Abstract
Swine act as both maintenance and incidental hosts of pathogenic Leptospira spp. Here, a serological test was performed on 131,660 pig sera collected between 2002 and 2017 from 4715 farms in Northern Italy. A positivity rate of 13.05% was determined. Australis was the most frequently identified serogroup (77.29%), followed by Pomona (18.47%), Tarassovi (1.51%) and Icterohaemorrhagie (1.40%). Culture isolation and real-time Polymerase chain reaction (PCR) were carried out on 347 kidneys and 470 clinical samples, respectively. Overall, 133 strains were cultured successfully and 43 randomly chosen isolates were identified as serogroup Pomona. Multi-locus sequence typing (MLST) revealed that 41 isolates and 8 DNA extracted from biological samples belonged to sequence type 140. Using a multiple-locus, variable-number tandem repeat analysis, 43 samples produced identical profiles but, after 2014, three new Leptospira interrogans serogroup Pomona genotypes were observed. Interestingly, two isolates showed new MLST profiles and an unclassified identification by monoclonal antibodies. The 16S rRNA gene sequencing clustered them into L. kirschneri species and a core genome MLST analysis revealed an allelic identity of 96% compared with Mozdok strains. Genotyping allowed us to discriminate leptospires and to identify new emerging strains. The accurate identification of infective strains is required for formulating preventive methods and intervention strategies.
1. Introduction
Leptospirosis is the most widespread zoonosis worldwide and it is caused by an infection with any of the pathogenic members of the genus Leptospira. While, in theory, any pathogenic Leptospira may infect any animal species, leptospirosis is a disease that shows a natural nidality, and each serovar tends to be maintained in specific maintenance hosts. For example, serovar Bratislava is associated with pigs and horses, Canicola is commonly found in dogs, Icterohaemorrhagiae in rats, Pomona in pigs and Hardjo in cattle [1,2,3]. Leptospires persist in the kidneys of carrier animals and are excreted in urine and genital fluids [3,4,5,6]. In this way, they are spread through the environment where they can survive for long periods, contaminating the surface water, soil and muddy areas [2,3,4,5,7]. Chronically infected animals may remain carriers over years and act as reservoirs for the infection of other animals and humans [3,5]. Despite rat being the main reservoir for these bacteria, many other mammals can act as carriers. Swine, for example, act as a maintenance host for leptospires and are a possible source of human and domestic animal infections [3,5]. Historically, pigs act as a maintenance host for the serovars Bratislava, Pomona and Tarassovi, while among the incidental serovars, the most important in pigs are those belonging to the Icterohaemorrhagiae, Canicola and Grippotyphosa serogroups [3,4,5].
In Italy, swine act commonly as carriers for serovar Bratislava, belonging to the Australis serogroup, and serovar Pomona, belonging to the Pomona serogroup. Additionally, serovar Tarassovi causes commonly incidental infections [8]. Endemic infections in swine herds generally remain subclinical, and the only clinical symptoms are reproductive disorders, such as late-term abortions, and increases in mummified, stillborn and weak piglets. However, leptospires can also cause severe diseases depending mainly on the infecting serovar and the age of the animal [4]. Once the infective agent has entered a farm, it spreads very easily, mostly among fattening pigs, both through direct (contaminated urine) and indirect (infected feed, water and environment) contact [8]. Vaccination, therapy and farm management can all be used to limit the spread of infection in a breeding herd.
Until 2010, in Italy, a trivalent vaccine against the serogroups Australis, Pomona and Tarassovi was available for swine. Nevertheless, very few breeders have adopted vaccination practices, primarily because the risk of leptospirosis is poorly understood by farmers and because the treatments and/or control strategies for other more virulent diseases are more important to the farm’s economy. In 2011, vaccinations were completely abandoned because no commercial vaccine was available for pigs. Starting from that moment, the main infection control strategy was the management of the breeding herd through the prevention of direct or indirect contact with free-living vectors or other domestic stock. Strong monitoring and surveillance systems are needed to better understand the disease epidemiology, and strict biosecurity tools should be applied to limit the transmission of the infective agent. Leptospirosis is not included in the Office International des Epizooties (OIE) list of notifiable terrestrial and aquatic animal diseases; however, in Italy, it is considered, by the current rules, a notifiable infection [9]. In cases of clinical suspicion, which are confirmed by a serological examination, the outbreak must be officially reported to the authorities. The farm is seized and sanitary measures, aimed at the eradication of the infective agent, are implemented. The interruption of transmission through the isolation of the infected animals is the first action that must be implemented. Only when all the animals become seronegative may the infection be considered eradicated, and the restrictive measures can be removed [9,10]. This can take months, or even years, resulting in significant economic losses.
In the laboratory, serological testing is the most widely used means for diagnosing leptospirosis, and the Microscopic Agglutination Test (MAT) is the serological gold standard method reported by the OIE [11]. Ideally, antigens selected for the MAT should include representative strains of all serogroups known to circulate in the study region as well as those known to be maintained elsewhere by the host species being analyzed.
The OIE indicates that strain isolation should be attempted from biological samples. Cultured strains can be studied in depth, and the serogroup and serovar status should be determined using polyclonal and Monoclonal Antibodies (mAbs), respectively [11]. Nevertheless, isolation does not contribute directly to acute diagnosis of leptospirosis because Leptospira grows slowly over weeks [12]. With the advent of molecular methods, the diagnosis of a Leptospira infection has become simpler and faster. Over the years, a number of real-time PCR-based methods have been described for the direct identification of Leptospira nucleic acids in biological samples, both for detection of pathogenic and environmental species [13,14,15,16,17,18,19,20]. Real-time PCR provides advantages over the classical conventional reference methods used to diagnose leptospirosis (such as MAT and isolation) including reduced turnaround and hands-on times, low carryover contamination risks and higher sensitivity and specificity levels.
Furthermore, in the last few years, genotypic classification has increasingly integrated traditional serological classification in a wide variety of bacterial species. For Leptospira, the classical classification into serogroups and serovars has been joined by molecular typing. Several molecular techniques, such as Multiple-Locus Variable-Number Tandem-Repeat (VNTR) Analysis (MLVA) and Multi-Locus Sequence Typing (MLST), aimed at identifying bacteria by examining individual genomic profiles, have been used to investigate the epidemiology of Leptospira. The MLVA method is a useful typing tool for identifying Leptospira genotypes by providing information on genetic relationships among isolates for the surveillance of Leptospira populations. Salaün and colleagues [21] developed an MLVA method that permits the discrimination of the three most common pathogenic species of Leptospira (L. interrogans, L. kirschneri and L. borgpetersenii) through the analysis of five loci of repetitive unit sequences (VNTR 4, 7, 10, Lb4 and Lb5).
In 2013, Boonsilp and colleagues [22] proposed an MLST scheme based on the sequencing of seven housekeeping genes. This protocol is optimized to work on isolate samples, but it is not effective on biological samples, owing to the poor bacterial load. Thanks to a new protocol published in 2016 by Weiss [23], it is now possible to perform MLST directly on DNA extracted from biological samples, overcoming the long and difficult isolation step. With Whole-Genome Sequencing (WGS), new genome-based analyses have been developed for bacterial typing. Very recently Guglielmini et al. [24] published a core genome MLST (cgMLST) scheme based on 545 highly conserved genes in Leptospira, increasing the discriminatory power necessary to distinguish isolates.
This study aims to provide a serological survey of the prevalence of Leptospira in pigs in Northern Italy and to characterize the circulating strains using innovative molecular techniques. Our goal is to understand the epidemiology of swine leptospirosis in this specific area and to identify possible new emerging strains to address preventive and control measures and to reduce the risk of infection in swine herds.
2. Results
2.1. Microscopic Agglutination Test (MAT)
Using MAT, 17,184 out of 131,660 sera collected from 4715 farms were identified as positive, with a seropositivity of 13.05% (cut-off ≥ 1:100). The minimum percentage of positive sera was observed in 2006 (8.46%), the maximum one in 2012 (18.31%). Among the positive samples, 13,809 (10.49% of the total samples and 80.36% of the positive samples) tested positive for one serogroup, while 3375 (2.56% of the total samples and 19.64% of the positive samples) were positive for more than one serogroup. Antibody titers against the serogroup Australis occurred in 8.11% of the total samples and 77.29% of the single positive samples, followed by Pomona (1.94% and 18.47%, respectively), Tarassovi (0.16% and 1.51%, respectively), Icterohaemorrhagiae (0.15% and 1.40%, respectively), Sejroe (0.07% and 0.67%, respectively), Ballum (0.03% and 0.32%, respectively), Canicola (0.02% and 0.22%, respectively) and Grippotyphosa (0.01% and 0.12%, respectively) (Table 1). Using the cut-off ≥ 1:200, the Australis-positive samples decreased to 66.75%, the Pomona-positive samples increased to 29.28%, and the other serogroups maintained similar values (data not shown).

Table 1.
Numbers and percentages of swine serum samples that tested positive by Microscopic Agglutination Test (MAT) for Leptospira serogroups from 2002 to 2017. Only positivity to one serogroup was included.
For positivity to more than one serogroup (Table 2), the predominant combination was represented by Australis-Pomona, having a prevalence of 54.72% (1847 out of 3375 multiple-positive sera) followed by Australis-Icterohaemorrhagiae-Pomona with 16.91% (571/3375), Australis-Tarassovi and Australis-Icterohaemorrhagiae (6.40% and 6.34%, respectively). The Icterohaemorrhagiae-Pomona combination was found in 2.75% of samples, while other combinations were revealed at levels < 2%. Australis was present in 3156 (93.51%), while Pomona was present in 2855 (84.59%), of 3375 multiple-positive sera.

Table 2.
Sera that react to more than one serogroup by Microscopic Agglutination Test (MAT).
The MAT titers of the single positive samples were generally low (1:100 or 1:200) for all the serogroups detected, except for the serogroup Pomona, which showed medium (1:400) or high titers (greater than 1:800) in more than 50% of the samples (Figure 1). In particular, more than 90% of the samples positive for the serogroup Australis showed low antibody titers, while those positive for Pomona showed mainly medium (17.99%) and high (38.79%) titers. Grippotyphosa-positive samples also showed high titers (≥1:800) in more than 35% of the samples (Figure 1). The detailed titer distributions of positive sera reacting to one serogroup are reported in Table 3.
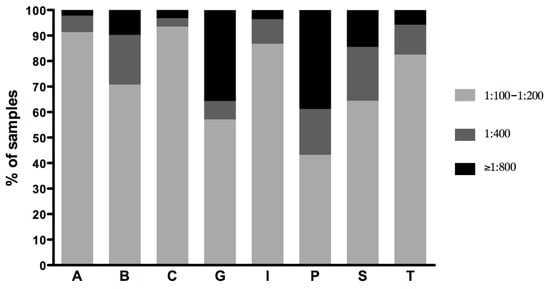
Figure 1.
Distribution of MAT titers of samples reacting to one serogroup. Titers were grouped as low (1:100–1:200), medium (1:400) and high (≥1:800). A, Australis; B, Ballum; C, Canicola; G, Grippotyphosa; I, Icterohaemorrhagiae; P, Pomona; S, Sejroe; T, Tarassovi.

Table 3.
Detailed MAT titers of samples reacting to one serogroup.
Out of 4715 tested farms, a mean of 53.62% resulted as positive for the presence of pathogenic Leptospira (Table 4). A farm was considered positive if at least one pig was positive by MAT to one or more Leptospira serogroups. The percentage of farms with positive test results ranged between 22.84% (n = 140) in 2006 and 77.52% (n = 231) in 2012.

Table 4.
Results of the MAT analysis of tested farms. Number (n) and percentage (%) of positive farms between 2002 and 2017.
Outbreaks caused by Australis were stable over the years, with a maximum value of 82.23% in 2012 and a minimum of 66.34% in 2002. This serogroup is responsible for a mean of 75.87% ± 0.15% (Confidence Interval 95%) of the outbreaks during the study period (Figure 2).
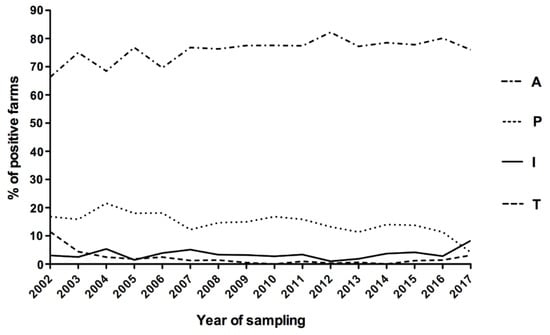
Figure 2.
Trend in the main observed outbreaks between 2002 and 2017 (only sera positive for a single serogroup were considered). A, Australis; B, Ballum; C, Canicola; G, Grippotyphosa; I, Icterohaemorrhagiae; P, Pomona; S, Sejroe; T, Tarassovi.
The second serogroup in terms of frequency of outbreak was Pomona, with a mean of 14.56% ± 0.31% (CI 95%) over the 16 years of the study with the maximum value observed in 2004 (21.58%) and the minimum one in 2017 (4.17%). Serogroup Icterohaemorrhagiae was responsible for a mean frequency of 3.52% ± 0.30% (CI 95%), with a peak of 8.33% in 2017. In 2017, an inversion occurred between the Pomona and Icterohaemorrhagiae serogroups: the first decreased to the minimum value (4.17%), while the latter increased to a new maximum level (8.33%). The mean frequency of Tarassovi was 2.10% ± 0.48% (CI 95%) but interestingly, its trend started from 11.49% in 2002 and became null in 2014. For Sejroe, a very low frequency was registered over the years, with values between 0% in 2014 and 2.84% in 2016 (mean of 1.64% ± 0.19%, CI 95%). For the other serogroups, the mean prevalence was less than 2% with rare exceptions during the considered time (Supplementary Table S1).
2.2. Molecular Detection, Isolation and Identification
Using real-time PCR, we detected pathogenic Leptospiral DNA in 102 out of 470 biological samples (21.70%). In total, 133 out of 347 samples (38.33%) were cultured successfully. The MAT carried out using the isolated strains as antigens against reference anti-sera showed that they were antigenically related to the Pomona serogroup. In total, 12 randomly chosen isolates (ID321, 331, 335, 340, 349, 354, 362, 385, 391, 430, 393 and 411) were subjected to serovar identification with mAbs. The reference strains Pomona and Mozdok 5621 reacted to mAb panels as expected (Table 5). Our isolates 321, 331, 335, 340, 349, 354, 362, 385, 391 and 430 reacted only to mAbs F43 C9 and F48 C6, suggesting that they belonged to the serovar Pomona of the L. interrogans species. The reaction profiles of isolates 393 and 411 were very similar to each other, but different from any other strain of the serogroup Pomona. If we consider an acceptable level of variability for the two dilutions used for the reaction titers, then all the mAbs, except for F48 C6 against which both isolates did not show any reactivity, indicated that they were related to the reference strain Mozdok 5621. The Tsaratsovo reference strain B 81/7 had reaction patterns for mAbs F48 C6 and F61 C7 that were similar to those of our isolates. However, the Tsaratsovo reaction patterns against mAbs F43 C9, F46 C9 and F58 C1 are unknown; therefore, they could not be compared with those of our isolates.

Table 5.
Agglutination titers of Monoclonal Antibodies (mAbs) against the Pomona serogroup reference strains and the isolated samples from pigs (titers in reciprocal).
2.3. Genotyping Analyses
2.3.1. MLST and MLVA Analyses
The genotypes of 43 randomly chosen isolates (including the 12 isolates tested for serovar determination) and the 8 biological samples (Supplementary Table S2) were determined. Using the MLST technique, 41 out of 43 isolates and all 8 DNAs extracted from biological samples were identified as ST140 and clustered with reference strain L. interrogans, Pomona st. Pomona (international reference strain) and st. Mezzano I (national reference strain). In total, 2 (ID 393 and 411) out of 43 isolates showed two new sequence types (STs), similar to ST117, typical of L. kirschneri Mozdok and also previously determined for reference strain 5621 present in the Istituto Zooprofilattico Sperimentale della Lombardia e dell’Emilia Romagna (IZSLER) collection. Compared with ST117 (pattern 13-25-15-22-33-18-23 for the loci glmU, pntA, sucA, tpiA, pfkB, mreA and caiB, respectively), the glmU gene of sample 393 showed the substitution 120 T > C, and the pntA gene had the substitution 410 A > G. The isolate 411, compared with ST117, showed the mutated pntA gene already found in isolate 393, while the glmU gene was the same as that of ST117 (allele 13). Both the sequences were submitted to the curators of the Leptospira database and alleles 75 and 85 were assigned to glmU and pntA, respectively. These new alleles defined two new STs, ST288 and ST289, for isolates 393 and 411, respectively.
As shown in Figure 3, 41 out of 43 isolates and all the DNA samples clustered together with the L. interrogans Pomona reference strains (Mezzano I and Pomona) having ST140, while the samples 393 and 411 clustered with L. kirschneri species near the reference strain 5621.
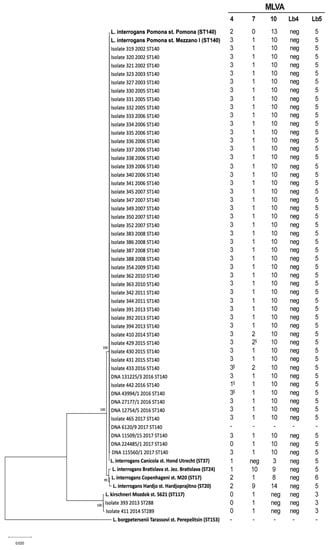
Figure 3.
Phylogenetic tree based on concatenated sequences of the seven gene of the multi-locus sequence typing scheme (3111 bp) [22]. Samples are indicated with their unique IDs, the isolation year and their sequence types (STs). The names of reference strains (in bold) include the Leptospira species serovar and strain. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. In the multiple-Loci Variable Tandem Repeat Analysis (MLVA) profile (on the right), the symbol “§” indicates the sequenced alleles.
Moreover, the MLVA analysis revealed that 37 out of 41 isolates, belonging to ST140, and 6 out of 8 DNA samples, had the MLVA profile 3-1-10-neg-5 for the VNTR loci 4, 7, 10, Lb4 and Lb5, respectively. This profile was identical to that of the Italian reference strain (Mezzano I), but different from the international reference strain (Pomona) profile (2-0-13-neg-5) at the loci 4, 7 and 10 (Figure 3). Three isolates belonging to ST140 (one each from 2014, 2015 and 2016) showed a new genotype (3-2-10-neg-5), which differed from strain Mezzano I and from the other samples at the VNTR7 locus. One ST140 isolate from 2017 (ID 442) harbored the new genotype 1-1-10-neg-5. One extract from 2017 (224485/1) and genotyped as ST140 showed a new VNTR profile that was 0-1-10-neg-5. The new alleles were confirmed by sequencing (Figure 3). No results were obtained for sample 6120/9, although many amplification attempts have been made for each of the five VNTR loci (Figure 3). The isolates 393 and 411 showed the same genotype found in strain Mozdok 5621, which is 0-1-neg-neg-3 for the loci 4, 7, 10, Lb4 and Lb5 respectively.
Sequencing the 16S rRNA gene confirmed that strains 393 and 411 belonged to L. kirschneri species, in agreement with the previous results.
2.3.2. Whole Genome Sequencing (WGS) and Core Genome MLST (cgMLST) Analysis
To clarify the genetic features of strains 393 and 411 belonging to the new STs, we performed WGS and a cgMLST analysis. Sequences of strain 393, 411, IZSLER 349/2007 and Mozdok 5621 were submitted to EBI database under the project number PRJEB36553 and are available under genome accession numbers from ERR4056312 to ERR4056315. The cgMLST analysis was performed on contigs of the strains 393, 411, IZSLER 349/2007 and Mozdok 5621 using the Bacterial Isolate Genome Sequence Database (BIGSdb) [25] (Table 6). The strains 393, 411 and IZSLER 349/2007 showed similarities with more than one cgST profile.

Table 6.
Core genome (cg) MLST analysis using the Bacterial Isolate Genome Sequence Database.
The closest cgSTs were queried against the BIGSdb to retrieve deposited isolates with the same genotypes. The results are reported in Table 7.

Table 7.
Metadata of isolates having the most similar allelic profiles to those of our samples retrieved from BIGSdb.
The identities of strains Mozdok 5621 and IZSLER 349/2007, included in the cgST analysis as controls, were confirmed (Table 6 and Table 7). The isolates 393 and 411 showed the greatest similarity (96%) with deposited strains classified as L. kirschneri Pomona Mozdok (Table 6 and Table 7). We performed a cgMLST comparison (BIGSdb Genome Comparator [25]) among genomes of sequenced isolates (strain 5621, 393 and 411) and cgST-related strains present in the database (134|Vehlefans_2, 140|Vehlefans_3, 251|61H and 252|M36/05) (Supplementary File S1).
In total, 17 and 19 out of 545 alleles were revealed as “new” in isolates 393 and 411, respectively. In total, 14 occurred in both isolates 393 and 411 (Supplementary Table S3). The distance matrix (Figure 4a) was used to build a distance tree using SplitsTree software ver. 4.15.1 [26] (Figure 4b).
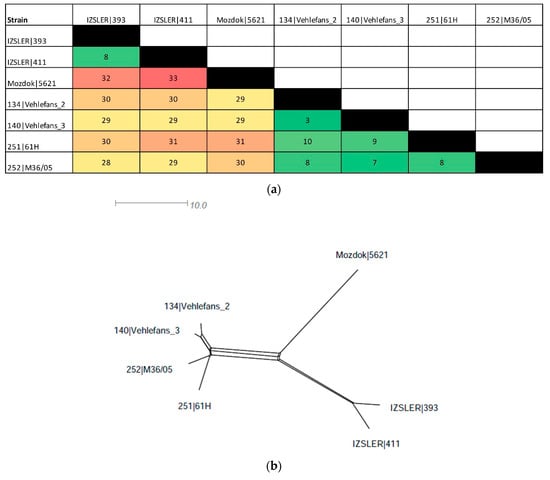
Figure 4.
Distance matrix of the cgMLST of IZSLER isolates and cgST-related strains in the BIGSdb (a) and SplitsTree networks constructed using the distance values (b). (a) The color level of the distance matrix is proportional to the similarity value between strains, from green (high similarity) to red (low similarity). Incomplete loci were ignored in pairwise comparisons unless the locus was missing in an isolate. (b) Distance matrix visualized as SplitsTree networks.
The strains 393 and 411 were very similar, having only eight allelic differences. Compared with Mozdok 5621, they had 32 and 33 allelic mismatches, respectively (Supplementary File S1 and Figure 4) and showed from 28 to 31 differences when compared with the other serovar Mozdok strains retrieved from the database (134|Vehlefans_2, 140|Vehlefans_3, 251|61H and 252|M36/05) (Supplementary File S1 and Figure 4a). The SplitsTree network revealed three different strain clusters: the first contained the Mozdok strains retrieved from the BIGSdb (3–10 differences between each other), the second contained the IZSLER strains 393 and 411 (eight differences between each other) and the last contained Mozdok 5621, which was located on a separated branch (Figure 4b).
3. Discussion
The main objective of this study was to evaluate the epidemiology of infective Leptospira serovars in fattening pigs from farms in Northern Italy (Lombardy and Emilia Romagna regions). This territory has a very large pig population, with a mean of 5,300,000 animals, and contained 12,900 farms from 2007 to 2017 (data for 2002–2006 are not available), which produce pork for the entire country. In this area, the control of swine pathogens responsible for zoonoses, like Leptospira, is crucial to limit human and domestic animal infections, as well as to reduce the related economic losses linked to such diseases on the farms. Furthermore, the identification of the strains responsible for leptospirosis in pigs and their specific genotypes are essential for epidemiological analyses needed to adopt proper preventive measures and to better understand the etiology of the disease.
In this study, MAT, considered the gold standard and the most commonly used test, was used for serological identification. We tested 131,660 pig sera collected for 16 years. In 2002, there were 18,096 collected samples but the number progressively decreased each year to 2505 samples in 2017, except for a peak of 9330 samples in 2012. This trend may result from the strictness of the Italian national rules [9,10], in which the infection associated with a disease outbreak is only considered eradicated when all the affected animals become seronegative. This could require months or even years, and during this period, the herd is subjected to restrictive measures that are very penalizing and economically disadvantageous to the farm. Moreover, a better sampling strategy may have contributed to the reduced numbers of samples over the last few years. In fact, in the past, sampling had been carried out in all the livestock sectors, but in the last few years, perhaps for economic reasons, the trend has been to perform targeted sampling, mainly among reproductive or symptomatic animals.
In the MAT analysis, to discriminate positive and negative sera, we considered a cut-off titer of 1:100, following the guidelines indicated by the OIE [11] and commonly used for international trade. More than 80% of the positive sera (relative to 10.49% of the total samples) tested positive for one serogroup, presumably indicating the real infecting serovar and the chronic stage of infection [7]. Instead, 19.64% of the positive sera reacted simultaneously with two or more serovars, indicating both cross-reactions of serovars of the same serogroup and the acute phase of infection [4,7]. In fact, paradoxical immune response, in which the predominant serogroup in MAT is unrelated to the infecting one, occurs in the acute phase of infection meaning that 2.56% (3375 out of 133,660 sera) of all pigs examined in this study were very likely in an acute stage of infection [7].
Considering both single positive sera and multiple positive sera, our results indicated a seropositivity of 13.05%, in agreement with other previous studies [27,28,29]. In particular, Bertelloni found a seroprevalence of 16.6% among slaughtered swine in North–Central Italy [27], Cerri reported a prevalence of 8.85% in swine sera collected in Italy from 1995 to 2001 [28], and Chiari and colleagues found an overall prevalence of 15.28% among wild boars in Northern Italy in a five-year period (2008–2013) [29]. In Brazil, the prevalence of seropositive pigs is 16.1% [30] and in Japanese herds it is 14.4% [31]. However, a very low prevalence (1.02%) has been found in the pig population of Poland [32].
Our data revealed that a mean of 53.6% of pig farms sampled in Northern Italy tested positive for leptospirosis, in agreement with the data reported by Bertelloni and colleagues [27], who observed a leptospirosis prevalence of 52.5% in farms located in North–Central Italy from September to December 2015. Interestingly, positivity among farms showed a wide variability, from a minimum of 22.8% in 2006 to a maximum of 77.5% in 2012, in accordance with the tendency over the last few years to perform targeted sampling on suspected cases.
The most common serovars associated with swine infections in Italy are Pomona (serogroup Pomona), Tarassovi (serogroup Tarassovi), Bratislava (serogroup Australis) and Muenchen (serogroup Australis) [8]. Our results indicated that Australis is still endemic in pigs in Northern Italy, where swine acts as reservoir host for this serogroup, as in many countries and regions worldwide [4]. More than 90% of Australis-infected animals had low antibody titers (≤1:200); this is in agreement with previous studies that described serogroup Australis, specifically serovar Bratislava, as pathogenic for swine, although it usually causes a weak immune response and most of the chronically infected pigs have titers less than 1:100 [33,34,35]. Infections caused by Bratislava are characterized by mild clinical signs and are frequently associated with subfertility and litter-size reduction [36,37]. Unfortunately, since any Bratislava strains were successfully isolated, there is no data that allow us to define the specific infective serovar present on Australis-positive farms. It is probable that these infections were largely caused by Leptospira Bratislava, which is widespread among hedgehogs (Erinaceus europaeus) [38] and was found in wild boars in Parma Apennines in Italy [39]. However, we cannot exclude a priori that there are other causal agents, such as the Muenchen and Lora serovars. In fact, these serovars, together with Bratislava, have been isolated from the genital tracts and kidneys of sows that have abortions, from aborted fetuses and from boars in the USA [40], the Netherlands [41] and Northern Ireland [42,43]. Serogroup Australis has emerged as the major swine-maintained cause of Leptospira infections, but its epidemiological role remains poorly understood owing to isolation difficulties [44]. The isolation of serovar Bratislava still represents a challenge in the diagnosis of leptospirosis and, in fact, in contrast to the high seroprevalence reported worldwide [3,45,46,47], L. interrogans Bratislava has only been recovered from swine in a few countries, such as the United Kingdom [48], Germany [44], the USA [40,49,50], Vietnam [51] and Brazil [36]. Even in this study, we did not isolate any Bratislava strains. This may result from the quality and composition of the medium used, as well as the low bacterial load in kidneys, the organ used for the culture isolation [52]. In fact, Ellis et al. [52] found a higher isolation frequency for the Australis serogroup (Bratislava and Muenchen) from genital organs compared with kidneys. Therefore, improvements in the quality and composition of the isolation medium, as well as sampling procedures, must be considered.
Pomona, the second most predominant serogroup in this work, is among the most common serogroup isolated from swine worldwide [53,54,55], and many strains of this serogroup are adapted to swine, which is recognized as a maintenance host [3]. Pigs infected with serogroup Pomona often experience abortions, stillbirths or the births of weak or ill piglets, and the infections result in subsequent decreased reproductive performances. Adult non-pregnant animals are usually asymptomatic carriers. Unfortunately, no data regarding the clinical symptoms of the analyzed pigs in this study were available. Swine infections caused by Pomona strains have been observed in Europe, particularly in its eastern and southern regions [56,57]. Recently, Bertelloni and colleagues [27] highlighted a recurrence of serogroup Pomona in pigs in North–Central Italy, revealing a Pomona serogroup seropositivity of 18% in the screened farms and of 8% in analyzed sera. Apart from swine, Pomona also causes clinically and economically significant infections in cattle, sheep and horses in many countries, resulting particularly in reproductive problems and equine uveitis [58]. A study from New Zealand reported that 31% of people working or residing on or within a 50-km radius of pig farms had microscopic agglutination titers of 1:24 or greater to one or more serovars of L. interrogans, with the majority being to Pomona [59]. Moreover, the ST140 genotype, the same as in our infected pigs, has been associated with symptomatic human leptospirosis in New Caledonia, Australia and Sri Lanka (this publication made use of the Leptospira MLST website https://pubmlst.org/Leptospira/ sited at the University of Oxford [25]. The development of this site has been funded by the Wellcome Trust).
For Tarassovi, historical data supported the widespread presence of this serogroup among the Italian pig population. In 1990, Tagliabue reported the isolation of four Tarassovi strains from swine kidneys [60], while in 1993, Scanziani and colleagues [37] described mild renal lesions in pigs affected by Tarassovi. Additionally, in 1995, Tagliabue and Farina reported a seroprevalence of 6.8% among the pig population, using a cut-off titer of 1:400 [38]. The identification of Tarassovi as the infective serogroup of swine in the past was corroborated by its presence in the vaccine formulation, indicating the need to protect hosts from this infective agent. In accordance with the observations already made by Tagliabue and Farina in 1995 [38], which noted a progressive decline in the Tarassovi frequency during the last few years, our study found that, in Italy, the infections caused by Tarassovi are decreasing, being responsible for the positivity of only 0.16% of the total samples tested. The infections caused by Tarassovi in swine have become incidental and probably result from contact with wild animals, including turtles, which act as a maintenance host for this serovar [61].
Among the minor serogroups found in tested pigs, we payed particular attention to some of them, discussed below. First of all, serogroup Icterohaemorrhagiae whose low prevalence could indicate the use of good rodent control measures inside and around the tested pig farms [62]. Moreover, the low prevalence observed for Grippotyphosa, Sejroe and Canicola were in agreement with historical data that described these serogroups as causes of incidental infection transmitted to pigs from wildlife hosts (serogroup Grippothyphosa [4]), from dogs (serogroup Sejroe serovar Sejroe [63] and serogroup Canicola serovar Canicola [64]), from bovines (serogroup Sejroe serovar Hardjo [4]) or small rodents (serogroup Sejroe serovar Saxkoebing [65]).
Through genotyping techniques we found that all the isolated and extracted samples belonged to the Pomona serogroup but, interestingly, from 2014, new Pomona genotypes started to circulate in Northern Italy. Unfortunately, the Sequence Type provided by MLST analysis (ST140) being associated with isolates characterized as belonging to serogroups Pomona serovar Pomona, serogroup Pyrogenes serovar Guaratuba and serogroup Grippothyphosa (data from BIGSdb) did not permit any discrimination at the serogroup and at the serovar level, this ST In this case, the information provided by MLST genotyping is limited to species discrimination (i.e., L. interrogans). However, we should distinguish two different situations: in the case of isolated strains, since serological analysis by polyclonal anti-sera and by mAbs were possible, we were able to distinguish their serovar (Pomona) and serogroup (Pomona). In the case of DNA samples, we cannot assume that they are definitively serovar Pomona based only on the MLST results. However, we can assume that six of the eight samples belonged to serogroup Pomona serovar Pomona because their MLVA profiles were identical to that of strain Mezzano I.
Interestingly, two isolates showed new mAb responses and new STs. Their reactions to the mAbs specific for the Pomona serogroup were very similar to each other but different from any of the known serogroup Pomona strain profiles. Their pattern was compatible with serovar Tsaratsovo, which was first isolated from harvest mice (Micromys minutus) in 1962, in the Plovdiv District of Bulgaria by Ivanov and later found in rodents in some parts of Europe [66]. Nevertheless, only mAbs F48 C6 and F61 C7, included in our panel, were tested on this serovar. Therefore, these data are insufficient to determine their serovars states. However, from an epidemiological point of view, this serovar has never been found in Italy; thus, it is very improbable that Tsaratsovo exists among the Italian pig population. Genotyping analyses of these strains indicated that they had the same VNTR pattern of Mozdok st. 5621 and belonged to ST288 and ST289, which had not been described previously, and are similar to ST117, which was found in L. kirschneri Mozdok st. 5621. In addition, the cgMLST analysis revealed that the most similar strains are classified as Leptospira, species kirschneri, serovar Mozdok. Definitely, MLST and cgMLST analyses excluded their belonging to the Tsaratsovo serovar, because strain Tsaratsovo B 81/7 was characterized as ST115 and cgST 245. We can also exclude Altodouro and Kunming, the other serovars included in L. kirschneri species and the Pomona serogroup, because they showed ST100 and ST70, respectively, with none of the seven loci being in common with those of ST288 and ST289. Therefore, we can assert that they are very similar to L. kirschneri Mozdok, but accurate descriptions of their genetic contents in comparison with serovar Mozdok are still being compiled. From an epidemiologic point of view, several studies have identified serovar Mozdok across Europe [67,68,69,70,71] in dogs [72] and in wild rodents [70,73,74]. It has been described also as a possible cause of abortions in cattle [75] and in pigs [69,73,74] and has been responsible of human leptospirosis in Cuba [76].
Owing to the advent of next-generation sequencing and the publication of the cgMLST scheme, the resolution powers of sequence-based analyses on Leptospira are increasing but the definitions of allelic distance cut-offs for the differentiation of strains are still open and in urgent need of cgMLST classification.
Our study, which provides information on the Leptospira serovars present in the Italian pig population, is also useful for implementing a successful diagnostic and vaccination program. Commercial Leptospira vaccines are available globally for cattle, pigs and dogs, but vaccination has proven to be only partially effective, owing in part to the serovar-restricted nature of vaccine-induced immunity and the potential presence of local serovars other than those included in the vaccines. In Italy, prior to 2010, a trivalent vaccine against Bratislava (serogroup Australis), Pomona (Pomona) and Hyos (Tarassovi) was available for swine. It was used in breeding boars, in non-pregnant sows and in pregnant sows before the octave week of gestation. An unpublished survey carried out by the National Centre of Leptospirosis of IZSLER revealed that among 333 pig farms in Northern Italy during the period February 2004–December 2005, only 19% of the sampled farms adopted the vaccination practice. The reasons for this limited use of vaccination against leptospirosis in pigs included the risk that this disease is poorly understood by farmers and that leptospirosis is a secondary problem in the farm life/economy compared with other more virulent and economically important diseases. Nevertheless, since MAT does not discriminate between vaccination titers and titers due to exposure and because the vaccination status of pigs tested in this study prior to 2010 were unknown, the risk of having an altered number of positive pigs due to the interference of vaccination should be considered. About this aspect, the previously mentioned survey reported that in the farms where vaccination against Leptospira was practiced, 41% of pigs resulted as seropositive, while where it was not practiced seropositive animals were equal to only 9.8%. However, when considering this situation, it highlighted various scenarios: in the case of Australis, the interference of vaccination immunity was significant because the percentage of seropositives increased from 6.1% (of unvaccinated farms) to 33.9% (of vaccinated farms), while for Pomona and Tarassovi, the interference level was lower, going from 2.4% to 5.4% and from 1.2% to 5.4% for unvaccinated and vaccinated farms, respectively (data not published). Based on these data, we can exclude the interference of vaccination on our seropositivity results, given that the overall seropositivity and the seropositivity levels related to Australis, Pomona and Tarassovi (13.1%, 8.1%, 1.9% and 0.2%, respectively) were in agreement with the data from the unvaccinated farms reported in the survey (9%, 6.1%, 2.4% and 1.2%). Furthermore, in this study the trends in the seropositivity of Australis, Pomona and Tarassovi did not reveal any remarkable changes after the end of vaccine use in Italy occurred in 2010, which provided further evidence for our statement.
Considering the serological data obtained and the frequency, in particular, of the Australis and Pomona serogroups, the use of vaccination in pig herds is highly advisable. In this regard, in March 2019, a vaccine has been registered in Italy. This vaccine contains nine serovars (Bananal, Bratislava, Canicola, Copenhageni, Gryppotyphosa, Icterohaemorrhagiae Pomona, Tarassovi, Vughia) and is active against six serogroups (Australis, Canicola, Gryppotyphosa, Icterohaemorrhagiae, Pomona, Tarassovi). As reported by Jacobs et al. [77], this vaccine can be safely used in gilts and sows and induces significant protection for the duration of at least one year. Furthermore, after a challenge with serovar Pomona, it induces protection against clinical signs, leptospiraemia and foetal death [78]. Its use can be considered a valid aid in limiting the infection among pig herds in our territory.
4. Materials and Methods
4.1. Sampling
A total of 133,660 sera, 347 kidneys and 470 biological samples (from kidneys and unspecified tissues, as well as urine) were included in this study. These samples were collected over 16 years (from 2002 to 2017) from 4715 pig farms located in Northern Italy (Lombardy and Emilia Romagna regions) during the routine diagnostic activity. In these farms, testing for Leptospira infections was previously included in the disease-monitoring programs. The samples were collected and analyzed by the National Reference Centre for Animal Leptospirosis at the Istituto Zooprofilattico Sperimentale della Lombardia ed Emilia Romagna (IZSLER) located in Brescia (Italy). In this study, data already published by Tagliabue [61] collected in 2010 and 2011, were also included. In Table 8 we report the numbers of collected serum samples and farms.

Table 8.
Numbers of serum samples analyzed by MAT and farms involved in this study.
Starting from 2008, 347 well-preserved samples were submitted for culture-based causal agent isolation. Since 2009, when molecular methods for the routine diagnosis of leptospirosis were introduced, 470 clinical samples were analyzed using a LipL32-based real-time PCR assay.
To cover the entire analysis period, 43 randomly isolated samples and DNA extracted from 8 biological samples were submitted to genotyping analysis (Supplementary Table S2). For the isolation and genotyping procedures, we selected only one positive sample per farm, assuming that all the positive animals of the same herd were infected with the same strain.
4.2. Serological Tests of Serum Samples
Sera were examined for the presence of antibodies against pathogenic Leptospira using the MAT in accordance with OIE standards [11]. For MAT testing, live cultures of eight reference strains of Leptospira were used (Table 9). They were cultivated at 30 ± 1 °C in EMJH (Ellinghausen, McCullough, Johnson, Harris) medium enriched with bovine serum albumin supplement (10% v/v) [79,80]. Sera were pretested at the final dilution of 1/100. Sera with 50% agglutination were retested to determine an endpoint using dilutions of sera beginning at 1/100 through to 1/6400. Serum samples with the widely accepted minimum significant titer of 100 (reciprocal of the final dilution of serum with 50% agglutination) were assessed positive.

Table 9.
Panel of eight Leptospira spp. used as live antigens for MAT.
4.3. DNA Extraction and Real-Time PCR Detection
DNA was extracted from 0.5–4.0 mL of urine or tissue homogenate using the PureLink Genomic DNA kit (Invitrogen, Paisley, UK) according to the manufacturer’s instructions. An internal control DNA (0.1 µL per µL of elution volume) was added to the digestion buffer. A Taqman-based PCR assay targeting the lipL32 gene was used to detect pathogenic leptospires using primers described previously [17]. The PCR was performed in a 25-µL final volume, using 5 µL of extracted DNA, 5 µL of 5× Mastermix Quantifast (Quantifast Pathogen + IC Kit, Qiagen, Hilden, Germany), 700 nM of primers and 200 nM of the probe. All extraction session included a negative control (water) and all amplification sessions included both a negative (water) and a positive control (DNA of Leptospira interrogans Pomona). The assay was performed on a Bio-Rad CFX96 System using the following thermal conditions: a holding stage of 95 °C for 5 min, and 45 cycles of 95 °C for 15 s and 60 °C for 30 s. Supplementary Table S4 reported the interpretation criteria of Real-time PCR results.
4.4. Isolation of Leptospira from Swine Kidneys
For isolation, 1 g of tissue was diluted, in a plastic bag, in 9 mL of EMJH added with 5-fluoruracil (selective EMJH) [79,80] and homogenized. This suspension was then filtered and collected in a sterile tube and diluted in a four ten-fold serial dilutions (10−2 to 10−5) performed in semisolid selective EMJH (added with 5-fluoruracil and agar). The cultures were incubated at 30 ± 1 °C and observed under dark-field microscopy weekly for up to 6 months. In case of contamination, the cultures were filtrated through a 0.22 μm sterile syringe filter and sub-cultured in fresh semisolid selective EMJH medium.
4.5. Characterization of Pig Isolates Using MAT
The serogroups of the isolates were determined by MAT using a panel of eight polyclonal anti-sera against the eight serovars described in Table 9. A high rate of agglutination with a particular antiserum was used to identify the presumptive serogroup of the strain [81].
Serovar classification was achieved by MAT using panels of mAbs directed against polysaccharide antigens, specific to the serogroup identified. A battery of five mAbs (F43 C9, F46 C9, F48 C6, F58 C1 and F61 C7), which react with serovars of the serogroup Pomona [82,83], were used to determine the serovar states of our isolates. These mAbs and the polyclonal anti-sera were previously purchased from the OIE Leptospirosis Reference Centre, Royal Tropical Institute (KIT), (Amsterdam, The Netherlands), and the protocol was in accordance to the standard serological methods used in this reference laboratory. The reference strains L. interrogans Pomona st. Pomona and L. kirschneri Mozdok st. 5621, provided by KIT, were used as controls for serovar identification. To interpret the results, we referred to the expected maximum dilution titer provided by the Leptospirosis Reference Centre of KIT, as reported in Table 10.

Table 10.
Maximum dilution titer patterns of the serovars in the Pomona serogroup against the available mAbs provided by the Leptospirosis Reference Centre, Royal Tropical Institute, Amsterdam, The Netherlands.
4.6. Genotyping
4.6.1. MLST
To genotype leptospires we used the scheme proposed by Bonsilp in 2013 [22], based on the seven housekeeping genes UDP-N-acetylglucosamine pyrophosphorylase (glmU), UDP-N-acetylglucosamine pyrophosphorylase (pntA), 2-oxoglutarate dehydrogenase E1 component (sucA), triosephosphate isomerase (tpiA), 1-phosphofructokinase (pfkB), rod shape-determining protein rodA (mreA) and acyl-CoA transferase/carnitine dehydratase (caiB). For the isolates, these loci were amplified using a KAPA2G Robust HotStart PCR kit (Kapa Biosystems Resnova, Roma, Italy) in a 25-µL total volume containing 0.4 µM each primer, 2.5 µL of boiled culture and MgCl2 at the following concentrations: 1.5 mM for mreA, pfkB, pntA, caiB and glmU; 2.5 mM for sucA; and 3.5 mM for tpiA. Temperature cycling was performed as follows: 1 cycle at 95 °C for 15 min, 35 amplification cycles of 95 °C, 55 °C for 30 s and 72 °C for 1 min, followed by a final elongation at 72 °C for 10 min. To genotype the DNA extracted from biological samples, we used the protocol described by Weiss and colleagues [23], consisting of one PCR reaction, similar to that described for isolates, but performed using 5 µL of extracted DNA, followed by a second amplification reaction, that used nested primers to improve the sensitivity. The nested PCR was performed in 25-μL reactions containing 5 pmol of each primer and 2 μL of the first-round PCR product. Cycling conditions were as follows: 95 °C for 10 min, 5 cycles of 95 °C for 30 s, 46 °C for 30 s and 72 °C for 30 s. In addition, 10 cycles, in which the annealing temperature increased by 1°C per cycle, and 20 cycles with an annealing temperature of 56 °C, were performed. A final extension step at 72 °C for 7 min was performed. The presence of PCR products was verified on 2% agarose gels stained with EuroSafe Nucleic Acid Stain (Euroclone, Milan, Italy). PCR products were purified using the NucleoSpin® Gel and PCR Clean-up (Macherey-Nagel, Düren, Germany) or with exonuclease I and FastAp Thermosensitive Phospahatase alkaline (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Cycle sequencing reactions were performed using the BigDye® Terminator Cycle Sequencing kit version 1.1 (Applied Biosystems, Foster City, CA, USA). Reactions were purified using a BigDye® XTerminatorTM Purification kit Thermo Fisher Scientific, Waltham, MA, USA) and sequenced on a Genetic Analyzer 3500xl sequencer (Thermo Fisher Scientific) according to the manufacturer’s instructions. Nucleotide sequences were assembled using the SeqMan module of the Lasergene sequencing analysis software package (DNASTAR, Inc., Madison, WI, USA). Assembled sequences were trimmed and aligned to allele reference sequences downloaded from the Bacterial Isolate Genome Sequence Database (BIGSdb) [25] (https://pubmlst.org/Leptospira/), to assign allele numbers to all seven loci. For strain identification, allelic profiles were queried against the Leptospira BIGSdb.
The phylogenetic tree was built using the concatemer of the seven MLST genes linked in the followed order: glmU-pntA-sucA-tpiA-pfkB-mreA-caiB using MEGA7 [84]. The phylogeny was inferred using the Neighbor-Joining method and by calculating genetic distances using the p-distance method.
4.6.2. VNTR Analysis
Five discriminatory loci (VNTR-4, VNTR-7, VNTR-10, VNTR-Lb4 and VNTR-Lb5) were used to characterize the isolates and the extracted DNA samples as described by Salaün and colleagues in 2006 [21]. Briefly, 5 µL of boiled isolate or extracted DNA was added to 20 µL of the KAPA2G Robust HotStart PCR kit (Kapa Biosystems Resnova) reaction mixture that contained 10 pmol of each primer. The PCR was carried out as follows: 95 °C for 15 min and 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min. An additional extension for 10 min at 72 °C was added to the end of the run. The PCR products were analyzed on 2% agarose gels stained with EuroSafe Nucleic Acid Stain (Euroclone), and the molecular weights were estimated by comparison with a 100-bp DNA ladder (Thermo Fisher Scientific). If the length of PCR product was unclear, we sequenced it using the same sequencing protocol described for the MLST analysis.
4.6.3. Genomic DNA Extraction from Cultured Strains
Genomic DNA of two Leptospira isolates from pigs belonging to new Sequence Types (ID 393 and 411) were extracted from the liquid EMJH cultures using the DNeasy Blood and Tissue kit (Qiagen), according to the manufacturer’s instructions. Extracted DNA was successfully used for species confirmation and for WGS. One strain isolated from pig previously identify as L. interrogans servar Pomona, “strain IZSLER 349/2007”, and the reference strain Mozdok 5621 were included as controls for the cgMLST analysis.
4.6.4. Species Confirmation
The species assignments of the isolates 393 and 411 were confirmed by sequencing portions of their 16S rRNA genes using the Microseq 500 16S rDNA PCR kit (Applied Biosystems, Foster City, CA, USA). The obtained sequences were compared with a controlled and validated reference database (MicroseqID; Applied Biosystems) and through BLAST-based analyses to determine the species of the Leptospira strains.
4.6.5. WGS and the cgMLST Analysis
WGS was performed on the four genomic DNAs extracted from cultured strains (ID 393, 411, IZSLER 349/2007 and strain Mozdok 5621) using an Illumina NextSeq (Illumina, San Diego, CA, USA) platform with a paired-end 150× 2-bp run starting from a genomic library prepared with a Nextera DNA Flex kit (Illumina). Raw reads were filtered using Trimmomatic ver. 0.38 [85], assembled using SPAdes Assembler ver. 3.9.0 [86] and evaluated using QUAST ver. 4.2 [87]. The resulting contigs were taxonomically classified using Kaiju software [88] and only the Leptospira contigs were used for further analyses. The draft genomes of samples 393, 411, IZSLER 349/2007 and strain Mozdok 5621 were used to perform cgMLST analyses with BIGSdb [25], using the cgMLST scheme recently developed by Guglielmini et al. [24].
Ethical statement: The study was exempt of ethical approval procedures because animal samplings were performed during the routinely diagnostic procedures in naturally infected farms.
5. Conclusions
In this study, an intensive serological survey evidenced that Australis and Pomona were the most serogroups causing leptospirosis in pigs in Italy. Molecular analyses revealed that a stable genotype of strains belonging to L. interrogans serogroup Pomona had been circulating for 10 years (from 2002 until 2013) and interestingly, L. interrogans strains collected from 2014 onwards had various heterogeneous genetic profiles. Furthermore, new strains belonging to L. kirschneri species have been identified for the first time. Since an accurate identification of the infective strain is crucial for an adequate vaccine formulation, these findings provide an important contribution for addressing prevention and intervention strategies to reduce infection risks on farms.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/5/332/s1, Table S1: Trends of outbreaks observed in 2002–2017 (only single positivities were considered), Table S2: Samples submitted for genotyping analysis, Table S3: Loci showing new alleles in samples 393 and 411. Table S4: Interpretation of Real-time PCR results. Common loci having the same new alleles in samples 393 and 411 are shown in bold, File S1: Comparison of allelic profiles resulting from the cgMLST analysis.
Author Contributions
Conceptualization, M.B.B., C.B. and M.D.; methodology, C.B., A.P. and E.S.; formal analysis, C.B.; investigation, C.B., M.B.B., M.D. and S.T.; resources, M.D. and M.B.B.; data curation, C.B. and M.D.; writing—original draft preparation, C.B. and M.B.B.; writing—review and editing, M.B.B., M.D., S.T. and E.S.; project administration, M.B.B. and M.D.; funding acquisition, M.B.B. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Italian Ministry of Health (project PRC2013006 and project PRC2017016).
Acknowledgments
We thank Arturo Scalvenzi, Marcello Fin and Eufrasia Peroni Ammaturo for their skilled technical assistance with the serology and bacterial isolation, and Anna Mangeli and Daniela Loda for their expert technical support for molecular and genotyping techniques. We are very grateful to Janjira Thaipadungpanit (Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand) for assigning new allelic numbers and sequence types to our Italian strains. Thank you also to Science Docs for the English language editing of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771. [Google Scholar] [CrossRef]
- Adler, B.; de la Pena Moctezuma, A. Leptospira and Leptospirosis. Vet. Microbiol. 2010, 140, iv–v. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A. Leptospirosis. In Diseases of Swine; Blackwell: Oxford, UK, 1999; pp. 483–554. [Google Scholar]
- Strutzberg-Minder, K.; Tschentscher, A.; Beyerbach, M.; Homuth, M.; Kreienbrock, L. Passive surveillance of Leptospira infection in swine in Germany. Porc. Heal. Manag. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Burnstein, T.; Baker, J.A. Leptospirosis in swine caused by Leptospira pomona. J. Infect. Dis. 1954, 94, 53–64. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Higa, N.; Okura, N.; Matsumoto, A.; Hermawan, I.; Yamashiro, T.; Suzuki, T.; Toma, C. Characterizing interactions of Leptospira interrogans with proximal renal tubule epithelial cells. BMC Microbiol. 2018, 18, 64. [Google Scholar] [CrossRef]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef]
- Nassuato, C.; Cominardi, P.; Tagliabue, S.; Pennelli, D. Gestione di un focolaio di Leptospira interrogans variante Pomona in un allevamento suino da ingrasso 1998. Osservatorio 2006, 9, 4–9. [Google Scholar]
- O.M. 4 September 1985, Profilassi delle leptospirosi animali. Gazz. Uff. 1985, 226. Available online: https://gestione.izsler.it/izs_bs/allegati/5753/1_OM_04091985.pdf (accessed on 25 April 2020).
- D.P.R. 320/1954. Regolamento di polizia veterinaria. Gazz. Uff. 1954, 142. Available online: http://www.salute.gov.it/imgs/C_17_normativa_925_allegato.pdf (accessed on 25 April 2020).
- Leptospirosis. In OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 8th ed.; World Organisation for Animal Health: Paris, France, 2018; Chapter 3.1.12; pp. 1–3, 503–516.
- Wuthiekanun, V.; Chierakul, W.; Limmathurotsakul, D.; Smythe, L.D.; Symonds, M.L.; Dohnt, M.F.; Slack, A.T.; Limpaiboon, R.; Suputtamongkol, Y.; White, N.J.; et al. Optimization of culture of Leptospira from humans with leptospirosis. J. Clin. Microbiol. 2007, 45, 1363–1365. [Google Scholar] [CrossRef]
- Smythe, L.D.; Smith, I.L.; Smith, G.A.; Dohnt, M.F.; Symonds, M.L.; Barnett, L.J.; McKay, D.B. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect. Dis. 2002, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N.; Morey, R.E.; Galloway, R.L.; Turner, D.E.; Steigerwalt, A.G.; Mayer, L.W. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol. 2005, 54, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, R.U.M.; Chang, Y.F.; Chang, C.F.; Pan, M.J.; Yang, C.W.; Harpending, P.; McDonough, S.P.; Dubovi, E.; Divers, T.; Qu, J.; et al. Evaluation of lig-based conventional and real time PCR for the detection of pathogenic leptospires. Mol. Cell. Probes 2005, 19, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Roczek, A.; Forster, C.; Raschel, H.; Hörmansdorfer, S.; Bogner, K.H.; Hafner-Marx, A.; Lepper, H.; Dobler, G.; Büttner, M.; Sing, A. Severe course of rat bite-associated Weil’s disease in a patient diagnosed with a new Leptospira-specific real-time quantitative LUX-PCR. J. Med. Microbiol. 2008, 57, 658–663. [Google Scholar] [CrossRef]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef]
- Slack, A.T.; Symonds, M.L.; Dohnt, M.F.; Smythe, L.D. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol. 2006, 6, 95. [Google Scholar] [CrossRef]
- Noubade, R.; Krishnamurthy, G.V.; Murag, S.; Venkatesha, M.D.; Krishnappa, G. Differentiation of pathogenic and saprophytic leptospires by polymerase chain reaction. Indian J. Med. Microbiol. 2002, 20, 33. [Google Scholar]
- Kositanont, U.; Rugsasuk, S.; Leelaporn, A.; Phulsuksombati, D.; Tantitanawat, S.; Naigowit, P. Detection and differentiation between pathogenic and saprophytic Leptospira spp. by multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2007, 57, 117–122. [Google Scholar] [CrossRef]
- Salaün, L.; Mérien, F.; Gurianova, S.; Baranton, G.; Picardeau, M. Application of Multilocus Variable-Number Tandem-Repeat Analysis for Molecular Typing of the Agent of Leptospirosis. J. Clin. Microbiol. 2006, 44, 3954–3962. [Google Scholar] [CrossRef]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.G.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef]
- Weiss, S.; Menezes, A.; Woods, K.; Chanthongthip, A.; Dittrich, S.; Opoku-Boateng, A.; Simuli, M.; Chalke, V. An Extended Multilocus Sequence Typing (MLST) Scheme for Rapid Direct Typing of Leptospira from Clinical Samples. PLoS Negl. Trop. Dis. 2016, 10, e0004996. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef]
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Bertelloni, F.; Turchi, B.; Vattiata, E.; Viola, P.; Pardini, S.; Cerri, D.; Fratini, F. Serological survey on Leptospira infection in slaughtered swine in North-Central Italy. Epidemiol. Infect. 2018, 146, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Cerri, D.; Ebani, V.V.; Fratini, F.; Pinzauti, P.; Andreani, E. Epidemiology of leptospirosis: Observations on serological data obtained by a “diagnostic laboratory for leptospirosis” from 1995 to 2001. New Microbiol. 2003, 26, 383–389. [Google Scholar]
- Chiari, M.; Figarolli, B.M.; Tagliabue, S.; Alborali, G.L.; Bertoletti, M.; Papetti, A.; D’Incau, M.; Zanoni, M.; Boniotti, M.B. Seroprevalence and risk factors of leptospirosis in wild boars (Sus scrofa) in northern Italy. Hystrix 2016, 27. [Google Scholar]
- Valença, R.M.B.; Mota, R.A.; Castro, V.; Anderlini, G.A.; Júnior, J.W.P.; Brandespim, D.F.; Valença, S.R.F.A.; Guerra, M.M.P. Prevalence and Risk Factors Associated with Leptospira spp. Infection in Technified Swine Farms in the State of Alagoas, Brazil Risk Factors Associated with Leptospira spp. in Swine Farms. Transbound. Emerg. Dis. 2013, 60, 79–86. [Google Scholar] [CrossRef]
- Naito, M.; Sakoda, Y.; Kamikawa, T.; Nitta, Y.; Hirose, K.; Sakashita, M.; Kurokawa, S.; Kida, H. Serological evidence of leptospiral infection in pig populations in different districts in Japan. Microbiol. Immunol. 2007, 51, 593–599. [Google Scholar] [CrossRef]
- Wasinski, B. Occurrence of Leptospira serovars in pigsin the years 2002–2003. Med. Weter. 2005, 61, 46–49. [Google Scholar]
- Higgins, R.; Champagne, M.J. La leptospirose porcine. Le Médicin Véterinaire du Québec 1993, 23, 7–12. [Google Scholar]
- Hanson, L.E. Bratislava in swine. In Proc Am Assoc Swine Pr. 1987, 85–91. [Google Scholar]
- Miller, D.A.; Wilson, M.A.; Owen, W.J.; Beran, G.W. Porcine Leptospirosis in Iowa. J. Vet. Diagnostic Investig. 1990, 2, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Hamond, C.; Martins, G.; Loureiro, A.P.; Bremont, S.; Medeiros, M.A.; Bourhy, P.; Lilenbaum, W. First isolation and characterization of Leptospira interrogans serogroup Australis from swine in Brazil. Pesqui. Veterinária Bras. 2015, 35, 6–8. [Google Scholar] [CrossRef]
- Scanziani, E.; Giusti, A.M.; Tagliabue, S.; Luini, M.; Conti, G.; Calzolai, C.; Galli, L. Indagine sulla diffusione della leptospirosi in suini da macello. Arch. Vet. Ital. 1993, 44, 161–165. [Google Scholar]
- Tagliabue, S.; Farina, R. Inchiesta sieroepidemiologica sulla diffusione della leptospirosi ta gli animali domestici ed alcune specie selvatiche. Sel. Vet. 1995, 36, 11–12. [Google Scholar]
- Tagliabue, S.; Raffo, A.; Foni, E.; Candotti, R.; Barigazzi, G. Anticorpi per Leptospira interrogans in sieri di cinghiale selvatico (Sus scrofa) nell’Appennino parmense. In Proceedings of the Convegno Nazionale di Ecopatologia della fauna selvatica, Bologna, Italy, 9–11 February 1995. [Google Scholar]
- Ellis, W.A.; Thiermann, A.B. Isolation of Leptospira interrogans serovar bratislava from sows in Iowa. Am. J. Vet. Res. 1986, 47, 1458–1460. [Google Scholar]
- Hartman, E.G.; Brummelman, B.; Dikken, H. Leptospirae of serotype lora of the serogroup Australis isolated for the first time from swine in the Netherlands. Tijdschr. Diergeneeskd. 1975, 100, 421–425. [Google Scholar]
- Ellis, W.A.; McParland, P.J.; Bryson, D.G.; Cassells, J.A. Prevalence of Leptospira infection in aborted pigs in Northern Ireland. Vet. Rec. 1986, 118, 63–65. [Google Scholar] [CrossRef]
- Ellis, W.A. Leptospira australis infection in pigs. Pig Vet. J. 1989, 22, 83–92. [Google Scholar]
- Schönberg, A.; Hahn-Hey, B.; Kämpe, U.; Schmidt, K.; Ellis, W.A. The isolation and identification of Leptospira interrogans serovar bratislava from a pig in Germany. Zentralbl. Veterinarmed. B 1992, 39, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Agunloye, C.A. Agglutinating antibodies to leptospires in slaughters pigs in Ibadan, Nigeria. Trop. Vet. 2001, 19, 188–190. [Google Scholar]
- Kikuchi, N.; Shikano, M.; Hatanaka, M.; Takahashi, T.; Mori, K.; Fujii, T.; Furuya, T. Prevalence of leptospiral antibody in sows in Japan. J. Vet. Epidemiol. 2013, 13, 95–99. [Google Scholar] [CrossRef]
- Choi, C.; Park, Y.C.; Park, M.A.; Yoo, C.K.; Park, M.Y.; Chae, C. Seroprevalence of Leptospira serovars in Korean sows. Vet. Rec. 2001, 148, 416. [Google Scholar] [CrossRef]
- Ellis, W.A.; Montgomery, J.M.; Thiermann, A.B. Restriction endonuclease analysis as a taxonomic tool in the study of pig isolates belonging to the Australis serogroup of Leptospira interrogans. J. Clin. Microbiol. 1991, 29, 957–961. [Google Scholar] [CrossRef]
- Bolin, C.A.; Cassels, J.A. Isolation of Leptospira interrogans server bratislava and hard from swine at slaughter. J. Vet. Diagnostic Investig. 1992, 4, 87–89. [Google Scholar] [CrossRef]
- Bolin, C.A.; Cassels, J.A. Isolation of Leptospira interrogans server bratislava from still born and weak pigs in Iowa. J. Am. Vet. Med. Assoc. 1990, 196, 1601–1604. [Google Scholar]
- Boqvist, S.; Montgomery, J.M.; Hurst, M.; Thu, H.T.V.; Engvall, E.O.; Gunnarsson, A.; Magnusson, U. Leptospira in slaughtered fattening pigs in southern Vietnam: Presence of the bacteria in the kidneys and association with morphological findings. Vet. Microbiol. 2003, 93, 361–368. [Google Scholar] [CrossRef]
- Ellis, W.A.; McParland, P.J.; Bryson, D.G.; Cassel, J.A. Boars as carriers of leptospires of the Australis serogroup on farms with an abortion problem. Vet. Rec. 1989, 118, 563. [Google Scholar] [CrossRef]
- Poonacha, K.B.; Smith, B.J.; Donahue, J.M.; Tramontin, R.R.; Tuttle, P.A.; Hong, C.B.; Giles, R.C. Leptospiral abortion in horses in central Kentucky. In Proceedings of the Annual Convention of the American Association of Equine Practitioners, Lexington, KY, USA, 2–5 December 1990; pp. 397–402. [Google Scholar]
- Miraglia, F.; Moreno, A.M.; Gomes, C.R.; Paixão, R.; Liuson, E.; Morais, Z.M.; Maiorka, P.; Seixas, F.K.; Dellagostin, O.A.; Vasconcellos, S.A. Isolation and characterization of Leptospira interrogans from pigs slaughtered in São Paulo State, Brazil. Braz. J. Microbiol. 2008, 39, 501–507. [Google Scholar] [CrossRef]
- Christmas, B.W.; Bragger, J.M.; Till, D.G. Dairy farm fever in New Zealand: Isolation of L pomona and L hardjo from a local outbreak. New Zealand Med. J. 1974, 79, 904–906. [Google Scholar] [PubMed]
- Firinu, A.; Ponti, M.N.; Patta, C.; Oggiano, A.; Ruiu, A.; Cabras, P.; Maestrale, C.; Cossu, P.; Pintore, A. Serologic survey on some transmissible diseases among wild boars and free ranging pigs in Sardinia. In Tradition and Innovation in Mediterranean Pig Production, Proceedings of the International Symposium on Mediterranean Pig, Evora, Portugal, 26–28 November 1998; Almeida, J.A., Tirapicos, N.J., Eds.; CIHEAM: Zaragoza, Spain, 2000; pp. 309–312. [Google Scholar]
- Żmudzki, J.; Jabłoński, A.; Nowak, A.; Zębek, S.; Arent, Z.; Bocian, Ł.; Pejsak, Z. First overall report of Leptospira infections in wild boars in Poland. Acta Vet. Scand. 2016, 58, 3. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A. Animal leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 99. [Google Scholar] [PubMed]
- Schollum, L.M.; Blackmore, D.K. Leptospirosis of pig farmers: The results of a serological survey. New Zealand Med. J. 1982, 95, 299–301. [Google Scholar] [PubMed]
- Tagliabue, S. Nuove acquisizioni nella diagnosi di leptospirosi. In Proceedings of the XLIV Convegno Nazionale della Società Italiana delle Scienze Veterinarie, Stresa, Italy, 27–29 September 1990. [Google Scholar]
- Tagliabue, S.; Figarolli, B.M.; D’Incau, M.; Foschi, G.; Gennero, M.S.; Giordani, R.; Natale, A.; Papa, P.; Ponti, N.; Scaltrito, D.; et al. Serological surveillance of Leptospirosis in Italy: Two-year national data (2010–2011). Vet. Ital. 2016, 52, 129–138. [Google Scholar]
- Delbem, Á.C.B.; de Freitas, J.C.; Bracarense, A.P.F.R.L.; Müller, E.E.; de Oliveira, R.C. Leptospirosis in slaughtered sows: Serological and histopathological investigation. Braz. J. Microbiol. 2002, 33, 174–177. [Google Scholar] [CrossRef]
- Scanziani, E.; Crippa, L.; Giusti, A.M.; Luini, M.; Pacciarini, M.L.; Tagliabue, S.; Cavalletti, E. Leptospira interrogans serovar sejroe infection in a group of laboratory dogs. Lab. Anim. 1995, 29, 300–306. [Google Scholar] [CrossRef]
- Paz-Soldán, S.V.; Dianderas, M.T.; Windsor, R.S. Leptospira interrogans serovar canicola: A causal agent of sow abortions in Arequipa, Peru. Trop. Anim. Health Prod. 1991, 23, 233–240. [Google Scholar] [CrossRef]
- Rühl-Fehlert, C.I.; Brem, S.; Feller, W.; Kopp, H.; Meyer, P.; Rinke, M. Clinical, microbiological and pathological observations in laboratory beagle dogs infected with leptospires of the serogroup sejroe. Exp. Toxicol. Pathol. 2000, 52, 201–207. [Google Scholar] [CrossRef]
- Collares-Pereira, M.; Korver, H.; Thi, B.V.C.; Santos-Reis, M.; Bellenger, E.; Baranton, G.; Terpstra, W.J. Analysis of Leptospira isolates from mainland Portugal and the Azores islands. FEMS Microbiol. Lett. 2000, 185, 181–187. [Google Scholar] [CrossRef]
- das Neves Paiva-Cardoso, M.; Arent, Z.; Gilmore, C.; Hartskeerl, R.; Ellis, W.A. Altodouro, a new Leptospira serovar of the Pomona serogroup isolated from rodents in northern Portugal. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2013, 13, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Rocha, T.; Ellis, W.A.; Montgomery, J.; Gilmore, C.; Regalla, J.; Brem, S. Microbiological and serological study of leptospirosis in horses at slaughter: First isolations. Res. Vet. Sci. 2004, 76, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Rocha, T. Isolation of Leptospira interrogans serovar mozdok from aborted swine fetuses in Portugal. Vet. Rec. 1990, 126, 602. [Google Scholar] [PubMed]
- Majetic, Z.S.; Galloway, R.; Sabljic, E.R.; Milas, Z.; Perko, V.M.; Habus, J.; Margaletic, J.; Pernar, R.; Turk, N. Epizootiological survey of small mammals as Leptospira spp. reservoirs in Eastern Croatia. Acta Trop. 2014, 131, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A. Control of canine leptospirosis in Europe: Time for a change? Vet. Rec. 2010, 167, 602–605. [Google Scholar] [CrossRef]
- Renaud, C.; Andrews, S.; Djelouadji, Z.; Lecheval, S.; Corrao-Revol, N.; Buff, S.; Demont, P.; Kodjo, A. Prevalence of the Leptospira serovars bratislava, grippotyphosa, mozdok and pomona in French dogs. Vet. J. 2013, 196, 126–127. [Google Scholar] [CrossRef]
- Pritchard, D.G.; Todd, N.; Barlow, A.; Little, S.A. Outbreak of Leptospira interrogans serovar mozdok in sows in Dorset, England. Isr. J. Vet. Med. 1987, 43, 343. [Google Scholar]
- Barlow, A.M. Reproductive failure in sows associated with Leptospira Mozdok from a wildlife source. Pig J. 2004, 54, 123–131. [Google Scholar]
- Hathaway, S.C.; Todd, J.N.; Headlam, S.A.; Jeffrey, M. Possible role of leptospires of the Pomona serogroup in sporadic bovine abortion in the south west of England. Vet. Rec. 1984, 115, 623–626. [Google Scholar] [CrossRef]
- Da Cunha, C.E.P.; Felix, S.R.; Neto, A.C.P.S.; Campello-Felix, A.; Kremer, F.S.; Monte, L.G.; Amaral, M.G.; Nobre, M.D.O.; Da Silva, É.F.; Hartleben, C.P.; et al. Infection with Leptospira kirschneri Serovar Mozdok: First Report from the Southern Hemisphere. Am. J. Trop. Med. Hyg. 2016, 94, 519–521. [Google Scholar] [CrossRef]
- Jacobs, A.; Harks, F.; Hoeijmakers, M.; Segers, R. A novel octavalent combined Erysipelas, Parvo and Leptospira vaccine provides (cross) protection against infection following challenge of pigs with 9 different Leptospira interrogans serovars. Porc. Heal. Manag. 2015, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.A.C.; Harks, F.; Hoeijmakers, M.; Collell, M.; Segers, R.P.A.M. Safety and efficacy of a new octavalent combined Erysipelas, Parvo and Leptospira vaccine in gilts against Leptospira interrogans serovar Pomona associated disease and foetal death. Vaccine 2015, 33, 3963–3969. [Google Scholar] [CrossRef] [PubMed]
- Ellinghausen, H.C.; Mccullough, W.G. Nutrition of Leptospira Pomona and growth of 13 other serotypes: A serum-free medium employing oleic albumin complex. Am. J. Vet. Res. 1965, 26, 39–44. [Google Scholar] [PubMed]
- Faine, S.; Adler, B.; Bolin, C.; Perolat, B. Leptospira and Leptospirosis; MediSci.: Melbourne Vic, Australia, 1999. [Google Scholar]
- Faine, S. Guidelines for the control of leptospirosis. In WHO Offset Publ No. 67; WHO: Geneva, Switzerland, 1982; Available online: https://apps.who.int/iris/handle/10665/37219 (accessed on 25 April 2020).
- Savio, M.L.; Pacciarini, M.L.; Cinco, M.; Tagliabue, S. Identification of Leptospira interrogans strains by monoclonal antibodies and genomic analysis. New Microbiol. 1993, 16, 315–321. [Google Scholar] [PubMed]
- Terpstra, W.J.; Korver, H.; Schoone, G.J.; von Leeuwen, J.; Schönemann, C.E.; de Jonge-Aglibut, S.; Kolk, A.H. Comparative classification of Leptospira serovars of the Pomona group by monoclonal antibodies and restriction-endonuclease analysis. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 1987, 266, 412–421. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamara, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).