H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host
Abstract
1. Introduction
1.1. Histone Modifications and Chromatin Architecture
1.2. Histone Methylation
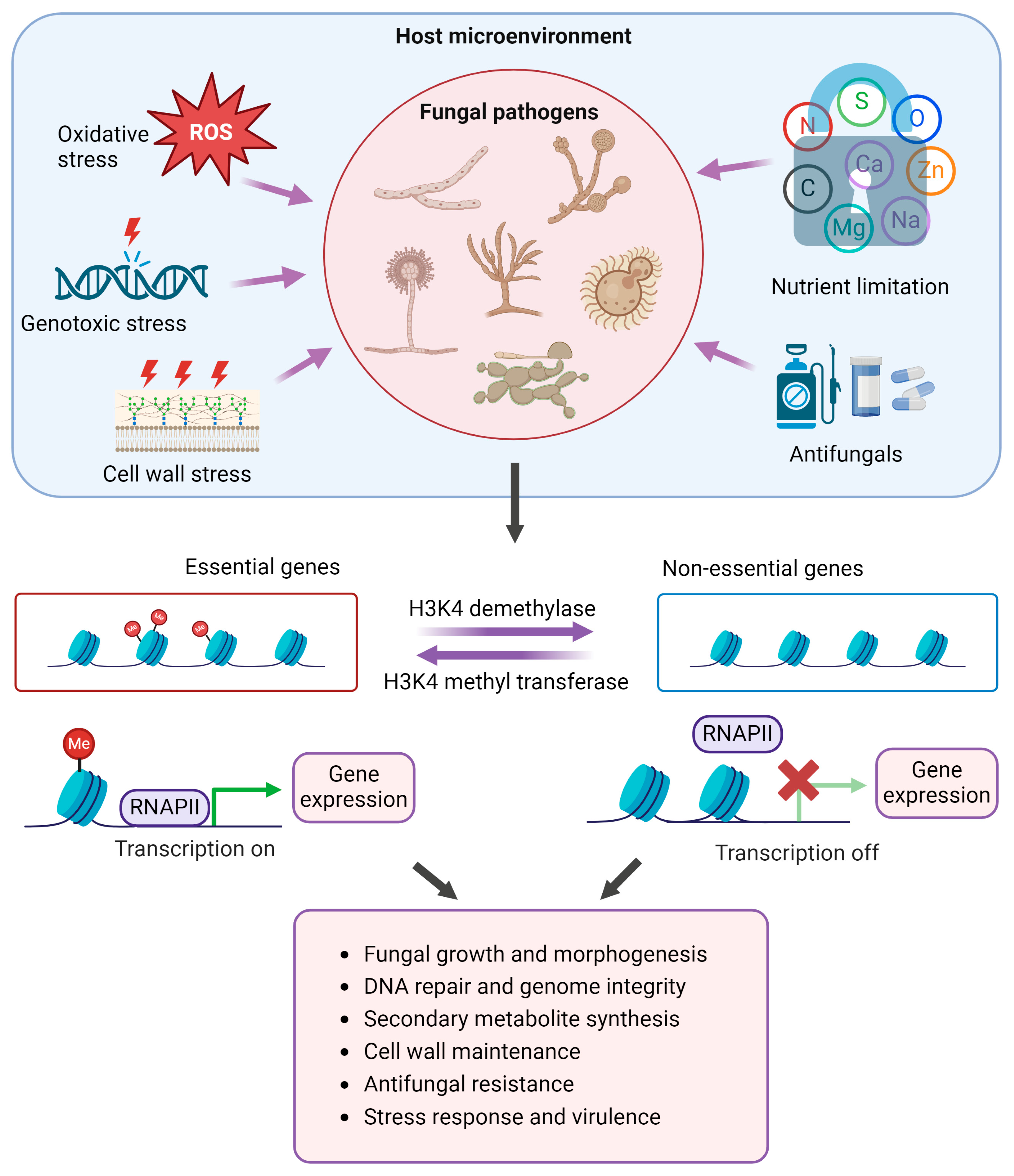
1.3. H3K4 Methylation and Demethylation
1.4. Roles of H3K4 Methylation and Demethylation in Pathogenic Adaptation in Fungi
- (a)
- Morphogenesis and development
- (b)
- Genome stability and DNA repair
- (c)
- Metabolic adaptation
- (d)
- Cell wall maintenance
- (e)
- Antifungal resistance
- (f)
- Stress response and virulence
2. Discussion and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Janbon, G.; Quintin, J.; Lanternier, F.; d’Enfert, C. Studying Fungal Pathogens of Humans and Fungal Infections: Fungal Diversity and Diversity of Approaches. Genes Immun. 2019, 20, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A. Evolution of the Human Pathogenic Lifestyle in Fungi. Nat. Microbiol. 2022, 7, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Benedict, K.; Richardson, M.; Vallabhaneni, S.; Jackson, B.R.; Chiller, T. Emerging Issues, Challenges, and Changing Epidemiology of Fungal Disease Outbreaks. Lancet Infect. Dis. 2017, 17, e403–e411. [Google Scholar] [CrossRef] [PubMed]
- Avery, S.V.; Singleton, I.; Magan, N.; Goldman, G.H. The Fungal Threat to Global Food Security. Fungal Biol. 2019, 123, 555–557. [Google Scholar] [CrossRef]
- Davies, C.R.; Wohlgemuth, F.; Young, T.; Violet, J.; Dickinson, M.; Sanders, J.-W.; Vallieres, C.; Avery, S.V. Evolving Challenges and Strategies for Fungal Control in the Food Supply Chain. Fungal Biol. Rev. 2021, 36, 15–26. [Google Scholar] [CrossRef]
- Xu, K.; Li, X.-Q.; Zhao, D.-L.; Zhang, P. Antifungal Secondary Metabolites Produced by the Fungal Endophytes: Chemical Diversity and Potential Use in the Development of Biopesticides. Front. Microbiol. 2021, 12, 689527. [Google Scholar] [CrossRef]
- Singh, Y.; Nair, A.M.; Verma, P.K. Surviving the Odds: From Perception to Survival of Fungal Phytopathogens under Host-Generated Oxidative Burst. Plant Commun. 2021, 2, 100142. [Google Scholar] [CrossRef]
- Brown, G.D. Innate Antifungal Immunity: The Key Role of Phagocytes. Annu. Rev. Immunol. 2011, 29, 1–21. [Google Scholar] [CrossRef]
- Romani, L. Immunity to Fungal Infections. Nat. Rev. Immunol. 2011, 11, 275–288. [Google Scholar] [CrossRef]
- Sethiya, P.; Rai, M.N.; Rai, R.; Parsania, C.; Tan, K.; Wong, K.H. Transcriptomic Analysis Reveals Global and Temporal Transcription Changes during Candida glabrata Adaptation to an Oxidative Environment. Fungal Biol. 2020, 124, 427–439. [Google Scholar] [CrossRef]
- Rai, M.N.; Balusu, S.; Gorityala, N.; Dandu, L.; Kaur, R. Functional Genomic Analysis of Candida glabrata-Macrophage Interaction: Role of Chromatin Remodeling in Virulence. PLoS Pathog. 2012, 8, e1002863. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S. To Be or Not To Be Pathogenic: Transcriptional Reprogramming Dictates a Fungal Pathogen’s Response to Different Hosts[OPEN]. Plant Cell 2020, 32, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Jenull, S.; Tscherner, M.; Mair, T.; Kuchler, K. ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. J. Fungi 2020, 6, 182. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.B.; van der Meer, J.W.M.; Kullberg, B.-J.; van de Veerdonk, F.L. Immune Defence against Candida Fungal Infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Quintin, J.; Saeed, S.; Martens, J.H.A.; Giamarellos-Bourboulis, E.J.; Ifrim, D.C.; Logie, C.; Jacobs, L.; Jansen, T.; Kullberg, B.-J.; Wijmenga, C.; et al. Candida Albicans Infection Affords Protection against Reinfection via Functional Reprogramming of Monocytes. Cell Host Microbe 2012, 12, 223–232. [Google Scholar] [CrossRef]
- Dubey, A.; Jeon, J. Epigenetic Regulation of Development and Pathogenesis in Fungal Plant Pathogens. Mol. Plant Pathol. 2017, 18, 887–898. [Google Scholar] [CrossRef]
- Rando, O.J.; Winston, F. Chromatin and Transcription in Yeast. Genetics 2012, 190, 351–387. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone Post-Translational Modifications—Cause and Consequence of Genome Function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.; Chen, H.V.; Liu, C.L.; Rahat, A.; Klien, A.; Soares, L.; Gudipati, M.; Pfeffner, J.; Regev, A.; Buratowski, S.; et al. Systematic Dissection of Roles for Chromatin Regulators in a Yeast Stress Response. PLoS Biol. 2012, 10, e1001369. [Google Scholar] [CrossRef] [PubMed]
- South, P.F.; Harmeyer, K.M.; Serratore, N.D.; Briggs, S.D. H3K4 Methyltransferase Set1 Is Involved in Maintenance of Ergosterol Homeostasis and Resistance to Brefeldin A. Proc. Natl. Acad. Sci. USA 2013, 110, E1016–E1025. [Google Scholar] [CrossRef]
- Lai, Y.; Cao, X.; Chen, J.; Wang, L.; Wei, G.; Wang, S. Coordinated Regulation of Infection-Related Morphogenesis by the KMT2-Cre1-Hyd4 Regulatory Pathway to Facilitate Fungal Infection. Sci. Adv. 2023, 6, eaaz1659. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Xie, R.; Zhang, F.; Wang, S.; Pan, X.; Wang, S.; Zhuang, Z. The Methyltransferase AflSet1 Is Involved in Fungal Morphogenesis, AFB1 Biosynthesis, and Virulence of Aspergillus Flavus. Front. Microbiol. 2020, 11, 234. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, X.; Sun, W.; Zhang, M.; Yin, Y.; Pan, S.; He, D.; Shen, M.; Yang, J.; Zheng, Q.; et al. The COMPASS-like Complex Modulates Fungal Development and Pathogenesis by Regulating H3K4me3-Mediated Targeted Gene Expression in Magnaporthe oryzae. Mol. Plant Pathol. 2021, 22, 422–439. [Google Scholar] [CrossRef]
- Pehrson, J.R.; Fuji, R.N. Evolutionary Conservation of Histone MacroH2A Subtypes and Domains. Nucleic Acids Res. 1998, 26, 2837–2842. [Google Scholar] [CrossRef]
- Phillips, E.O.N.; Gunjan, A. Histone Variants: The Unsung Guardians of the Genome. DNA Repair 2022, 112, 103301. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Kuo, M.-H.; Allis, C.D. Roles of Histone Acetyltransferases and Deacetylases in Gene Regulation. BioEssays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Kurdistani, S.K.; Grunstein, M. Histone Acetylation and Deacetylation in Yeast. Nat. Rev. Mol. Cell Biol. 2003, 4, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Separovich, R.J.; Wilkins, M.R. Ready, SET, Go: Post-Translational Regulation of the Histone Lysine Methylation Network in Budding Yeast. J. Biol. Chem. 2021, 297, 100939. [Google Scholar] [CrossRef] [PubMed]
- Separovich, R.J.; Pang, C.N.I.; Wilkins, M.R. Controlling the Controllers: Regulation of Histone Methylation by Phosphosignalling. Trends Biochem. Sci. 2020, 45, 1035–1048. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Colabardini, A.C.; Wang, F.; Miao, Z.; Pardeshi, L.; Valero, C.; de Castro, P.A.; Akiyama, D.Y.; Tan, K.; Nora, L.C.; Silva-Rocha, R.; et al. Chromatin Profiling Reveals Heterogeneity in Clinical Isolates of the Human Pathogen Aspergillus fumigatus. PLoS Genet. 2022, 18, e1010001. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Humphrey, E.L.; Erlich, R.L.; Schneider, R.; Bouman, P.; Liu, J.S.; Kouzarides, T.; Schreiber, S.L. Methylation of Histone H3 Lys 4 in Coding Regions of Active Genes. Proc. Natl. Acad. Sci. USA 2002, 99, 8695–8700. [Google Scholar] [CrossRef]
- Kouzarides, T. Histone Methylation in Transcriptional Control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [Google Scholar] [CrossRef]
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3K4me3 Instructive for Transcription Activation? Bioessays 2017, 39, 1–12. [Google Scholar] [CrossRef]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Targeted Recruitment of Set1 Histone Methylase by Elongating Pol II Provides a Localized Mark and Memory of Recent Transcriptional Activity. Mol. Cell 2003, 11, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.-W.; Allis, C.D. Ubiquitination of Histone H2B Regulates H3 Methylation and Gene Silencing in Yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Dover, J.; Schneider, J.; Tawiah-Boateng, M.A.; Wood, A.; Dean, K.; Johnston, M.; Shilatifard, A. Methylation of Histone H3 by COMPASS Requires Ubiquitination of Histone H2B by Rad6. J. Biol. Chem. 2002, 277, 28368–28371. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.; Bryk, M. Diverse and Dynamic Forms of Gene Regulation by the S. cerevisiae Histone Methyltransferase Set1. Curr. Genet. 2023, 69, 91–114. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Whetstine, J.R. Dynamic Regulation of Histone Lysine Methylation by Demethylases. Mol. Cell 2007, 25, 1–14. [Google Scholar] [CrossRef]
- Liang, G.; Klose, R.J.; Gardner, K.E.; Zhang, Y. Yeast Jhd2p Is a Histone H3 Lys4 Trimethyl Demethylase. Nat. Struct. Mol. Biol. 2007, 14, 243–245. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Pokhrel, S.; Palani, S.; Pflueger, C.; Parnell, T.J.; Cairns, B.R.; Bhaskara, S.; Chandrasekharan, M.B. Counteracting H3K4 Methylation Modulators Set1 and Jhd2 Co-Regulate Chromatin Dynamics and Gene Transcription. Nat. Commun. 2016, 7, 11949. [Google Scholar] [CrossRef]
- Dallery, J.-F.; Adelin, É.; Le Goff, G.; Pigné, S.; Auger, A.; Ouazzani, J.; O’Connell, R.J. H3K4 Trimethylation by CclA Regulates Pathogenicity and the Production of Three Families of Terpenoid Secondary Metabolites in Colletotrichum higginsianum. Mol. Plant Pathol. 2019, 20, 831–842. [Google Scholar] [CrossRef]
- Liu, R.; Chen, X.; Zhao, F.; Jiang, Y.; Lu, Z.; Ji, H.; Feng, Y.; Li, J.; Zhang, H.; Zheng, J.; et al. The COMPASS Complex Regulates Fungal Development and Virulence through Histone Crosstalk in the Fungal Pathogen Cryptococcus neoformans. J. Fungi 2023, 9, 672. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, N.; Yin, Y.; Chen, Y.; Jiang, J.; Ma, Z. Histone H3K4 Methylation Regulates Hyphal Growth, Secondary Metabolism and Multiple Stress Responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4615–4630. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Li, B.; Tian, S. Histone H3K4 Methyltransferase PeSet1 Regulates Colonization, Patulin Biosynthesis, and Stress Responses of Penicillium expansum. Microbiol. Spectr. 2023, 11, e03545-22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Fang, W.; Tong, J.; Liu, S.; Wu, H.; Shi, J. Metarhizium robertsii as a Promising Microbial Agent for Rice in Situ Cadmium Reduction and Plant Growth Promotion. Chemosphere 2022, 305, 135427. [Google Scholar] [CrossRef]
- Meng, S.; Shi, H.; Lin, C.; Wu, Z.; Lin, F.; Tao, Z.; Kou, Y. UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens. J. Fungi 2022, 8, 553. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.-W.; Ahn, S.H. Role of Yeast JmjC-Domain Containing Histone Demethylases in Actively Transcribed Regions. Biochem. Biophys. Res. Commun. 2011, 410, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Faucher, D.; Wellinger, R.J. Methylated H3K4, a Transcription-Associated Histone Modification, Is Involved in the DNA Damage Response Pathway. PLoS Genet. 2010, 6, e1001082. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.Y.; Cutler, S.; Lin, J.-J.; Tsai, C.-H.; Tsai, H.-K.; Biggins, S.; Tsukiyama, T.; Lo, Y.-C.; Kao, C.-F. H3K4 Methylation at Active Genes Mitigates Transcription-Replication Conflicts during Replication Stress. Nat. Commun. 2020, 11, 809. [Google Scholar] [CrossRef]
- Choudhary, N.K.; Jain, A.K.; Soni, R.; Gahlot, N. Mucormycosis: A Deadly Black Fungus Infection among COVID-19 Patients in India. Clin. Epidemiol. Glob. Health 2021, 12, 100900. [Google Scholar] [CrossRef]
- Osorio-Concepción, M.; Lax, C.; Lorenzo-Gutiérrez, D.; Cánovas-Márquez, J.T.; Tahiri, G.; Navarro, E.; Binder, U.; Nicolás, F.E.; Garre, V. H3K4 Methylation Regulates Development, DNA Repair, and Virulence in Mucorales. IMA Fungus 2023, 15, 6. [Google Scholar] [CrossRef]
- Ren, K.; Mou, Y.-N.; Tong, S.-M.; Ying, S.-H.; Feng, M.-G. DIM5/KMT1 Controls Fungal Insect Pathogenicity and Genome Stability by Methylation of Histone H3K4, H3K9 and H3K36. Virulence 2021, 12, 1306–1322. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Human and Plant Fungal Pathogens: The Role of Secondary Metabolites. PLoS Pathog. 2014, 10, e1003859. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Bachleitner, S.; Sørensen, J.L.; Gacek-Matthews, A.; Sulyok, M.; Studt, L.; Strauss, J. Evidence of a Demethylase-Independent Role for the H3K4-Specific Histone Demethylases in Aspergillus nidulans and Fusarium graminearum Secondary Metabolism. Front. Microbiol. 2019, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Bok, J.W.; Lee, S.; Dagenais, T.R.T.; Andes, D.R.; Kontoyiannis, D.P.; Keller, N.P. Loss of CclA, Required for Histone 3 Lysine 4 Methylation, Decreases Growth but Increases Secondary Metabolite Production in Aspergillus fumigatus. PeerJ 2013, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Gacek-Matthews, A.; Berger, H.; Sasaki, T.; Wittstein, K.; Gruber, C.; Lewis, Z.A.; Strauss, J. KdmB, a Jumonji Histone H3 Demethylase, Regulates Genome-Wide H3K4 Trimethylation and Is Required for Normal Induction of Secondary Metabolism in Aspergillus nidulans. PLoS Genet. 2016, 12, e1006222. [Google Scholar] [CrossRef]
- Baker, K.M.; Hoda, S.; Saha, D.; Gregor, J.B.; Georgescu, L.; Serratore, N.D.; Zhang, Y.; Cheng, L.; Lanman, N.A.; Briggs, S.D. The Set1 Histone H3K4 Methyltransferase Contributes to Azole Susceptibility in a Species-Specific Manner by Differentially Altering the Expression of Drug Efflux Pumps and the Ergosterol Gene Pathway. Antimicrob. Agents Chemother. 2022, 66, e0225021. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.; Kwon, S.; Lee, E.-J.; Lee, J.-S. Set1-Mediated H3K4 Methylation Is Required for Candida albicans Virulence by Regulating Intracellular Level of Reactive Oxygen Species. Virulence 2021, 12, 2648–2658. [Google Scholar] [CrossRef]
- Gacek-Matthews, A.; Noble, L.M.; Gruber, C.; Berger, H.; Sulyok, M.; Marcos, A.T.; Strauss, J.; Andrianopoulos, A. KdmA, a Histone H3 Demethylase with Bipartite Function, Differentially Regulates Primary and Secondary Metabolism in Aspergillus nidulans. Mol. Microbiol. 2015, 96, 839–860. [Google Scholar] [CrossRef]
- Choi, Y.-H.; Lee, M.-W.; Shin, K.-S. The Lysine Demethylases KdmA and KdmB Differently Regulate Asexual Development, Stress Response, and Virulence in Aspergillus fumigatus. J. Fungi 2022, 8, 590. [Google Scholar] [CrossRef]
- Meng, S.; Huang, S.; Liu, J.; Gai, Y.; Li, M.; Duan, S.; Zhang, S.; Sun, X.; Yang, Q.; Wang, Y.; et al. Histone Methylation Is Required for Virulence, Conidiation, and Multi-Stress Resistance of Alternaria alternata. Front. Microbiol. 2022, 13, 924476. [Google Scholar] [CrossRef]
- Hou, J.; Feng, H.-Q.; Chang, H.-W.; Liu, Y.; Li, G.-H.; Yang, S.; Sun, C.-H.; Zhang, M.-Z.; Yuan, Y.; Sun, J.; et al. The H3K4 Demethylase Jar1 Orchestrates ROS Production and Expression of Pathogenesis-Related Genes to Facilitate Botrytis cinerea Virulence. New Phytol. 2020, 225, 930–947. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wang, L.; Zheng, W.; Wang, S. Regulatory Roles of Histone Modifications in Filamentous Fungal Pathogens. J. Fungi 2022, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fan, Z.; Shliaha, P.V.; Miele, M.; Hendrickson, R.C.; Jiang, X.; Helin, K. H3K4me3 Regulates RNA Polymerase II Promoter-Proximal Pause-Release. Nature 2023, 615, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the Roles of Histone-Modifying Enzymes and Their Associated Chromatin Modifications in Transcriptional Regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Pham, K.T.M.; Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe oryzae) Regulates Global Gene Expression during Infection-Related Morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Wozniak, G.G.; Strahl, B.D. Hitting the “Mark”: Interpreting Lysine Methylation in the Context of Active Transcription. Biochim. Biophys. Acta 2014, 1839, 1353–1361. [Google Scholar] [CrossRef]
- Yaseen, I.; White, S.A.; Torres-Garcia, S.; Spanos, C.; Lafos, M.; Gaberdiel, E.; Yeboah, R.; El Karoui, M.; Rappsilber, J.; Pidoux, A.L.; et al. Proteasome-Dependent Truncation of the Negative Heterochromatin Regulator Epe1 Mediates Antifungal Resistance. Nat. Struct. Mol. Biol. 2022, 29, 745–758. [Google Scholar] [CrossRef]
- Patra, S.; Raney, M.; Pareek, A.; Kaur, R. Epigenetic Regulation of Antifungal Drug Resistance. J. Fungi 2022, 8, 875. [Google Scholar] [CrossRef]
- Moirangthem, R.; Kumar, K.; Kaur, R. Two Functionally Redundant FK506-Binding Proteins Regulate Multidrug Resistance Gene Expression and Govern Azole Antifungal Resistance. Antimicrob. Agents Chemother. 2021, 65, 10–1128. [Google Scholar] [CrossRef]
- Torres-Garcia, S.; Yaseen, I.; Shukla, M.; Audergon, P.N.C.B.; White, S.A.; Pidoux, A.L.; Allshire, R.C. Epigenetic Gene Silencing by Heterochromatin Primes Fungal Resistance. Nature 2020, 585, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.B.; Nguyen, M.H.; Zhang, Z.; Cheng, S.; Jia, H.Y.; Weisner, N.; Iczkowski, K.; Clancy, C.J. Candida albicans SET1 Encodes a Histone 3 Lysine 4 Methyltransferase That Contributes to the Pathogenesis of Invasive Candidiasis. Mol. Microbiol. 2006, 60, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yang, W.; Liu, Y.; Li, W.; Wang, Y.; Liu, N.; Sheng, C. Jumonji Histone Demethylase Inhibitor JIB-04 as a Broad-Spectrum Antifungal Agent. ACS Infect. Dis. 2022, 8, 1316–1323. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, M.N.; Rai, R. H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host. Pathogens 2024, 13, 1080. https://doi.org/10.3390/pathogens13121080
Rai MN, Rai R. H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host. Pathogens. 2024; 13(12):1080. https://doi.org/10.3390/pathogens13121080
Chicago/Turabian StyleRai, Maruti Nandan, and Rikky Rai. 2024. "H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host" Pathogens 13, no. 12: 1080. https://doi.org/10.3390/pathogens13121080
APA StyleRai, M. N., & Rai, R. (2024). H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host. Pathogens, 13(12), 1080. https://doi.org/10.3390/pathogens13121080





