Genetic Changes in Experimental Populations of a Hybrid in the Cryptococcus neoformans Species Complex
Abstract
1. Introduction
2. Results
2.1. Patterns of Heterozygosity of the Experimentally Evolved Lines
2.2. Genotype on Chromosome 1 in the F-series Lines
2.3. Fluconazole MIC of the Mutation Accumulation Lines
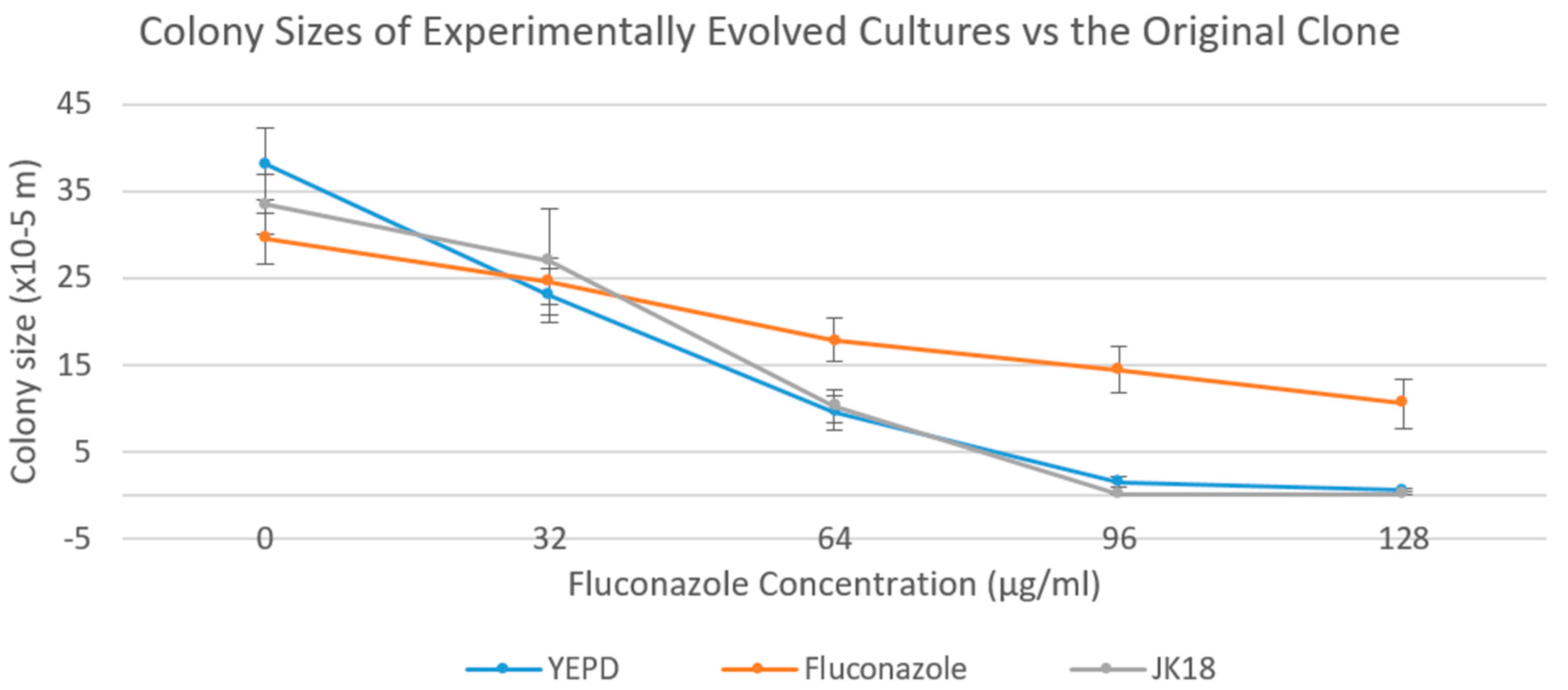
2.4. Colony Sizes of the Experimentally Evolved Cultures on YEPD Media Supplemented with Various Concentrations of Fluconazole
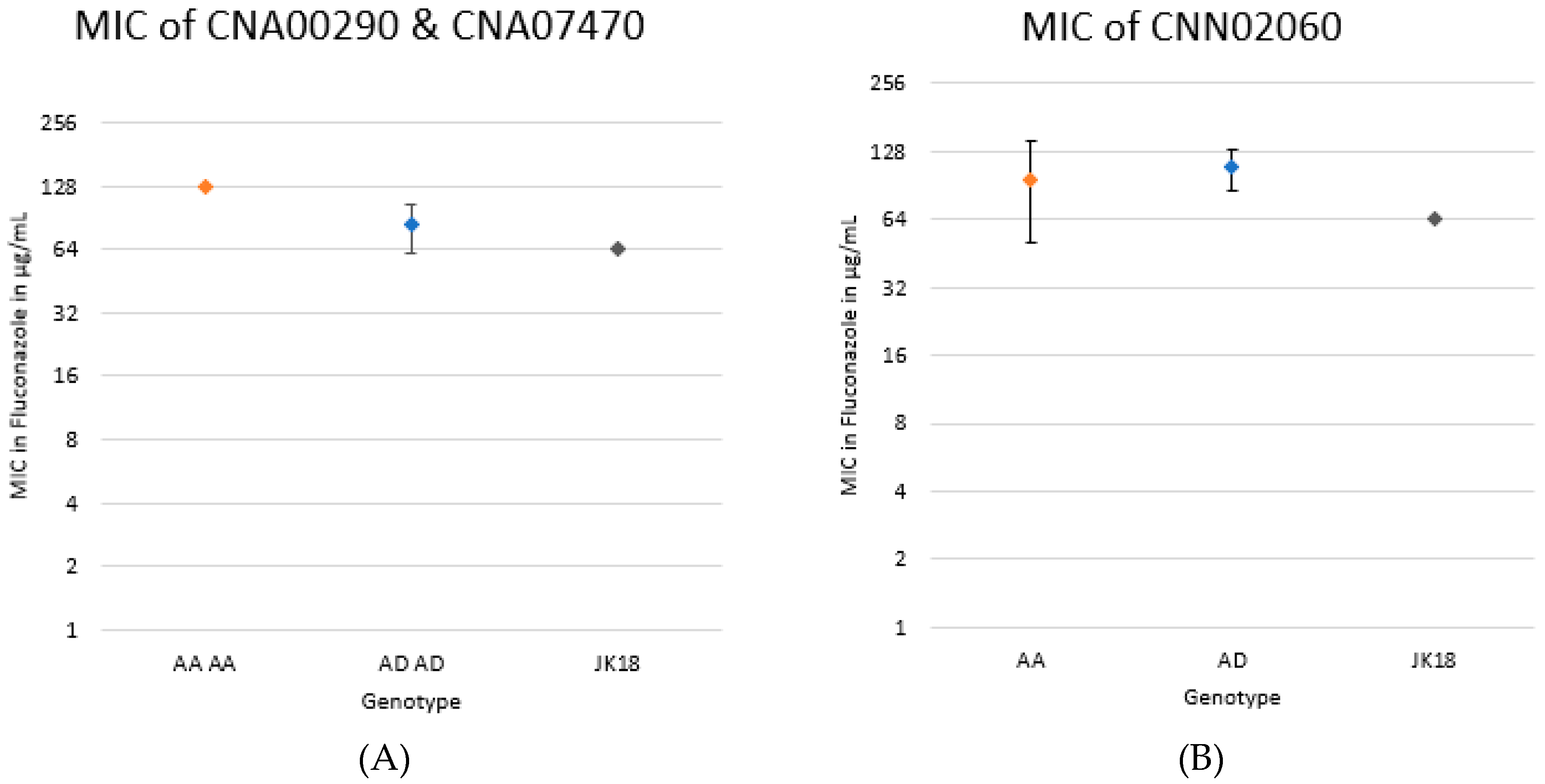
2.5. Genotypic Contribution to MIC
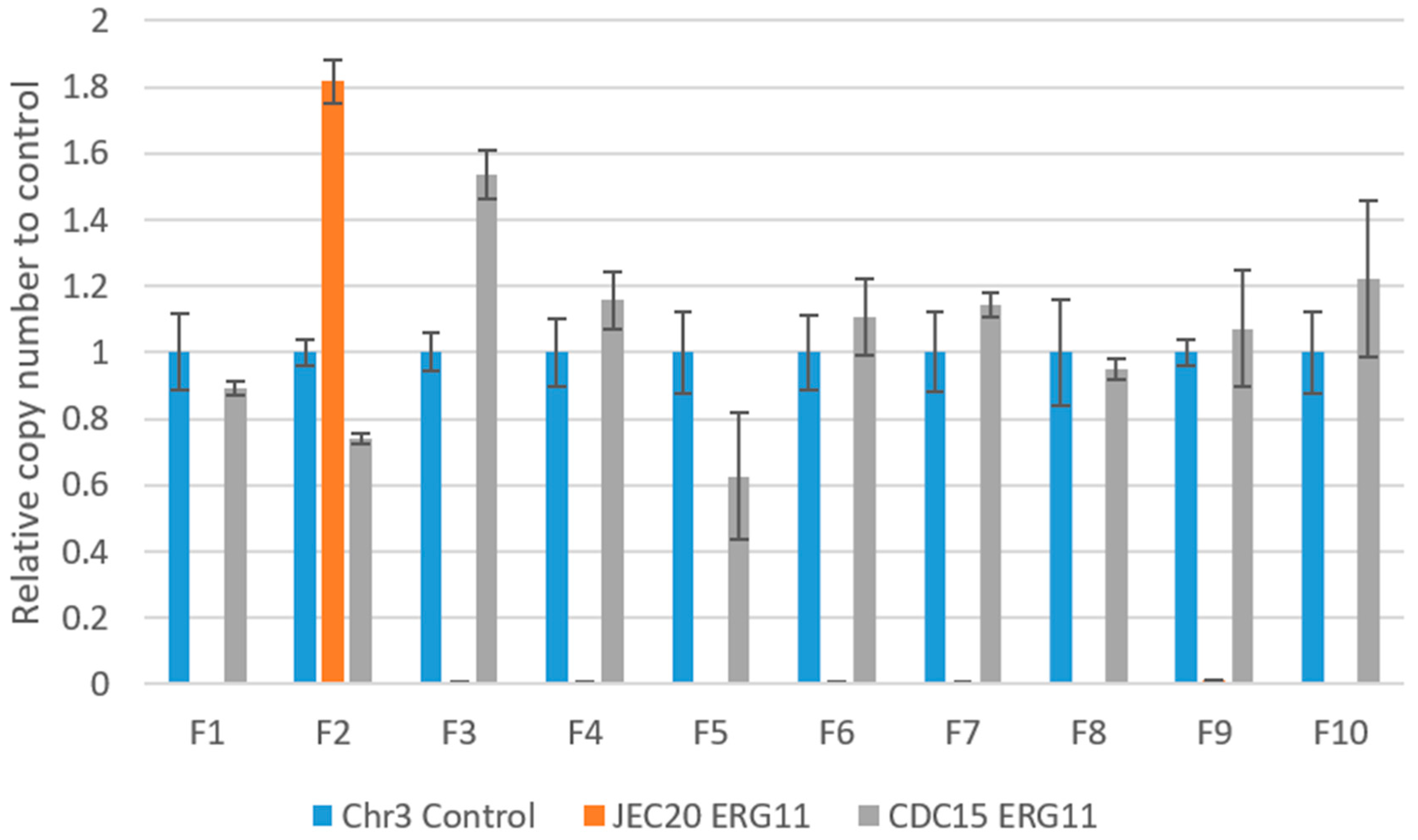
2.6. Copy Number of ERG11 in the Experimentally Evolved Cultures
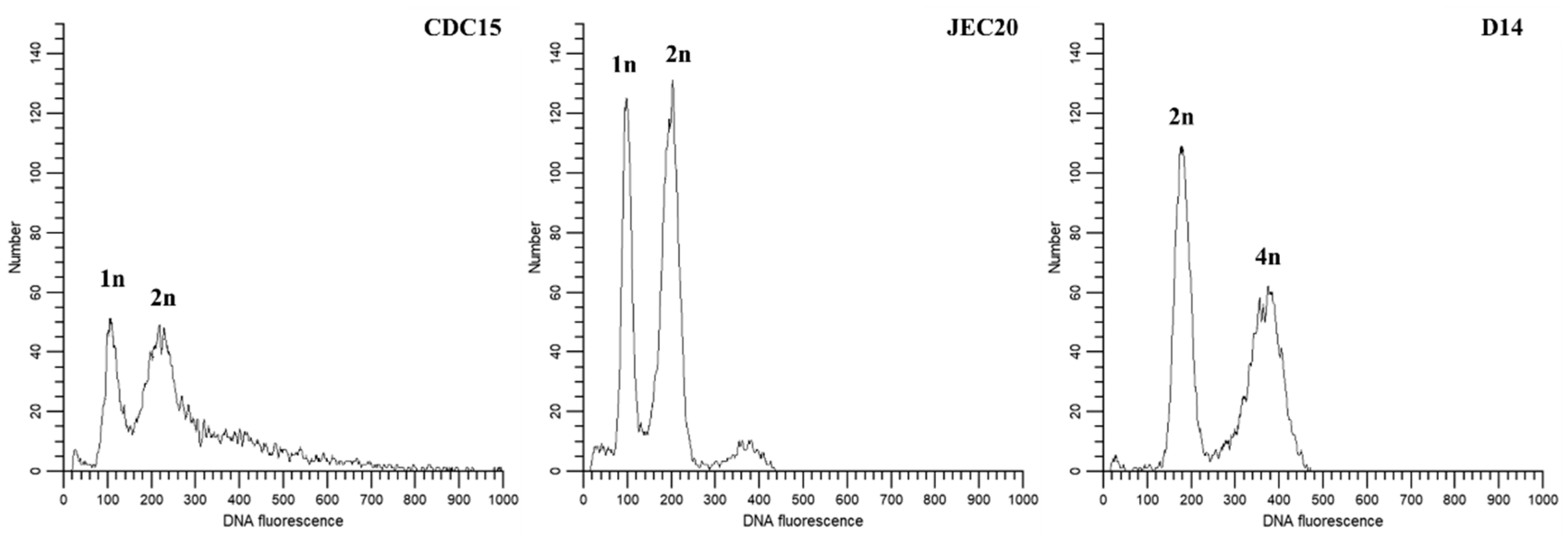
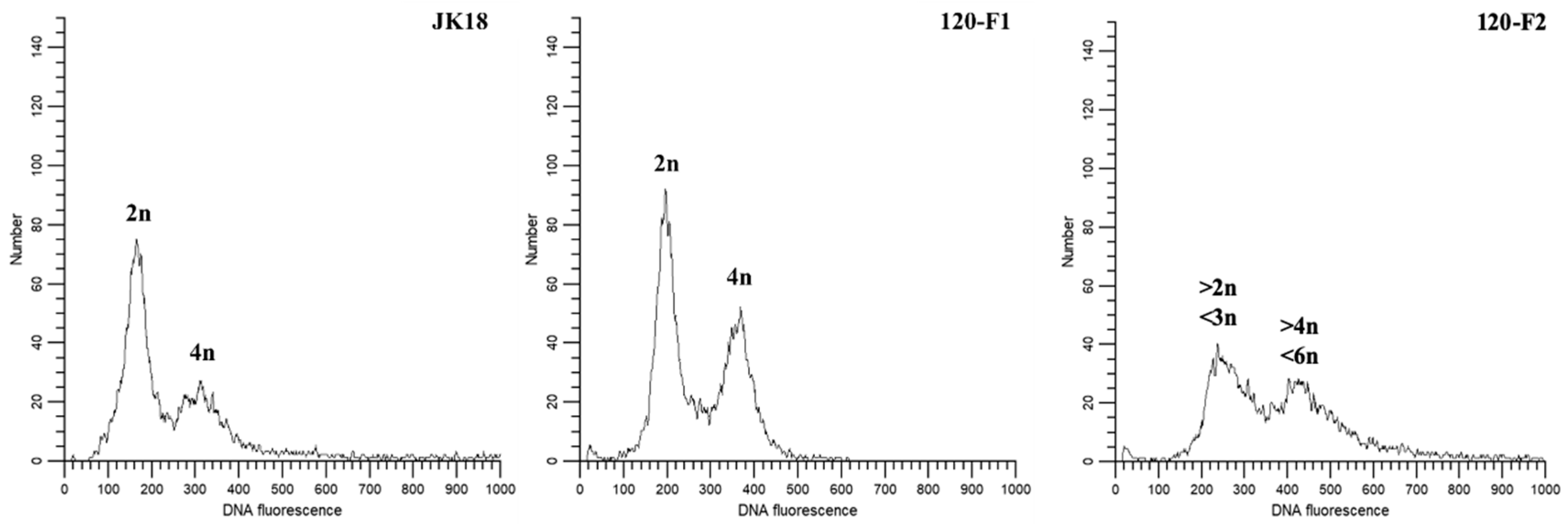
2.7. Ploidy Level
3. Discussion
3.1. Relationship Between Genetic Changes in Chromosome 1 and Susceptibility to Fluconazole
3.2. Biased Loss of Alleles on Other Chromosomes
3.3. Implications in Evolutionary Biology and Clinical Practice
4. Materials and Methods
4.1. Strains
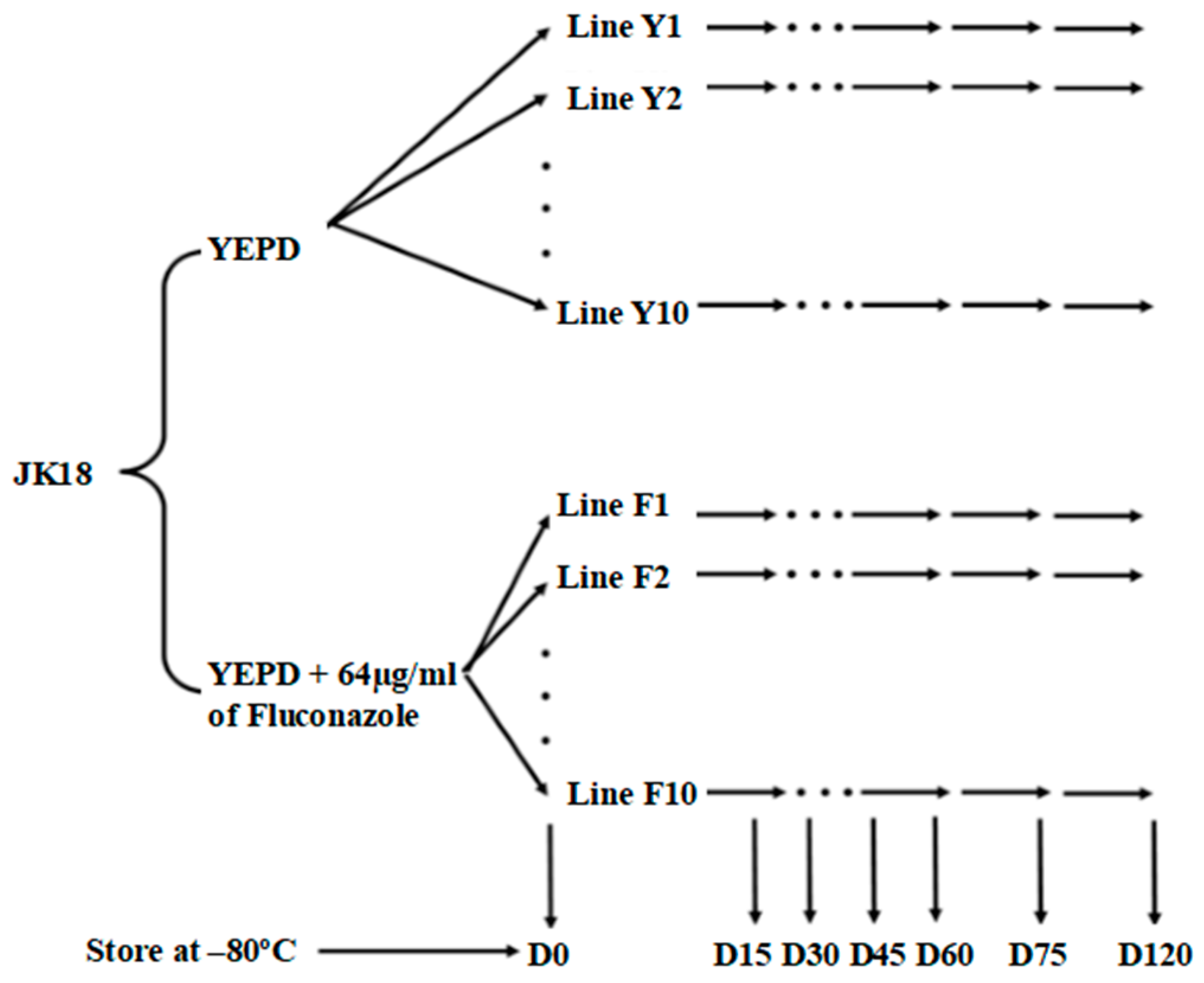
4.2. Mutation Accumulation Lines
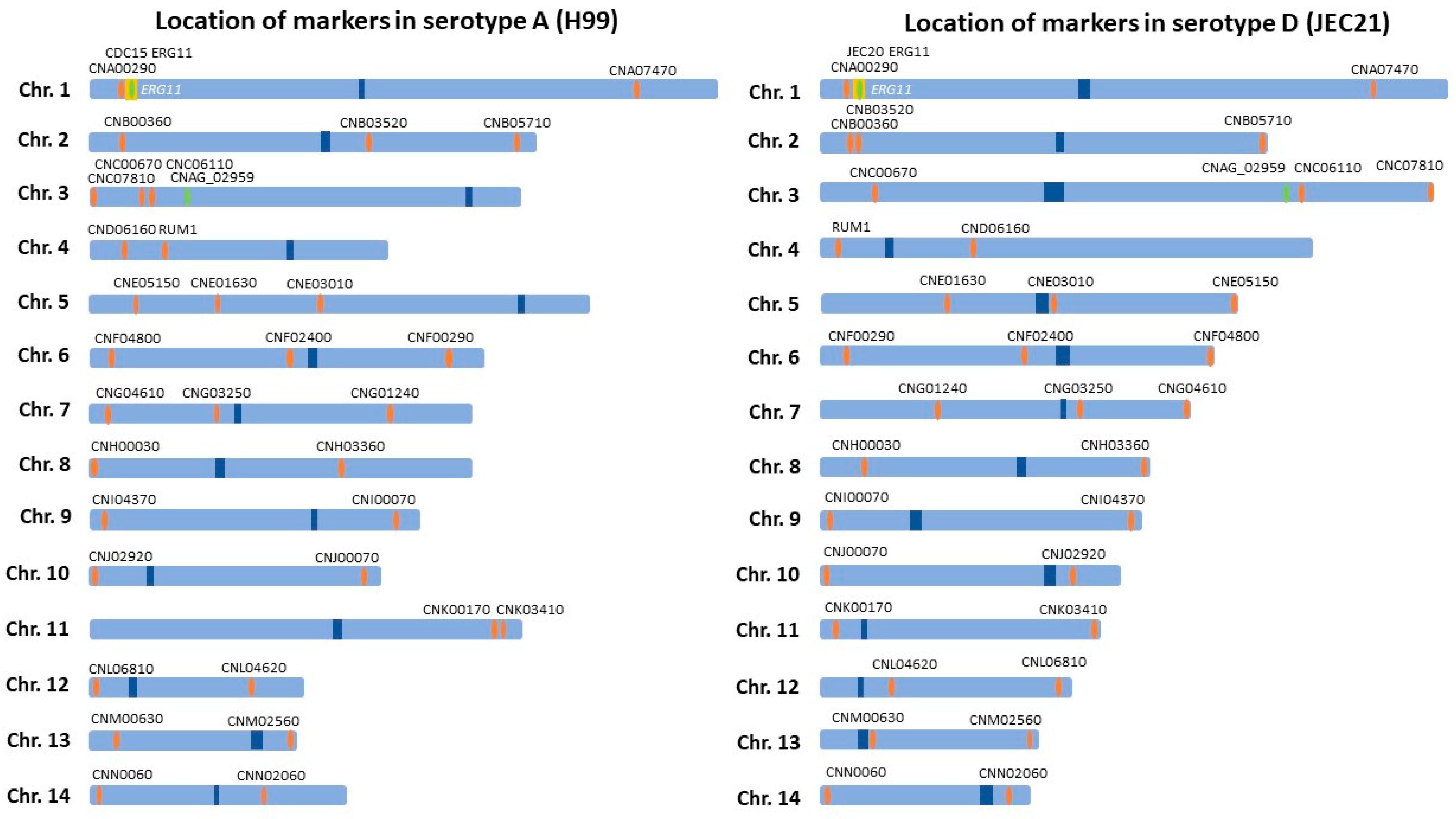
4.3. Genotyping
4.4. Minimum Inhibitory Concentration for Fluconazole
4.5. qPCR Copy Number Variation of the ERG11 Gene
4.6. Ploidy Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Del Valle, L.; Piña-Oviedo, S. HIV disorders of the brain: Pathology and pathogenesis. Front. Biosci. 2006, 11, 718–732. [Google Scholar] [CrossRef] [PubMed]
- Lengeler, K.B.; Cox, G.M.; Heitman, J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 2001, 69, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Samarasinghe, H.; Xu, J. Hybrids and hybridization in the Cryptococcus neoformans and Cryptococcus gattii species complexes. Infect. Genet. Evol. 2018, 66, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.C.; Kuzmin, A.; Otto, S.P. Loss-of-heterozygosity facilitates passage through Haldane’s sieve for Saccharomyces cerevisiae undergoing adaptation. Nat. Commun. 2014, 5, 3819. [Google Scholar] [CrossRef] [PubMed]
- Dunkel, N.; Blass, J.; Rogers, P.D.; Morschhäuser, J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 2008, 69, 827–840. [Google Scholar] [CrossRef]
- Forche, A.; Abbey, D.; Pisithkul, T.; Weinzierl, M.A.; Ringstrom, T.; Bruck, D.; Petersen, K.; Berman, J. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2011, 2, e00129-11. [Google Scholar] [CrossRef]
- Lancaster, S.M.; Payen, C.; Smukowski Heil, C.; Dunham, M.J. Fitness benefits of loss of heterozygosity in Saccharomyces hybrids. Genome Res. 2019, 29, 1685–1692. [Google Scholar] [CrossRef]
- Sun, S.; Xu, J. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics 2007, 177, 1475–1486. [Google Scholar] [CrossRef]
- Sun, S.; Xu, J. Chromosomal rearrangements between serotype A and D strains in Cryptococcus neoformans. PLoS ONE 2009, 4, e5524. [Google Scholar] [CrossRef]
- Harrison, B.D.; Hashemi, J.; Bibi, M.; Pulver, R.; Bavli, D.; Nahmias, Y.; Wellington, M.; Sapiro, G.; Berman, J. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 2014, 12, e1001815. [Google Scholar] [CrossRef]
- Vogan, A.A.; Khankhet, J.; Samarasinghe, H.; Xu, J. Identification of QTLs Associated with Virulence Related Traits and Drug Resistance in Cryptococcus neoformans. G3 (Bethesda) 2016, 6, 2745–2759. [Google Scholar] [CrossRef] [PubMed]
- Sionov, E.; Lee, H.; Chang, Y.C.; Kwon-Chung, K.J. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010, 6, e1000848. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Vilgalys, R.; Mitchell, T.G. Colony size can be used to determine the MIC of fluconazole for pathogenic yeasts. J. Clin. Microbiol. 1998, 36, 2383–2385. [Google Scholar] [PubMed]
- Revankar, S.G.; Fu, J.; Rinaldi, M.G.; Kelly, S.L.; Kelly, D.E.; Lamb, D.C.; Keller, S.M.; Wickes, B.L. Cloning and characterization of the lanosterol 14alpha-demethylase (ERG11) gene in Cryptococcus neoformans. Biochem. Biophys. Res. Commun. 2004, 324, 719–728. [Google Scholar] [CrossRef]
- Sionov, E.; Chang, Y.C.; Garraffo, H.M.; Kwon-Chung, K.J. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 2009, 53, 2804–2815. [Google Scholar] [CrossRef]
- Bennett, R.J.; Forche, A.; Berman, J. Rapid mechanisms for generating genome diversity: Whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb. Perspect. Med. 2014, 4, a019604. [Google Scholar] [CrossRef]
- Gerstein, A.C.; Fu, M.S.; Mukaremera, L.; Li, Z.; Ormerod, K.L.; Fraser, J.A.; Berman, J.; Nielsen, K. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 2015, 6, e01340-15. [Google Scholar] [CrossRef]
- Mitchell, T.G.; Perfect, J.R. Cryptococcosis in the era of AIDS—100 Years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 1995, 8, 515–548. [Google Scholar] [CrossRef]
- Smukowski Heil, C.S.; Large, C.R.L.; Patterson, K.; Hickey, A.S.-M.; Yeh, C.-L.C.; Dunham, M.J. Temperature preference can bias parental genome retention during hybrid evolution. PLoS Genet. 2019, 15, e1008383. [Google Scholar] [CrossRef]
- Baker, D.J.; Jin, F.; Jeganathan, K.B.; van Deursen, J.M. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 2009, 16, 475–486. [Google Scholar] [CrossRef]
- Vogan, A.A.; Xu, J. Evidence for genetic incompatibilities associated with post-zygotic reproductive isolation in the human fungal pathogen Cryptococcus Neoformans. Genome 2014, 57, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Sia, R.A.; Lengeler, K.B.; Heitman, J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet. Biol. 2000, 29, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Vogan, A.A.; Khankhet, J.; Xu, J. Evidence for mitotic recombination within the basidia of a hybrid cross of Cryptococcus neoformans. PLoS ONE 2013, 8, e62790. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.J.; Vogan, A.A.; Xu, J. Genotypic and phenotypic analyses of two “isogenic” strains of the human fungal pathogen Cryptococcus neoformans var. neoformans. Mycopathologia 2019, 184, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Skosireva, I.; James, T.Y.; Sun, S.; Xu, J. Mitochondrial inheritance in haploid × non-haploid crosses in Cryptococcus neoformans. Curr. Genet. 2010, 56, 163–176. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Marker | Ch. | Y1 | Y2 | Y3 | Y4 | Y5 | Y6 | Y7 | Y8 | Y9 | Y10 | Ho | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | Ho |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CNA00290 | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | A | 0.9 | A | AD | A | A | A | A | A | A | A | A | 0.1 |
| CNA07470 | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | A | 0.9 | A | AD | A | A | A | A | A | A | A | A | 0.1 |
| CNB00360 | 2 | AD | AD | AD | D | AD | D | AD | AD | AD | AD | 0.8 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNB03520 | 2 | AD | AD | AD | AD | AD | AD | AD | AD | A | AD | 0.9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNB05710 | 2 | AD | AD | AD | AD | AD | AD | AD | AD | A | AD | 0.9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNC00670 | 3 | AD | AD | D | AD | AD | AD | AD | AD | AD | AD | 0.9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNC06110 | 3 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNC07810 | 3 | AD | AD | AD | D | D | AD | AD | AD | AD | A | 0.7 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CND06160 | 4 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| RUM1 | 4 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNE01630 | 5 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNE03010 | 5 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNE05150 | 5 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | D | AD | AD | AD | AD | AD | 0.9 |
| CNF00290 | 6 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNF02400 | 6 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNF04800 | 6 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNG01240 | 7 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNG03250 | 7 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNG04610 | 7 | AD | D | AD | AD | AD | AD | AD | AD | AD | AD | 0.9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNH00030 | 8 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | D | AD | AD | AD | AD | 0.9 |
| CNH03360 | 8 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNI00070 | 9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | A | 0.9 |
| CNI04370 | 9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | A | 0.9 |
| CNJ00070 | 10 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNJ02920 | 10 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNK00170 | 11 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNK03410 | 11 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNL04620 | 12 | AD | AD | AD | AD | AD | AD | AD | AD | AD | D | 0.9 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 |
| CNL06810 | 12 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | 1 | AD | A | AD | AD | AD | AD | AD | AD | AD | AD | 0.9 |
| CNM00630 | 13 | AD | AD | AD | AD | AD | A | AD | AD | AD | AD | 0.9 | AD | AD | AD | D | AD | AD | AD | AD | AD | AD | 0.9 |
| CNM02560 | 13 | AD | AD | AD | AD | AD | A | AD | AD | AD | AD | 0.9 | AD | AD | AD | D | AD | AD | AD | AD | AD | AD | 0.9 |
| CNN0060 | 14 | AD | A | AD | AD | AD | AD | AD | AD | AD | A | 0.8 | AD | A | AD | AD | A | AD | AD | AD | AD | AD | 0.8 |
| CNN02060 | 14 | AD | A | AD | AD | AD | AD | AD | AD | AD | AD | 0.9 | AD | A | AD | AD | A | A | AD | AD | AD | AD | 0.7 |
| Ho | 1 | 0.9 | 1 | 1 | 1 | 0.9 | 1 | 1 | 0.9 | 0.8 | 1 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | 0.9 | |
| MIC | 32 | 32 | 32 | 16 | 32 | 16 | 32 | 32 | 32 | 128 | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | >128 | |||
| Y10 | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 0 | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD | AD |
| Day 15 | AD | A | AD | A | A | AD | AD | A | AD | AD | A |
| Day 30 | AD | A | AD | A | A | AD | AD | A | A | A | A |
| Day 45 | A | A | AD | A | A | A | AD | A | A | A | A |
| Day 60 | A | A | AD | A | A | A | A | A | A | A | A |
| Day 75 | A | A | AD | A | A | A | A | A | A | A | A |
| Day 120 | A | A | AD | A | A | A | A | A | A | A | A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, K.; You, M.; Xu, J. Genetic Changes in Experimental Populations of a Hybrid in the Cryptococcus neoformans Species Complex. Pathogens 2020, 9, 3. https://doi.org/10.3390/pathogens9010003
Dong K, You M, Xu J. Genetic Changes in Experimental Populations of a Hybrid in the Cryptococcus neoformans Species Complex. Pathogens. 2020; 9(1):3. https://doi.org/10.3390/pathogens9010003
Chicago/Turabian StyleDong, Kelly, Man You, and Jianping Xu. 2020. "Genetic Changes in Experimental Populations of a Hybrid in the Cryptococcus neoformans Species Complex" Pathogens 9, no. 1: 3. https://doi.org/10.3390/pathogens9010003
APA StyleDong, K., You, M., & Xu, J. (2020). Genetic Changes in Experimental Populations of a Hybrid in the Cryptococcus neoformans Species Complex. Pathogens, 9(1), 3. https://doi.org/10.3390/pathogens9010003






