location are required to understand the virulence and genetic factors that allow the Ewingella group to be versatile and adapt to a broad niche. This work suggests the optimization of commercial mushroom production processes to minimize the use of antibiotics, including whole-genome sequencing techniques, as routine testing for mushroom quality and safety to minimize the potential risk to human health.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/5/330/s1, Figure S1: The quality of genome from Quast, Figure S2: The pan-genome analysis of five Ewingella americana strains, Figure S3: Phylogenetic analysis of the five strains of Ewingella americana and other closely related neighbors in the family Yer-siniaceae, Table S1: Physiological and biochemical characteristics of Bacterium causing brown rot disease of Flammulina filiformis, Table S2: List of Bacterial species used for 16S rRNA gene phylogenetic tree, Table S3: The List of Bacterial Species Used in Phylogenetic family tree, Table S4: The ANI and dDDH values for the Ewingella americana strains and other closely related species, Table S5: The Tetra Correlation Search (TCS) calculations for E4, Table S6: The number core, accessory, and unique genes in the Ewingella americana pan-genome, Table S7: The number of putative antibiotic resistant genes for the Ewingella americana strains identified from search against the CARD database, Table S8: Evaluation standards for antibiotic sensitivity, Table S9: The number of Class I integron found in the genomes of the Ewingella americana strains, Table S10: The type of insertion sequences in the genomes of the Ewingella americana strains, Table S11: SwissProt annotation of plasmids in Ewingella americana B6-1, Table S12: The number of toxin-antitoxin system-related genes found in the genomes of Ewingella americana strains; Table S13: The number of putative virulence factor genes for the Ewingella americana strains identified from search against the VFDB database, Table S14: The number of macromolecular secretion system genes for the Ewingella americana strains, Table S15: The number of stress response related genes found in the genomes of Ewingella americana strains, Table S16: List of secondary metabolites found in the genomes of Ewingella americana strains using antiSMASH.
Author Contributions
Conceptualization, Y.L., and F.L.S.; Data curation, Z.L., and F.L.S.; Formal analysis, Z.L., and F.L.S.; Funding acquisition, Y.L.; Methodology, Z.L., and F.L.S.; Software, Z.L., F.L.S. and B.A.O.; Supervision, Y.L.; Validation, Z.L., F.L.S., B.A.O., and H.S.; Writing—original draft, Z.L., and F.L.S.; Writing—review & editing, Z.L., F.L.S., B.A.O., and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Changchun Science and Technology Project (No. 15SS11); Special Fund for Agro-scientific Research in the Public Interest (No. 201503137); International Cooperation Research Center of China (No. 2017B01011); the “111” Project (No. D17014) and Creation of Ganoderma Germplasm Resources and Breeding and Development of New Varieties (No. GF20190034).
Acknowledgments
Thanks to Deputy Director Yan Piao of Changchun Gaorong Biotechnology Co., Ltd. for allowing us to collect samples to investigate diseases of needle mushrooms in China. We are also grateful to the Genome Evolution Laboratory, University of Wisconsin, Madison, USA for the Ewingella americana ATCC 33852; the Public Health England, Pacific Biosciences and the Wellcome Sanger Institute through the NCTC 3000 project for the Ewingella americana NCTC12157 genome sequence; the Culture Collection, University of Gothenburg (CCUG) and the Chun Laboratory for the Ewingella americana CCUG 14506T genome sequence; the Hudson Laboratory, Rochester Institute of Technology, New York, USA, for the Ewingella americana RIT713 genome sequence and Lyle Whyte, Ianina Altshuler, and the University of Laval, Canada, for the E4 genome sequence.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| B6-1 | Ewingella americana B6-1 |
| ATCC | Ewingella americana ATCC 33852 |
| CCUG | Ewingella americana CCUG 14506T |
| NCTC | Ewingella americana NCTC12157 |
| RIT713 | Ewingella americana RIT713 |
| E4 | E4 |
| E. americana | Ewingella americana |
| ANI | Average Nucleotide Identity |
| CAZyme | Carbohydrate-active enzymes |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| dDDH | digital DNA-DNA hybridization |
| GC | Guanine-cytosine |
| MGE | Mobile genetic elements |
| NCBI | National Center for Biotechnology Information |
| SMs | Secondary metabolites |
| SNP | Single-nucleotide polymorphism |
| R-M system | Restriction-modification system |
| TA system | Toxin-antitoxin system |
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Grimont, P.A.D.; Farmer, J.J.; Grimont, F.; Asbury, M.A.; Brenner, D.J.; Deval, C. Ewingella americana gen.nov., sp. nov., a new Enterobacteriaceae isolated from clinical specimens. Annales de l’Institut Pasteur Microbiologie 1983, 134, 39–52. [Google Scholar] [CrossRef]
- Esposito, S.; Miconi, F.; Molinari, D.; Savarese, E.; Celi, F.; Marchese, L.; Valloscuro, S.; Miconi, G.; Principi, N. What is the role of Ewingella americana in humans? A case report in a healthy 4-year-old girl. BMC Infect. Dis. 2019, 19, 386. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.E.; Fanning, G.R.; Brenner, D.J. Isolation of Ewingella americana from mollusks. Curr. Microbiol. 1995, 31, 287–290. [Google Scholar] [CrossRef]
- Hamilton-Miller, J.; Shah, S. Identity and antibiotic susceptibility of enterobacterial flora of Lettuce vegetables. Int. J. Antimicrob. Agents 2001, 18, 81–83. [Google Scholar] [CrossRef]
- Wei, M.; Zhao, Z.B.; Liu, H.Y.; Fu, C.H.; Yu, L.J. First Report of Ewingella americana Causing Bacterial Shot Hole on Anoectochilus roxburghii in China. Plant Dis. 2020. [Google Scholar] [CrossRef]
- Helps, C.; Harbour, D.; Corry, J. PCR-based 16S ribosomal DNA detection technique for Clostridium estertheticum causing spoilage in vacuum-packed chill-stored beef. Int. J. Food Microbiol. 1999, 52, 57–65. [Google Scholar] [CrossRef]
- Lyon, W.J.; Milliet, J.B. Microbial Flora Associated with Louisiana Processed Frozen and Fresh Nutria (Myocastor coypus) Carcasses. J. Food Sci. 2000, 65, 1041–1045. [Google Scholar] [CrossRef]
- Inglis, P.W.; Burden, J.L.; Peberdy, J.F. Evidence for the association of the enteric bacterium Ewingella americana with internal stipe necrosis of Agaricus bisporus. Microbiology 1996, 142, 3253–3260. [Google Scholar] [CrossRef][Green Version]
- Liu, Z.H.; Sossah, F.L.; Li, Y.; Fu, Y.P. First Report of Ewingella americana Causing Bacterial Brown Rot Disease on Cultivated Needle Mushroom (Flammulina velutipes) in China. Plant Dis. 2018, 102, 2633. [Google Scholar] [CrossRef]
- Inglis, P.W.; Peberdy, J.F. Isolation of Ewingella americana from the Cultivated Mushroom, Agaricus bisporus. Curr. Microbiol. 1996, 33, 334–337. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.R.; Pay, J.; Braithwaite, M. Isolation, identification and ecology of Ewingella americana (the causal agent of internal stipe necrosis) from cultivated mushrooms in New Zealand. Australas. Plant Pathol. 2007, 36, 424–428. [Google Scholar] [CrossRef]
- Hassan, S.; Amer, S.; Mittal, C.; Sharma, R. Ewingella americana: An emerging true pathogen. Case Rep. Infect. Dis. 2012, 2012, 2. [Google Scholar] [CrossRef]
- Reyes, J.E.; Venturini, M.E.; Oria, R.; Blanco, D. Prevalence of Ewingella americana in retail fresh cultivated mushrooms (Agaricus bisporus, Lentinula edodes and Pleurotus ostreatus) in Zaragoza (Spain). FEMS Microbiol. Ecol. 2004, 47, 291–296. [Google Scholar] [CrossRef]
- Bukhari, S.; Hussain, W.A.; Fatani, M.; Ashshi, A. Multi-drug resistant Ewingella americana. Saudi Med. J. 2008, 29, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Madbouly, A.K.; Elshatoury, E.; Abouzeid, M. Etiology of stipe necrosis of cultivated mushrooms (Agaricus bisporus) in Egypt. Phytopathol. Mediterr. 2014, 53, 124–129. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Yin, Z.; Yuan, C.; Du, Y.; Yang, P.; Qian, C.; Wei, Y.; Zhang, S.; Huang, D.; Liu, B. Comparative genomic analysis of the Hafnia genus reveals an explicit evolutionary relationship between the species alvei and paralvei and provides insights into pathogenicity. BMC Genom. 2019, 20, 768. [Google Scholar] [CrossRef]
- Urbaniak, C.; van Dam, P.; Zaborin, A.; Zaborina, O.; Gilbert, J.A.; Torok, T.; Wang, C.C.C.; Venkateswaran, K. Genomic Characterization and Virulence Potential of Two Fusarium oxysporum Isolates Cultured from the International Space Station. mSystems 2019, 4, e00345-18. [Google Scholar] [CrossRef]
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the Virulence Plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–774. [Google Scholar] [CrossRef]
- Leopold, S.R.; Goering, R.V.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial Whole-Genome Sequencing Revisited: Portable, Scalable, and Standardized Analysis for Typing and Detection of Virulence and Antibiotic Resistance Genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Pound, M.W.; Tart, S.B.; Okoye, O. Multidrug-Resistant Ewingella americana: A Case Report and Review of the Literature. Ann. Pharmacother. 2007, 41, 2066–2070. [Google Scholar] [CrossRef]
- Naito, M.; Pawlowska, T.E. The role of mobile genetic elements in evolutionary longevity of heritable endobacteria. Mob. Genet. Elem. 2016, 6, e1136375. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.; da Costa, M.; Rooney, A.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Venter, S.N.; Palmer, M.; Beukes, C.W.; Chan, W.-Y.; Shin, G.; van Zyl, E.; Seale, T.; Coutinho, T.A.; Steenkamp, E.T. Practically delineating bacterial species with genealogical concordance. Antonie van Leeuwenhoek 2017, 110, 1311–1325. [Google Scholar] [CrossRef]
- Jensen, R.A. Orthologs and paralogs—We need to get it right. Genom. Biol. 2001, 2. [Google Scholar] [CrossRef]
- Segerman, B. The genetic integrity of bacterial species: The core genome and the accessory genome, two different stories. Front. Cell. Infect. Microbiol. 2012, 2. [Google Scholar] [CrossRef]
- Cotty, P.J.; Bayman, P.; Egel, D.S.; Elias, K.S. Agriculture, aflatoxins and Aspergillus. In The Genus Aspergillus; Powell, K.A., Renwick, A., Peberdy, J.F., Eds.; Springer: New York, NY, USA, 1994. [Google Scholar]
- Lackner, G.; Partida-Martinez, L.P.; Hertweck, C. Endofungal bacteria as producers of mycotoxins. Trends Microbiol. 2009, 17, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K. Cultivation of Mushrooms in Plastic Bottles and Small Bags. In Edible and Medicinal Mushrooms; Diego, C.Z., Pardo-Giménez, A., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 309–338. [Google Scholar] [CrossRef]
- Chhotaray, C.; Wang, S.; Tan, Y.; Ali, A.; Shehroz, M.; Fang, C.; Liu, Y.; Lu, Z.; Cai, X.; Hameed, H.M.A.; et al. Comparative Analysis of Whole-Genome and Methylome Profiles of a Smooth and a Rough Mycobacterium abscessus Clinical Strain. G3 (Bethesda) 2020, 10, 13–22. [Google Scholar] [CrossRef]
- Lehtinen, S.; Blanquart, F.; Lipsitch, M.; Fraser, C.; The Maela Pneumococcal Collaboration. On the evolutionary ecology of multidrug resistance in bacteria. PLoS Pathog. 2019, 15, e1007763. [Google Scholar] [CrossRef]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Duarte, I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics 2019, 8, 23. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug Resistance in Bacteria. Ann. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef]
- Aminov, R.I. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011, 2, 158. [Google Scholar] [CrossRef]
- Van der Beek, S.L.; Zorzoli, A.; Çanak, E.; Chapman, R.N.; Lucas, K.; Meyer, B.H.; Evangelopoulos, D.; de Carvalho, L.P.S.; Boons, G.-J.; Dorfmueller, H.C.; et al. Streptococcal dTDP-L-rhamnose biosynthesis enzymes: Functional characterization and lead compound identification. Mol. Microbiol. 2019, 111, 951–964. [Google Scholar] [CrossRef]
- Kati, C.; Bibashi, E.; Kokolina, E.; Sofianou, D. Case of peritonitis caused by Ewingella americana in a patient undergoing continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 1999, 37, 3733–3734. [Google Scholar] [CrossRef]
- Repizo, G.D.; Espariz, M.; Blancato, V.S.; Suárez, C.A.; Esteban, L.; Magni, C. Genomic comparative analysis of the environmental Enterococcus mundtii against enterococcal representative species. BMC Genom. 2014, 15, 489. [Google Scholar] [CrossRef]
- Gomez, A.; Ladire, M.; Marcille, F.; Nardi, M.; Fons, M. Characterization of ISRgn1, a Novel Insertion Sequence of the IS3 Family Isolated from a Bacteriocin-Negative Mutant of Ruminococcus gnavus E1. Appl. Environ. Microbiol. 2002, 68, 4136–4139. [Google Scholar] [CrossRef][Green Version]
- Nicolas, E.; Oger, C.A.; Nguyen, N.; Lambin, M.; Draime, A.; Leterme, S.C.; Chandler, M.; Hallet, B.F.J. Unlocking Tn3-family transposase activity in vitro unveils an asymetric pathway for transposome assembly. Proc. Natl. Acad. Sci. USA 2017, 114, E669–E678. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de la Cruz, F. Mobility of Plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, H.; Ortega-Polo, R.; Zaheer, R.; Goji, N.; Amoako, K.K.; Brown, R.S.; Majury, A.; Liss, S.N.; McAllister, T.A. Comparative genomics of multidrug-resistant Enterococcus spp. isolated from wastewater treatment plants. BMC Microbiol. 2020, 20, 20. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Pawluk, A.; Maxwell, K.L.; Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2013, 493, 429–432. [Google Scholar] [CrossRef]
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef]
- Negri, A.; Jąkalski, M.; Szczuka, A.; Pryszcz, L.P.; Mruk, I. Transcriptome analyses of cells carrying the Type II Csp231I restriction-modification system reveal cross-talk between two unrelated transcription factors: C protein and the Rac prophage repressor. Nucleic Acids Res. 2019, 47, 9542–9556. [Google Scholar] [CrossRef]
- Donahue, J.P.; Peek, R.M., Jr. Restriction and Modification Systems. In Helicobacter Pylori: Physiology and Genetics; Mobley, H.L.T., Mendz, G.L., Hazell, S.L., Eds.; ASM Press: Washington, DC, USA, 2001. [Google Scholar]
- Wang, X.; Wood, T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Park, J.-H.; Inouye, M. Toxin-Antitoxin Systems in Bacteria and Archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.-Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef]
- Wen, Y.; Behiels, E.; Devreese, B. Toxin–Antitoxin systems: Their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 2014, 70, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Broxterman, S.E.; Schols, H.A. Interactions between pectin and cellulose in primary plant cell walls. Carbohydr. Polym. 2018, 192, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, I.R.; Brouillet, S.; Soulié, M.-C.; Gribaldo, S.; Sirven, C.; Charron, N.; Boccara, M.; Choquer, M. Genome-wide analyses of chitin synthases identify horizontal gene transfers towards bacteria and allow a robust and unifying classification into fungi. BMC Evol. Biol. 2016, 16, 252. [Google Scholar] [CrossRef]
- Killiny, N.; Martinez, R.H.; Dumenyo, C.K.; Cooksey, D.A.; Almeida, R.P.P. The Exopolysaccharide of Xylella fastidiosa Is Essential for Biofilm Formation, Plant Virulence, and Vector Transmission. Mol. Plant-Microbe Interact. 2013, 26, 1044–1053. [Google Scholar] [CrossRef][Green Version]
- Looi, H.K.; Toh, Y.F.; Yew, S.M.; Na, S.L.; Tan, Y.-C.; Chong, P.-S.; Khoo, J.-S.; Yee, W.-Y.; Ng, K.P.; Kuan, C.S. Genomic insight into pathogenicity of dematiaceous fungus Corynespora cassiicola. PeerJ 2017, 5, e2841. [Google Scholar] [CrossRef][Green Version]
- Sangwan, N.S.; Jadaun, J.S.; Tripathi, S.; Mishra, B.; Narnoliya, L.K.; Sangwan, R.S. Chapter 9—Plant Metabolic Engineering. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 143–175. [Google Scholar] [CrossRef]
- Shankar-Sinha, S.; Valencia, G.A.; Janes, B.K.; Rosenberg, J.K.; Whitfield, C.; Bender, R.A.; Standiford, T.J.; Younger, J.G. The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect. Immun. 2004, 72, 1423–1430. [Google Scholar] [CrossRef]
- Abreu, A.G.; Barbosa, A.S. How Escherichia coli Circumvent Complement-Mediated Killing. Front. Immunol. 2017, 8, 452. [Google Scholar] [CrossRef]
- Albelda-Berenguer, M.; Monachon, M.; Joseph, E. Chapter Five—Siderophores: From natural roles to potential applications. In Advances in Applied Microbiology; Gadd, G.M., Sariaslani, S., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 106, pp. 193–225. [Google Scholar]
- Telford, J.R.; Raymond, K.N. Amonabactin: A family of novel siderophores from a pathogenic bacterium. JBIC J. Biol. Inorg. Chem. 1997, 2, 750–761. [Google Scholar] [CrossRef]
- Grammbitter, G.L.C.; Schmalhofer, M.; Karimi, K.; Shi, Y.-M.; Schöner, T.A.; Tobias, N.J.; Morgner, N.; Groll, M.; Bode, H.B. An Uncommon Type II PKS Catalyzes Biosynthesis of Aryl Polyene Pigments. J. Am. Chem. Soc. 2019, 141, 16615–16623. [Google Scholar] [CrossRef]
- Schöner, T.A.; Gassel, S.; Osawa, A.; Tobias, N.J.; Okuno, Y.; Sakakibara, Y.; Shindo, K.; Sandmann, G.; Bode, H.B. Aryl Polyenes, a Highly Abundant Class of Bacterial Natural Products, Are Functionally Related to Antioxidative Carotenoids. ChemBioChem 2016, 17, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Zeth, K.; Römer, C.; Patzer, S.I.; Braun, V. Crystal Structure of Colicin M, a Novel Phosphatase Specifically Imported by Escherichia coli. J. Biol. Chem. 2008, 283, 25324–25331. [Google Scholar] [CrossRef]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial activities of bacteriocins: Application in foods and pharmaceuticals. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Whole-Genome Sequence of Ewingella americana ATCC 33852. Available online: https://www.ncbi.nlm.nih.gov/genome/11412?genome_assembly_id=204696 (accessed on 8 January 2020).
- Draft Genome Sequence of Ewingella americana CCUG 14506T. Available online: https://www.ncbi.nlm.nih.gov/genome/11412?genome_assembly_id=694515 (accessed on 8 January 2020).
- Genome Sequence of Ewingella americana NCTC12157. Available online: https://www.ncbi.nlm.nih.gov/genome/11412?genome_assembly_id=392147 (accessed on 8 January 2020).
- Wong, N.H.; Rosato, A.J.; Rose, Y.M.; Penix, T.S.; Fung, J.B.; Vanitski, A.L.; Goossen, C.J.; Bradshaw, S.G.; Lopp, S.M.; Pennington, A.D.; et al. Isolation and Whole-Genome Sequencing of 12 Mushroom-Associated Bacterial Strains: An Inquiry-Based Laboratory Exercise in a Genomics Course at the Rochester Institute of Technology. Microbiol. Resour. Announc. 2020, 9, e01457-19. [Google Scholar] [CrossRef]
- Altshuler, I.; Hamel, J.; Turney, S.; Magnuson, E.; Levesque, R.; Greer, C.W.; Whyte, L.G. Species interactions and distinct microbial communities in high Arctic permafrost affected cryosols are associated with the CH4 and CO2 gas fluxes. Environ. Microbiol. 2019, 21, 3711–3727. [Google Scholar] [CrossRef]
- Mergaert, J.; Verdonck, L.; Kersters, K. Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the Genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., Respectively, and Description of Pantoea stewartii subsp. indologenes subsp. nov. Int. J. Syst. Evol. Microbiol. 1993, 43, 162–173. [Google Scholar] [CrossRef]
- Ivanovic, N.; Minic, R.; Djuricic, I.; Radojevic Skodric, S.; Zivkovic, I.; Sobajic, S.; Djordjevic, B. Active Lactobacillus rhamnosus LA68 or Lactobacillus plantarum WCFS1 administration positively influences liver fatty acid composition in mice on a HFD regime. Food Funct. 2016, 7, 2840–2848. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.; Abrams, N.; Jackson, J.; Jacobs, A.; Kiryutin, B.; Koonin, E.; Krylov, D.; Mazumder, R.; Mekhedov, S.; Nikolskaya, A.; et al. The COG Database: An Updated Version Includes Eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.; Auch, A.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R.; Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2015, 32. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.; Roos, D. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Chiou, C.-S.; Chen, C.-C. PGAdb-builder: A web service tool for creating pan-genome allele database for molecular fine typing. Sci. Rep. 2016, 6, 36213. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Hudzicki, J. Kirby-bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. Protoc. 2009. Available online: https://www.asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf.pdf (accessed on 8 January 2020).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Approved Standard; 25th Informational Supplement; CLSI Document M100-S25; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Moura, A.; Soares, M.; Pereira, C.; Leitão, N.; Henriques, I.; Correia, A. INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics (Oxf. Engl.) 2009, 25, 1096–1098. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Dandan, Z.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2018, 47. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, S.; Larsen, M.; Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Abby, S.; Néron, B.; Ménager, H.; Touchon, M.; Rocha, E. MacSyFinder: A Program to Mine Genomes for Molecular Systems with an Application to CRISPR-Cas Systems. PLoS ONE 2014, 9, e110726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).