Abstract
Pathogenomic analysis was performed on a novel carbapenem-resistant Citrobacter freundii isolate (H2730R) from a rectal swab of an adult male patient admitted to a tertiary hospital, Durban, South Africa. H2730R was identified using selective media and API 20e kit. Confirmatory identification and antibiotic susceptibility testing were performed using the VITEK II. H2730R was whole-genome sequenced on the Illumina MiSeq platform. H2730R was resistant to all tested antibiotics except tigecycline and was defined as ST498 by the C. freundii multilocus sequence typing (MLST) database. The estimated pathogenic potential predicted a higher probability (Pscore ≈ 0.875), supporting H2730R as a human pathogen. H2730R harbored 25 putative acquired resistance genes, 4 plasmid replicons, 4 intact prophages, a class 1 integron (IntI1), 2 predominant insertion sequences (IS3 and IS5), numerous efflux genes, and virulome. BLASTn analysis of the blaNDM-1 encoding contig (00022) and its flanking sequences revealed the blaNDM-1 was located on a plasmid similar to the multireplicon p18-43_01 plasmid reported for the spread of carbapenem resistance in South Africa. Phylogenomic analysis showed clustering of H2730R with CF003/CF004 strains in the same clade, suggesting a possible association between C. freundii strains/clones. Acquiring the p18-43_01 plasmid containing blaNDM-1, the diversity, and complex resistome, virulome, and mobilome of this pathogen makes its incidence very worrying regarding mobilized resistance. This study presents the background genomic information for future surveillance and tracking of the spread of carbapenem-resistant Enterobacteriaceae in South Africa.
Keywords:
Pathogenomics; Novel; Citrobacter freundii; blaNDM-1; Mobilome; South Africa; Carbapenemase 1. Introduction
Citrobacter freundii (C. freundii) is a rod-shaped, Gram-negative, aerobic member of the Enterobacterales family that is an intestinal commensal in humans and animals [1,2]. It is a known cause of nosocomial infections associated with the biliary tract, respiratory tract, urinary system, and central nervous system, including meningitis, neonatal sepsis, and infectious diarrhea [3,4]. C. freundii can persist in their hosts for a long time and acquire resistance mechanisms, making the treatment of their infections challenging [4]. The emergence and dissemination of drug-resistant C. freundii in humans, animals, and the environment have previously been described, making this pathogen a potential reservoir for the spread of antimicrobial-resistant genes [1,2,4,5,6,7].
Carbapenem resistance is a growing concern in C. freundii, as carbapenems are regarded as antimicrobial agents of last resort used to treat life-threatening infections caused by these pathogens [8]. The easy mobilization of carbapenemase encoding genes, due to their locations on mobile genetic elements (MGEs), has favored the acquisition of carbapenem resistance by diverse Enterobacterales [9]. These MGEs, specifically plasmids, prophages, insertion sequences (IS), and integrons, are known to influence microbial virulence and pathogenicity via horizontal gene transfer (HGT) [10,11]. HGT allows new genomic traits to be acquired from other unrelated bacteria and remains the most effective means of bacterial evolution, causing the dissemination of antibiotic resistance and associated disease pathogenesis [12,13]. In this study, we present the emergence of a novel carbapenem-resistant sequence type (ST498) of C. freundii isolated from a rectal swab of an adult male patient admitted to a tertiary healthcare facility in Durban, South Africa, harboring a diverse resistome, virulome, and mobilome. This investigation forms part of a broader study specifically designed to isolate carbapenemase-producing Enterobacteriaceae (CPE) for patients at admission in a public hospital in the province. We also show the phylogenomic relationship between this novel isolate (H2730R) and all the deposited C. freundii genomes from South Africa at the Pathosystems Resource Integration Center (PATRIC) online platform.
2. Results
2.1. Isolation, Phenotypic Confirmation of Carbapenem Resistance, and Antibiotic Susceptibility Testing (AST)
The carbapenem-resistant C. freundii strain (H2730R) was isolated using the selective media and confirmed using the VITEK II AST-N255 automated platform. Antibiotic susceptibility testing (AST) revealed that H2730R was extensively drug-resistant (XDR) (Table 1).

Table 1.
Susceptibility Pattern of Isolate H2730R.
2.2. General Genomic Features of H2730R
The CG Viewer Server was used for the genomic visualization of the isolate (Figure A1; Appendix A). The C. freundii (H2730R) genomic features are shown in Table A1. The size, GC content, number of contigs, N50, and L50 of the H2730R genome were 5.29 Mbp, 51.80%, 58, 518368, and 4, respectively. Annotation with RAST and PGAP resulted in 5006 protein-coding genes, 70 RNAs, and 12 tRNAs (Table A1). The protein-coding genes (CDSs) and non-protein-coding genes were assigned to 27 clusters of orthologous groups (COG) and functional categories (Table A1).
2.3. WGS-Based Confirmation and Multilocus Sequence Typing (MLST)
The identification of H2730R isolate was confirmed with generated genomic data via the Global Platform for Genomic Surveillance (Pathogenwatch). In silico determination of the classical sequence type from WGS data using the C. freundii MLST scheme resulted in a previously undescribed MLST comprising a new allelic combination for arca_5, aspc_16, clpx_14, dnag_54, fadd_103, lysp_5, mdh_15. Allele sequences were submitted for curation and assigned to the new ST 498 by the C. freundii PubMLST database. BURST (Based Upon Related Sequence Types) analysis identified the novel ST498 as a satellite clone (more distantly related strain) with no single-locus variant (SLV) or double-locus variant (DLV) of globally curated C. freundii STs. Of note, the new ST had different allelic profiles from the five deposited C. freundii genomes from South Africa.
2.4. WGS Analysis of Resistance Genes and Genetic Support
WGS analysis via the different databases revealed 25 putative acquired antibiotic resistance genes responsible for resistance to the various antibiotics found in the isolate, which corroborated the phenotypic profile (Table 2). Chromosomal mutation in GyrA [S83I] and the plasmid-mediated quinolone resistance genes (aac (6')-Ib-cr and qnrB1) were found. Most of the resistance genes were plasmid-borne (Table 2). Resistance to carbapenems was linked to the New Delhi metallo-β-lactamase-1 (blaNDM-1) belonging to the sub-class B1. The BLASTn analysis of the blaNDM-1 carbapenemase containing contig (00022) and its flanking sequences matched the 212.3 Kbp multireplicon plasmid p18-43_01 (GenBank accession number CP023554.1). The PlasmidFinder v1.3 identified four plasmid replicons (Inc A/C2, Inc FIB(pB171), Inc FII(Yp), and Inc Q1).

Table 2.
Resistance gene profile and efflux pump systems of Isolate H2730R.
The PHAge Search Tool (PHAST) detected four intact phages (Escher_HK639, Entero_c_1, Salmon_RE_2010, and Salmon_SJ46). The insertion sequences in the genomes belonging to the IS3 family (IS2) and IS5 family (IS903) were predicted by BLAST searches against contigs on the ISFinder database. In silico analysis identified the class 1 integron-integrase gene (IntI1) in H2730R. The relationship between the IntI1 integrase in H2730R and other closely related IntI1 integrases in diverse species obtained from the RAST Server was shown as a phylogenetic tree using the MAFFT online tool (Figure 1). The Intl1 of the H2730R strain had the closest similarity with the Salmonella enterica subsp. enterica serovar Choleraesuis plasmid pOu7519, as depicted in Figure 1.
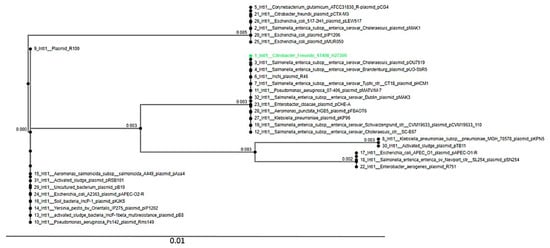
Figure 1.
A phylogenetic tree showing the association between Integron_Integrase_IntI1 of C. freundii H2730R and other predicted closely related IntI1 integrases in diverse species.
2.5. Pathogenicity, Defence Systems Mechanisms, and Virulome Predictions
H2730R had a 0.875 probability of being pathogenic to humans and was found to match with 110 pathogenic families. All the 110 pathogenic families were linked to members of the Enterobacterales, of which Citrobacter koseri ATCC BAA-895 was the organism with the highest possible pathogen linkage. The CRISPRCasFinder predicted two clustered, regularly interspaced, short palindromic repeat (CRISPR) arrays with no Cas systems. Of note, the in silico analysis revealed a Type II restriction-modification (R-M) defense system consisting of Eco128I (restriction enzyme) and M. EcoRII (methyltransferase)]. The whole-genome virulome analysis predicted virulence-encoded pathogenesis-associated proteins predominantly made up of adherence determinants, regulation, toxin, motility, antiphagoctyosis, invasion, and biofilm formation (Table 3).

Table 3.
Genomic characterization of putative virulence factors in the isolate H2730R*.
2.6. Phylogenomic Analysis and Metadata Insights of Reported Citrobacter Freundii Isolates from South Africa
The phylogenetic relationship and epidemiological distribution of all deposited C. freundii genomes on GenBank from South Africa are depicted in Figure 2 in which isolates of the same cluster are highlighted in the same color. The tree analysis, coupled with metadata, revealed two clusters (A and B) of C. freundii with insights into the diversity of clones in South Africa (Table A2). Of note, the H2730R was related to the CF003 and CF004 strains (Cluster B). All the isolates were from human hosts.
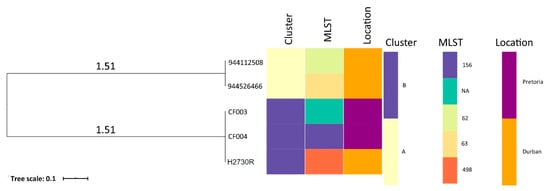
Figure 2.
The whole-genome phylogenomic branch and metadata [MLST and Demographics (Location and Host)] coupled using Phandango showing the relationship between H2730R and all deposited Citrobacter freundii strains (n = 4) from South Africa. All the isolates were from a human host.
3. Discussion
Carbapenem resistance in Enterobacteriaceae has become a major public health threat that requires urgent attention. In this study, we use whole-genome sequencing (WGS) and bioinformatics analysis to describe the emergence of a novel carbapenem-resistant sequence type (ST498, satellite clone) of C. freundii isolated from the rectal swab of an adult male patient admitted to a tertiary healthcare facility in Durban, South Africa, harboring a diverse resistome, virulome, and mobilome.
H2730R was categorized as XDR due to its phenotypic resistance to all tested antibiotics except tigecycline [14] (Table 1). The carbapenem phenotypes of the strain, as identified on selective chromogenic screening medium, were confirmed by the presence of carbapenemase (blaNDM-1) (Table 1 and Table 2). The 100% concordance between phenotypes and predicted resistome annotation data reiterated the reliability of WGS for the accurate prediction of antibiotic resistance [15,16]. The C. freundii H2730R genome harbored a total of 25 putative resistance genes, which were predominantly plasmid-borne (Table 2), indicating an acquisition and the potential for spread via horizontal resistome transfer between H2730R and other species. Resistance to ciprofloxacin was mediated by GyrA [S83I] chromosomal mutation, plasmid-mediated quinolone resistance genes (PMQR) (aac(6')-Ib-cr and qnrB1), and efflux genes (Table 2) [17]. Sekyere and Amoako et al. (2017) [15] reported that high-level fluroquinolone resistance was mediated by efflux, PMQR genes, and gyrA mutations in Enterobacterales, including Citrobacter freundii, which supported our results. More so, efflux genes belonging to three main efflux pump systems (Table 2) reported to play vital roles in antibiotic resistance in Enterobacterales were also identified [18,19]. However, the mere presence of these efflux genes does not directly implicate resistance, and hence further studies on their expression levels will be needed to ascertain the possibility of efflux pump hyperexpression [18]. The diverse resistance mechanisms (plasmid-borne, chromosomal, and efflux) involved in H2730R resistance indicate the various processes the organism expresses to survive the effects of antibiotics and other biocides (Table 2).
The National Center for Biotechnology Information (NCBI) microbial nucleotide BLAST search of the blaNDM-1 carbapenemase containing contig (00022) and its flanking sequences in the isolate revealed the blaNDM-1 was located on a plasmid similar to the 212.3 Kbp multireplicon p18-43_01 plasmid (accession no. CP023554.1) reported for the spread of carbapenemases in Enterobacterales (including K. pneumoniae [n = 4], Citrobacter freundii [n = 1], Serratia marcescens [n = 11], and Enterobacter family complex [n = 7]) in the private [20] and public healthcare sector [21] in Durban, South Africa. This implicates a possible blaNDM-1 acquisition as well as nosocomial spread and development of an XDR genetic lineage via this local plasmid across sectors in the KZN province, which is very worrisome. The four plasmid replicon genes (Inc A/C2, Inc FIB(pB171), Inc FII(Yp), and Inc Q1) identified via the PlasmidFinder supported the multireplicon plasmid nature of the p18-43_01. This global spread of carbapenem resistance through the acquisition of plasmid encoding carbapenemase has been highly documented in the literature [22,23,24].
Moreover, H2730R contained two insertion sequences (IS3 family and IS5 family) whose genetic transposition could help in the plasticity and concomitant adaptability of H2730R phenotypic traits, including catabolism, pathogenicity, resistance to antibacterial agents, and virulence [25]. The IS2 (IS3 family) has been reported to cause erythromycin and fluoroquinolone resistance through the formation of hybrid promoters [25]. Additionally, the IS930 (IS5 family) has been implicated in carbapenem and metal ion (silver) resistance in Enterobacter aerogenes [26] and E. coli, respectively. Further analysis of H2730R revealed prophage with possible origination from diverse sources (E. coli and Salmonella) [27]. For instance, the two prophages Salmon_RE_2010 and Salmon_SJ46 were initially identified in Salmonella enterica subsp. enterica serovar Enteritidis and Salmonella enterica subsp. enterica serovar Indiana, respectively [28]. This supports a possible exchange of genetic traits via horizontal transfer between Enterobacterales. Class 1 integrons, reported to be associated with multiple classes of antibiotic resistome, including β-lactams, quinolones, and aminoglycosides worldwide, were found [29]. However, the full mobile gene cassettes and integron genetic structure could not be determined as they were truncated into different contigs during the assembly process.
The pathogenic potential (Pscore) is a machine learning algorithm used for the in silico prediction of the possibility of a strain being a human pathogen with the probability ranging from 0 to 1. This estimation of the pathogenic potential using trained algorithms to differentiate between pathogenic or commensal strains predicted a higher probability (Pscore ≈0.875) suggesting H2730R as a potential human pathogen. Further, in silico analysis of H2730R predicted the presence of two CRISPR arrays and a Type II restriction–modification (R-M) defense system which offers bacterial protection against viral attacks and thus increases its survival and adaptability in the microbial landscape [30,31].
Moreover, the genomic virulome analysis revealed a battery of different virulence encoded proteins from diverse sources. These virulence determinants were predominantly made up of adherence determinants which aid in the complement-independent attachment to host mammalian cells [32]. H2730R possessed toxins to induce cell membrane damage, trigger cytokine formation, and reduce or kill neutrophils [33]. Interestingly, the strain also harbored the ibeB and pgaC genes, which contribute to invasion, biofilm formation, and adhesion to eukaryotic host cells. Additionally, H2730R harbored the two-component regulation system (PhoPQ) activated by low divalent cations magnesium (Mg2+) and calcium (Ca2+) levels and required for intracellular survival, cationic antimicrobial peptides (CAMPs) resistance, and stimulation of cytokine secretion [34]. The higher virulence factor homology existing between H2730R and other Gram-negative species (Table 3) suggests that they were possibly obtained horizontally, necessitating further studies on transconjugation and/or transcriptomics to ascertain this assertion. The detection of the virulome can inform vaccine development by delineating possible bacterial protein targets.
Phylogenomic analysis, coupled with metadata, showed clustering of H2730R (Durban) with CF003/CF004 strains in the same cluster (B) suggesting a possible association of C. freundii strains/clones. However, not much inference could be drawn from the tree analysis due to the small number of deposited C. freundii genomes from South Africa. It is thus recommended that more studies be carried out in all the sectors (human, animal, and environment) to monitor the emergence and spread of XDR C. freundii using high throughput technologies such as whole-genome sequencing (WGS). Moreover, there was limited data on the clinical information, history and/or clinical examination of the patient, and all deposited C. freundii genomes obtained from the human host, which could be very useful in comparative phylogenomic analysis. Ultimately, a combination of demographics, clinical information, and WGS data coupled with graphical analysis should be applied to offer insights into the spread of pathogens and increase confidence during molecular epidemiological investigations [35].
4. Materials and Methods
4.1. Ethical Approval
This study was approved by the Biomedical Research Ethics Committee (approval no: BE599/16, a substudy of BCA444/16), College of Health Sciences, University of KwaZulu-Natal (UKZN).
4.2. Identification of the Isolate
Selective chromogenic screening medium, CHROMID® CARBA (BioMérieux, Marcy l'Étoile, France), was used for the isolation of carbapenemase-producing Enterobacteriaceae from a screening rectal swab obtained from an adult patient. The CHROMID® CARBA (BioMérieux, Marcy l'Étoile, France) agar plate was inoculated with the following control strains: carbapenemase-negative K. pneumoniae ATCC 700603 and carbapenemase-positive K. pneumoniae ATCC BAA-1705.
4.3. Antibiotic Susceptibility Testing (AST)
Confirmatory phenotypic microbial identification and antibiotic susceptibility testing (AST) was performed on the VITEK II AST-N255 automated platform (BioMérieux, Marcy l'Étoile, France). The universal antimicrobial test panel included: penicillin, ampicillin, amoxicillin-clavulanate, ceftriaxone, cefepime, cefuroxime, cefoxitin, ceftazidime, imipenem, meropenem, ertapenem, piperacillin-tazobactam, amikacin, gentamicin, nitrofurantoin, trimethoprim/sulfamethoxazole, ciprofloxacin, and tigecycline. The isolate was characterized as susceptible or resistant using CLSI-approved breakpoints [36].
4.4. DNA Purification, Genome Sequencing, and Preprocessing of Sequence Data
The isolate was streaked onto a nutrient agar (NA) (Sigma-Aldrich, St. Louis, USA) plate and incubated at 37 °C for 24 h. Following incubation, genomic DNA was extracted from a visibly pure culture using the Quick-DNA™ Fungal/Bacterial Miniprep Kit (Zymogen Research, USA, cat. no. D6005), quantified by Nanodrop 8000 (Thermo Scientific, Waltham, MA) and Qubit, and verified by agarose gel electrophoresis. A paired-end library (2 × 300 bp) was prepared using Illumina Nextera XT DNA Sample Preparation Kit and sequenced on a MiSeq machine (Illumina, USA). The generated sequenced reads were quality assessed and trimmed using the Skesa assembler (version 2.3). Default parameters were used for all software unless otherwise specified. The CheckM tool [37] was used to verify that the sequence reads were not from mixed species using lineage-specific marker sets from other genetically well-characterized, closely related C. freundii strains.
4.5. Bioinformatic Analysis
4.5.1. Genome Visualization and Annotation
The raw reads were de novo assembled using the Skesa (version 2.3) assembler [38]. The genomes of the strains were visualized using the CG Viewer Server (Figure A1; Appendix A). The National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP; version 4.3) [39] and Rapid Annotation using Subsystem Technology (RAST) Server (version 2.0) [40] were used for annotation of the size, GC content, number of contigs, N50, L50, average coverage, number of RNAs, and protein-coding sequences of the isolate.
4.5.2. WGS-Based Confirmation and Molecular Typing
The generated contigs from the WGS data were used to confirm the identity of the isolate on the pathogen watch platform [41]. The assembled genome was submitted to the C. freundii MLST database, which predicted the allelic profiles of the seven housekeeping genes to assign the new sequence type [42]. An eBURST [43] algorithmic analysis was performed in the MLST database (https://pubmlst.org/cfreundii/) to ascertain whether the novel sequence type (ST) was a single-locus variant (SLV) or double-locus variant (DLV) or satellite (SAT, more distantly related strain) of known STs. The allelic profiles and STs of all deposited C. freundii genomes from South Africa in the PATRIC database were also predicted using the MLST database.
4.5.3. Genomic Identification of the Antibiotic Resistome and Mobile Genetic Elements (MGEs)
The bacterial analysis pipeline of GoSeqIt via ResFinder [44], Antibiotic Resistance Gene-Annotation database (ARG-ANNOT) [45], and the comprehensive antibiotic resistance database (CARD) [46] tools were also used to annotate and identify antibiotic resistance genes. To determine if the resistance genes and their associated MGEs were plasmid-borne or chromosomal, the contigs bearing these resistance genes were BLASTed on BLASTn to determine if the closest nucleotide homologies were chromosomal or plasmids. Plasmid replicons were predicted through PlasmidFinder [47]. The PHAge Search Tool [48] server was used for the identification, annotation, and visualization of prophage sequences. Phage regions were extracted and analyzed in the NCBI nucleotide BLASTn service to confirm their identity. Insertion sequences and transposase in the genomes were predicted by blasting contigs on the ISFinder database [49] to find the common insertion sequences. Integrons in the genomes predicted by PGAP and RAST subsystems were blasted on the INTEGRALL database to find the actual integrons. Additionally, using the nucleotide sequences, the MAFFT multiple alignment program [50] was used to show the phylogenetic relationship between predicted and other closely related integrases in diverse species obtained from the RAST Server.
4.5.4. Pathogenicity, Defence Systems, and Virulome Predictions
PathogenFinder [51] was used to predict pathogenicity towards human hosts. The CRISPRCasFinder was used to identify the putative CRISPR loci and Cas cluster in the draft genomes [52]. Restriction Modification Finder predicted the R-M system in the isolate [53]. Virulence determinants (sequences and functions) corresponding to different major bacterial virulence factors associated with C. freundii were searched for virulence factors with the pathogenic bacteria database, VFanalyzer [54], and validated by blasting assembled genomes to a pseudomolecule generated by concatenating a set of target genes using the NCBI in-house BLASTN tool.
4.6. Phylogenomic Analyses of C. freundii Isolates from South Africa
The genome sequences of all five available genomes were downloaded from GenBank and PATRIC for genomic comparison and phylogenomic analysis via CSI Phylogeny-1.4 [55], an online service which identifies SNPs from WGS data, filters and validates the SNP positions, and then infers phylogeny based on concatenated SNP profiles [55]. A bootstrapped indicator with 100 replicates was applied to identify recombined regions and provide the phylogenetic accuracy in groups with little homoplasy. Figtree was used to edit and visualize the phylogenetic tree. The phylogeny was visualized alongside annotations for isolate demographics and WGS in silico molecular typing metadata using Phandango [56].
4.7. Data Availability
This whole-genome shotgun project has been deposited in DDBJ/ENA/GenBank under the accession no. VWTQ00000000. The version described in this paper is version VWTQ01000000.
5. Conclusions
This study reports the comprehensive pathogenomics of an XDR C. freundii strain isolated from South Africa. The multireplicon plasmid p18-43_01 encoding the blaNDM-1 reported for carbapenem resistance coupled with the diversity of the resistome, virulome, and mobilome of this novel pathogen makes this incidence very worrying. This necessitates continuous resistance surveillance programs, including rectal screening, stringent infection control measures, and antibiotic stewardship policies to curb further emergence and spread of these antibiotic-resistant bacteria.
Author Contributions
Conceptualization, Y.R., K.P.M., D.G.A., A.L.K.A., M.A., A.I., R.S., and S.Y.E; methodology, Y.R., K.P.M., D.G.A., A.A.L.K., and S.Y.E.; validation, D.G.A., M.A., and A.I.; formal analysis, Y.R., D.G.A., M.A.; investigation, Y.R.; resources, K.P.M., R.S., and S.Y.E; data curation, D.G.A., M.A., A.L.K.A., and A.I.; writing—original draft preparation, Y.R.; writing—review and editing, Y.R., K.P.M., M.A., D.G.A., A.L.K.A., A.I., R.S., and S.Y.; supervision, K.P.M. and S.Y.E.; project administration, Y.R., K.P.M., and S.Y.E.; funding acquisition, Y.R., K.P.M., and S.Y.E. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Antimicrobial Research Unit and College of Health Science of the University of KwaZulu-Natal (Durban, South Africa) and the Institut Merieux. The National Research Foundation (NRF) provided funding for this study via the NRF Incentive Funding for Rated Researchers [Grant no. 85595], the NRF Competitive Grant for Rated Researchers [Grant no. 106063], the Department of Science and Technology (DST-NRF South African Research Chair in Antibiotic Resistance and One Health [Grant no. 98342], and the South African Medical Research Council Self-Initiated Research Grant. The authors also thank the University of KwaZulu-Natal for funding support through the University Capacity Development Programme (UCDP). The APC was funded by the L'Oréal-UNESCO For Women in Science Awards.
Acknowledgments
The authors acknowledge Piotr Majewski and the team of curators at the Citrobacter freundii MLST database on PubMLST for curating the genomic data and making them publicly available.
Conflicts of Interest
Sabiha Y. Essack is the chairperson of the Global Respiratory Infection Partnership sponsored by an unconditional education grant from Reckitt and Benckiser. All other authors declare no competing interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Appendix A
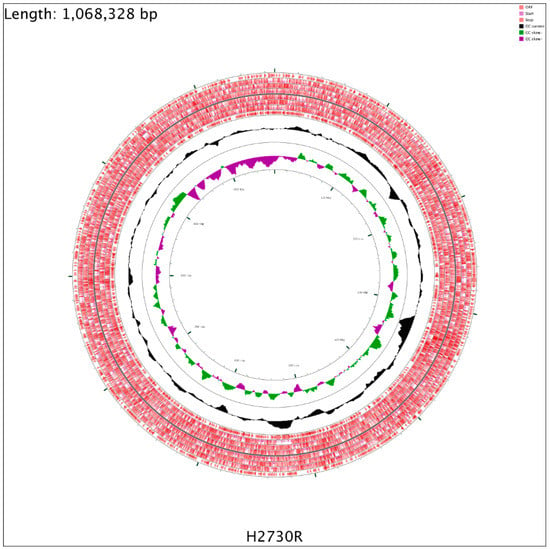
Figure A1.
Graphic depiction of the H2730R genome belonging to ST498. The two outer circles show the open reading frame (ORF–in mauve). The inner circle shows the GC skew, with green and purple indicating positive and negative values, respectively. The GC content is indicated in black. This genome map was visualized using the CGView Server.
Appendix B

Table A1.
Genomic features of Isolate H2730R.
Table A1.
Genomic features of Isolate H2730R.
| Attribute | Value |
|---|---|
| Sequencing platform | Illumina MiSeq machine |
| Assembler | Skesa (version 2.3) |
| Assembly accession | VWTQ00000000 |
| No. of Contigs | 58 |
| Genome size (bp) | 5,299,408 |
| DNA G + C (%) | 51.8 |
| Genome coverage (X) | 99.0 |
| Number of RNAs genes | 70 |
| Number of tRNAs genes | 12 |
| 23S rRNAs | 7 |
| 5S rRNAs | 5 |
| N50 | 518,368 |
| L50 | 4 |
| Number of subsystems | 396 |
| Number of CDSs | 5135 |
| Genes assigned to COGs | 5006 |
| Pseudo Genes | 129 |

Table A2.
A table showing the diversity of MLST (ST types) and allelic profiles of the seven housekeeping genes of all deposited Citrobacter freundii strains (n = 5) from South Africa.
Table A2.
A table showing the diversity of MLST (ST types) and allelic profiles of the seven housekeeping genes of all deposited Citrobacter freundii strains (n = 5) from South Africa.
| Isolate | MLST | arcA | aspC | clpX | dnaG | fadD | lysP | mdh |
|---|---|---|---|---|---|---|---|---|
| 944112508 | ST156 | 7 | 15 | 63 | 54 | 13 | 5 | 12 |
| 944526466 | NA | 33 | 49 | 50 | 45 | 57 | 16 | 47 |
| CF003 | ST62 | 32 | 48 | 10 | 6 | 14 | 45 | 13 |
| CF004 | ST63 | 33 | 49 | 50 | 45 | 57 | 16 | 47 |
| H2730R | ST498 | 5 | 16 | 14 | 54 | 103 | 5 | 15 |
NA, Not Available.
References
- Liu, L.; Chen, D.; Liu, L.; Lan, R.; Hao, S.; Jin, W.; Sun, H.; Wang, Y.; Liang, Y.; Xu, J. Genetic Diversity, Multidrug Resistance, and Virulence of Citrobacter freundii From Diarrheal Patients and Healthy Individuals. Front. Microbiol. 2018, 8, 233. [Google Scholar] [CrossRef]
- Heo, G.-J.; Hossain, S.; Wimalasena, S. Virulence Factors and Antimicrobial Resistance Pattern of Citrobacter freundii Isolated from Healthy Pet Turtles and their Environment. Asian J. Anim. Veter- Adv. 2017, 12, 10–16. [Google Scholar] [CrossRef]
- Mohanty, S.; Singhal, R.; Sood, S.; Dhawan, B.; Kapil, A.; Das, B.K. Citrobacter infections in a tertiary care hospital in Northern India. J. Infect. 2007, 54, 58–64. [Google Scholar] [CrossRef]
- Pepperell, C.; Kus, J.V.; Gardam, M.A.; Humar, A.; Burrows, L.L. Low-Virulence Citrobacter Species Encode Resistance to Multiple Antimicrobials. Antimicrob. Agents Chemother. 2002, 46, 3555–3560. [Google Scholar] [CrossRef]
- Gupta, N.; Yadav, A.; Choudhary, U.; Aropa, D.R. Citrobacter Bacteremia in a Tertiary Care Hospital. Scand. J. Infect. Dis. 2003, 35, 765–768. [Google Scholar] [CrossRef]
- Divyashree, M.; Kumar, D.V.; Ballamoole, K.K.; Shetty, V.; Chakraborty, A.; Karunasagar, I. Occurrence of antibiotic resistance among gram-negative bacteria isolated from effluents of fish processing plants in and around Mangalore. Int. J. Environ. Heal. Res. 2019, 1–8. [Google Scholar] [CrossRef]
- Park, Y.-J.; Park, S.Y.; Oh, E.-J.; Park, J.-J.; Lee, K.-Y.; Woo, G.-J.; Lee, K. Occurrence of extended-spectrum β-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn. Microbiol. Infect. Dis. 2005, 51, 265–269. [Google Scholar] [CrossRef]
- Majewski, P.; Wieczorek, P.; Łapuć, I.; Ojdana, D.; Sieńko, A.; Sacha, P.; Kłoczko, J.; Tryniszewska, E. Emergence of a multidrug-resistant Citrobacter freundii ST8 harboring an unusual VIM-4 gene cassette in Poland. Int. J. Infect. Dis. 2017, 61, 70–73. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Liang, B.; Hu, X.; Li, J.; Xie, J.; Yang, C.; Hao, R.; Wang, L.; Jia, L.; et al. Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci. Rep. 2018, 8, 10653. [Google Scholar] [CrossRef]
- Aminov, R.I. Horizontal Gene Exchange in Environmental Microbiota. Front. Microbiol. 2011, 2, 1–19. [Google Scholar] [CrossRef]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.E.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Sekyere, J.O.; Amoako, D.G. Genomic and phenotypic characterisation of fluoroquinolone resistance mechanisms in Enterobacteriaceae in Durban, South Africa. PLOS ONE 2017, 12, e0178888. [Google Scholar] [CrossRef]
- Mbelle, N.M.; Maningi, N.E.; Tshisevhe, V.; Modipane, L.; Amoako, D.G.; Sekyere, J.O. Draft Genome Sequence of a Clinical Enterococcus faecium Sequence Type 18 Strain from South Africa. Genome Announc. 2017, 5, e01381-17. [Google Scholar] [CrossRef]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Amoako, D.G.; Allam, M.; Janice, J.; Pedersen, T.; Sundsfjord, A.; Essack, S. Genomic characterization of multidrug-resistant ESBL-producing Klebsiella pneumoniae isolated from a Ghanaian teaching hospital. Int. J. Infect. Dis. 2019, 85, 117–123. [Google Scholar] [CrossRef]
- Sekyere, J.O.; Amoako, D.G. Carbonyl Cyanide m-Chlorophenylhydrazine (CCCP) Reverses Resistance to Colistin, but Not to Carbapenems and Tigecycline in Multidrug-Resistant Enterobacteriaceae. Front. Microbiol. 2017, 8, 228. [Google Scholar] [CrossRef]
- Mbelle, N.M.; Sekyere, J.O.; Amoako, D.G.; Maningi, N.E.; Modipane, L.; Essack, S.Y.; Feldman, C. Genomic analysis of a multidrug-resistant clinical Providencia rettgeri (PR002) strain with the novel integron ln1483 and an A/C plasmid replicon. Ann. New York Acad. Sci. 2019. [Google Scholar] [CrossRef]
- Pedersen, T.; Sekyere, J.O.; Govinden, U.; Moodley, K.; Sivertsen, A.; Samuelsen, Ø.; Essack, S.Y.; Sundsfjord, A. Spread of Plasmid-Encoded NDM-1 and GES-5 Carbapenemases among Extensively Drug-Resistant and Pandrug-Resistant Clinical Enterobacteriaceae in Durban, South Africa. Antimicrob. Agents Chemother. 2018, 62, e02178-17. [Google Scholar] [CrossRef]
- Ramsamy, Y.; Mlisana, K.P.; Allam, M.; Amoako, D.G.; Abia, A.L.K.; Ismail, A.; Singh, R.; Kisten, T.; Essack, S.Y. Genomic Analysis of Carbapenemase-Producing Extensively Drug-Resistant Klebsiella pneumoniae Isolates Reveals the Horizontal Spread of p18–43 _ 01 Plasmid Encoding bla NDM-1 in South Africa. Microorganisms 2020, 8, 137. [Google Scholar] [CrossRef]
- Hartl, R.; Kerschner, H.; Gattringer, R.; Lepuschitz, S.; Allerberger, F.; Sorschag, S.; Ruppitsch, W.; Apfalter, P. Whole-Genome Analysis of a Human Enterobacter mori Isolate Carrying a blaIMI-2 Carbapenemase in Austria. Microb. Drug Resist. 2019, 25, 94–96. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Hansen, F.; Olesen, B.; Struve, C.; Holzknecht, B.J.; Andersen, P.S.; Thye, A.-M.; Jakobsen, L.; Røder, B.L.; Stegger, M.; et al. Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J. Glob. Antimicrob. Resist. 2015, 3, 219–221. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- Chen, Y.-G.; Zhang, Y.; Yu, Y.-S.; Qu, T.-T.; Wei, Z.-Q.; Shen, P.; Li, L.-J. In vivo development of carbapenem resistance in clinical isolates of Enterobacter aerogenes producing multiple β-lactamases. Int. J. Antimicrob. Agents 2008, 32, 302–307. [Google Scholar] [CrossRef]
- Fortier, L.-C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef]
- Mottawea, W.; Duceppe, M.-O.; Dupras, A.A.; Usongo, V.; Jeukens, J.; Freschi, L.; Emond-Rheault, J.-G.; Hamel, J.; Kukavica-Ibrulj, I.; Boyle, B.; et al. Salmonella enterica Prophage Sequence Profiles Reflect Genome Diversity and Can Be Used for High Discrimination Subtyping. Front. Microbiol. 2018, 9, 836. [Google Scholar] [CrossRef]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef]
- Shabbir, M.A.B.; Hao, H.; Shabbir, M.Z.; Hussain, H.I.; Iqbal, Z.; Ahmed, S.; Sattar, A.; Iqbal, M.; Li, J.; Yuan, Z. Survival and Evolution of CRISPR–Cas System in Prokaryotes and Its Applications. Front. Immunol. 2016, 7, 511. [Google Scholar] [CrossRef]
- Pleška, M.; Qian, L.; Okura, R.; Bergmiller, T.; Wakamoto, Y.; Kussell, E.; Guet, C.C. Bacterial Autoimmunity Due to a Restriction-Modification System. Curr. Boil. 2016, 26, 404–409. [Google Scholar] [CrossRef]
- Clements, A.; Young, J.C.; Constantinou, N.; Frankel, G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012, 3, 71–87. [Google Scholar] [CrossRef]
- Amoako, D.G.; Somboro, A.M.; Abia, A.L.K.; Allam, M.; Ismail, A.; Bester, L.A.; Essack, S.Y. Genome Mining and Comparative Pathogenomic Analysis of An Endemic Methicillin-Resistant Staphylococcus Aureus (MRSA) Clone, ST612-CC8-t1257-SCCmec_IVd(2B), Isolated in South Africa. Pathogens 2019, 8, 166. [Google Scholar] [CrossRef]
- Johnson, L.; Horsman, S.R.; Charron-Mazenod, L.; Turnbull, A.L.; Mulcahy, H.; Surette, M.G.; Lewenza, S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2013, 13, 115. [Google Scholar] [CrossRef]
- Amoako, D.G.; Somboro, A.M.; Abia, A.L.K.; Allam, M.; Ismail, A.; Bester, L.; Essack, S.Y. Genomic analysis of methicillin-resistant Staphylococcus aureus isolated from poultry and occupational farm workers in Umgungundlovu District, South Africa. Sci. Total Environ. 2019, 670, 704–716. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 27th Edition Informational Supplement M100-S27; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Boil. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; A Best, A.; DeJongh, M.; Disz, T.; A Edwards, R.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Neher, R.A.; Bedford, T. Real-Time Analysis and Visualization of Pathogen Sequence Data. J. Clin. Microbiol. 2018, 56, 11. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. eBURST: Inferring Patterns of Evolutionary Descent among Clusters of Related Bacterial Genotypes from Multilocus Sequence Typing Data. J. Bacteriol. 2004, 186, 1518–1530. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: a fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Xu, Z. Study the Features of 57 Confirmed CRISPR Loci in 38 Strains of Staphylococcus aureus. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Cosentino, S.; Larsen, M.V.; Aarestrup, F.M.; Lund, O. PathogenFinder - Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLOS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinformat. 2009, 10, 421. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Ahrenfeldt, J.; Skaarup, C.; Hasman, H.; Pedersen, A.G.; Aarestrup, F.M.; Lund, O. Bacterial whole genome-based phylogeny: construction of a new benchmarking dataset and assessment of some existing methods. BMC Genom. 2017, 18, 19. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; AbuDahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2017, 34, 292–293. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).