From Stress Tolerance to Virulence: Recognizing the Roles of Csps in Pathogenicity and Food Contamination
Abstract
1. Introduction
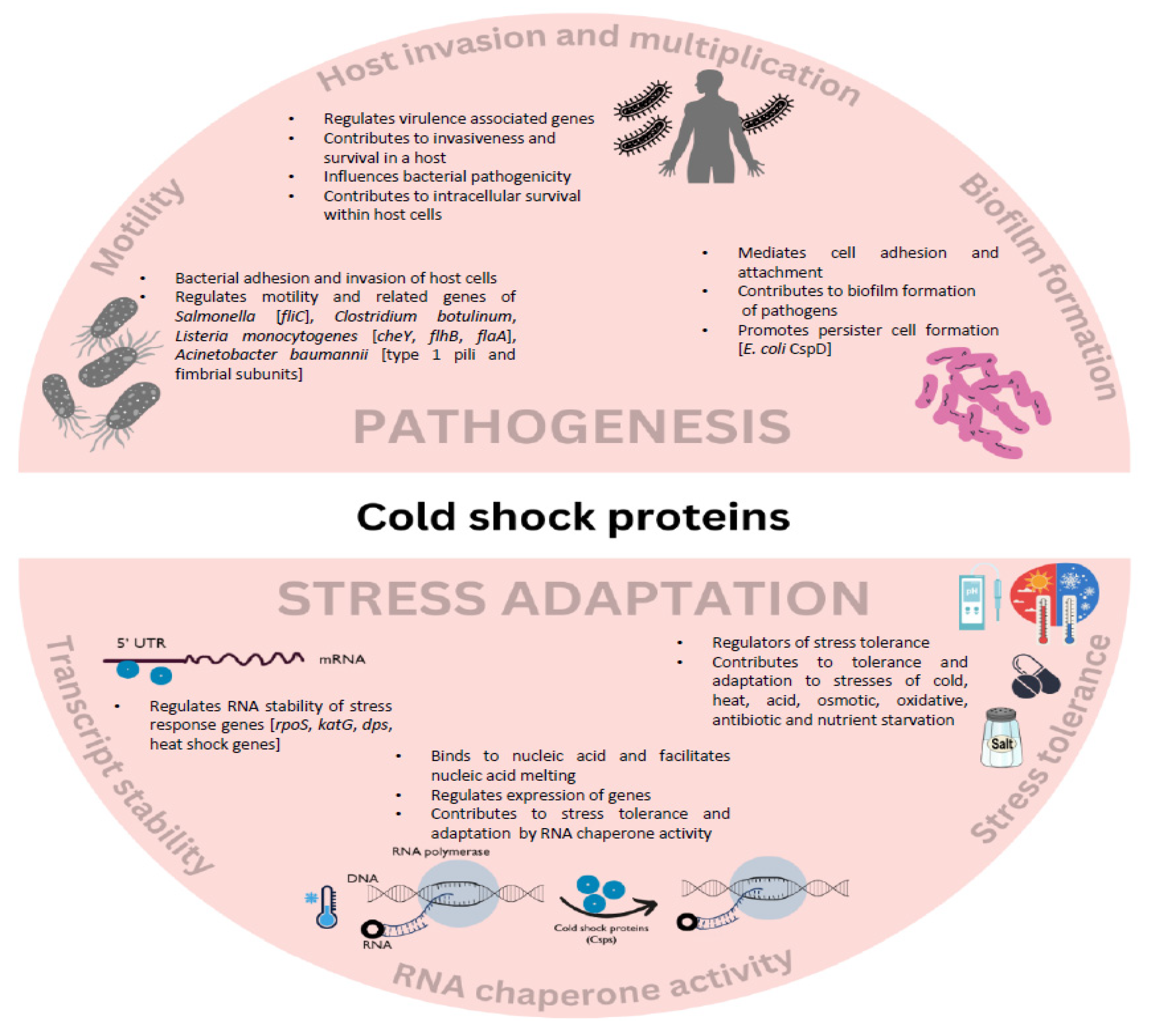
2. Cold Shock Proteins in Virulence and Infection
2.1. Csps Mediate Virulence via Regulation of Stress Tolerance
2.2. Csps Influence Invasiveness of Pathogens
2.3. Csps Regulate Motility-Related Factors and Biofilm Formation
2.4. Csps of Plant Pathogens
3. Involvement of Csps in Food Contamination
3.1. Csps Impact Bacterial Survival under Food Preservation and Disinfection Strategies
3.2. Csps Influence Pathogenesis by Means of Stress Adaptation
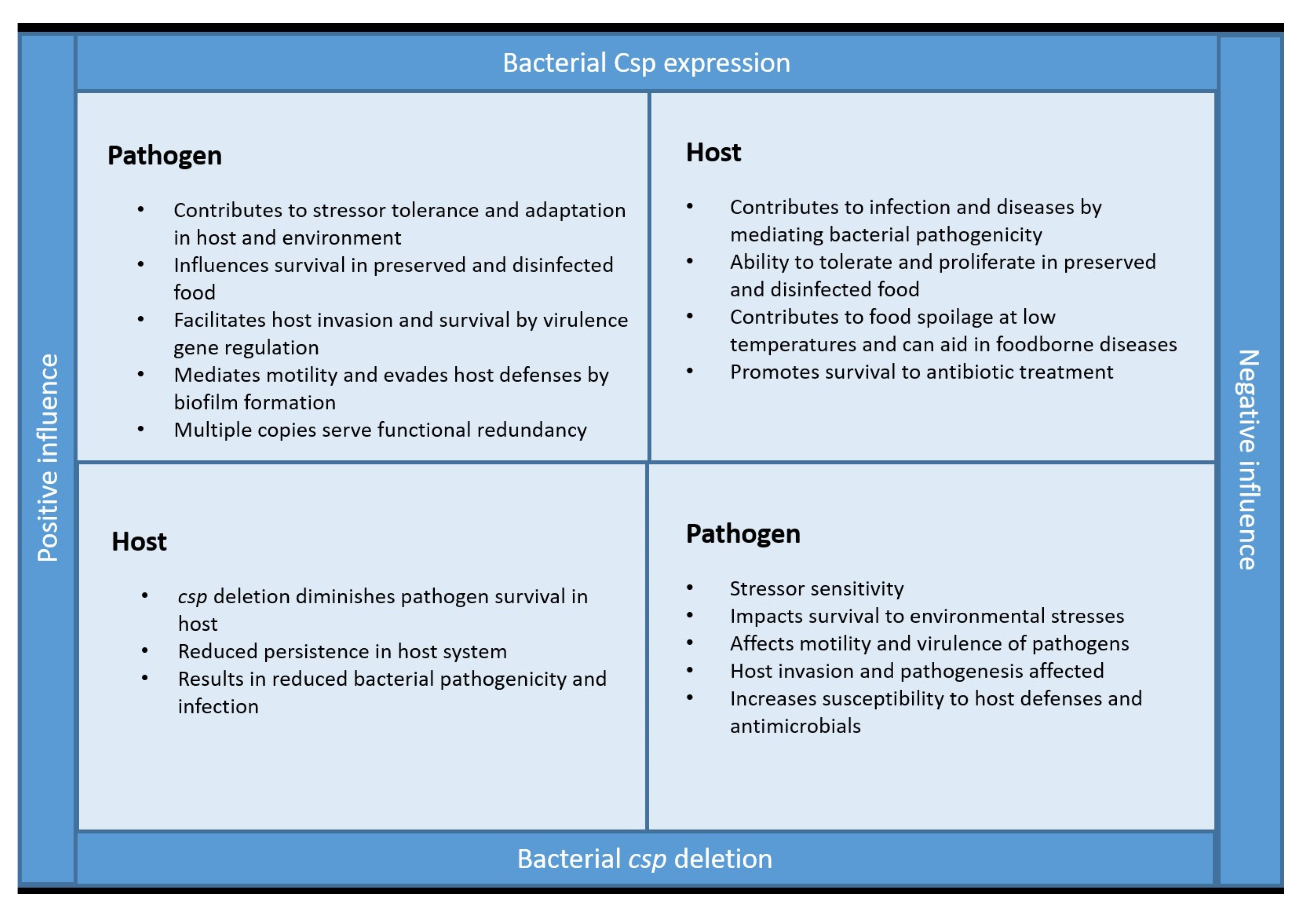
4. Are Csps the Good or Bad Guys?
5. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Graumann, P.; Marahiel, M.A. Some like it cold: Response of microorganisms to cold shock. Arch. Microbiol. 1996, 166, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of Proteins in Response to Low Temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Giuliodori, A.M.; Pon, C.L. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003, 331, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Horn, G.; Hofweber, R.; Kremer, W.; Kalbitzer, H.R. Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 2007, 64, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Keto-Timonen, R.; Jiang, X.; Virtanen, J.P.; Korkeala, H. Insights into the phylogeny and evolution of cold shock proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria. Int. J. Mol. Sci. 2019, 20, 4059. [Google Scholar] [CrossRef] [PubMed]
- Graumann, P.; Marahiel, M.A. A case of convergent evolution of nucleic acid binding modules. BioEssays 1996, 18, 309–315. [Google Scholar] [CrossRef]
- Schindelin, H.; Jiang, W.; Inouye, M.; Heinemann, U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 5119–5123. [Google Scholar] [CrossRef]
- Newkirk, K.; Feng, W.; Jiang, W.; Tejero, R.; Emerson, S.D.; Inouye, M.; Montelione, G.T. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: Identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. USA 1994, 91, 5114–5118. [Google Scholar] [CrossRef]
- Phadtare, S.; Tyagi, S.; Inouye, M.; Severinov, K. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002, 277, 46706–46711. [Google Scholar] [CrossRef]
- Phadtare, S.; Inouye, M.; Severinov, K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002, 277, 7239–7245. [Google Scholar] [CrossRef]
- Jiang, W.; Hou, Y.; Inouye, M. CspA, the Major Cold-shock Protein of Escherichia coli, Is an RNA Chaperone. J. Biol. Chem. 1997, 272, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K. Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1999, 1, 193–202. [Google Scholar] [PubMed]
- Herschlag, D. Minireview RNA Chaperones and the RNA Folding Problem. J. Biol. Chem. 1995, 270, 20871–20874. [Google Scholar] [CrossRef]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018, 16, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Graumann, P.L.; Marahiel, M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998, 23, 286–290. [Google Scholar] [CrossRef]
- Heinemann, U.; Roske, Y. Cold-Shock Domains—Abundance, Structure, Properties, and Nucleic-Acid Binding. Cancers 2021, 13, 190. [Google Scholar] [CrossRef]
- Budkina, K.S.; Zlobin, N.E.; Kononova, S.V.; Ovchinnikov, L.P.; Babakov, A.V. Cold Shock Domain Proteins: Structure and Interaction with Nucleic Acids. Biochemistry 2020, 85, S1–S19. [Google Scholar] [CrossRef]
- Nakaminami, K.; Karlson, D.T.; Imai, R. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. USA 2006, 103, 10122–10127. [Google Scholar] [CrossRef]
- Chaikam, V.; Karlson, D.T. Comparison of structure, function and regulation of plant cold shock domain proteins to bacterial and animal cold shock domain proteins. BMB Rep. 2010, 43, 1–8. [Google Scholar] [CrossRef]
- Kohno, K.; Izumi, H.; Uchiumi, T.; Ashizuka, M.; Kuwano, M. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays 2003, 25, 691–698. [Google Scholar] [CrossRef]
- Sasaki, K.; Imai, R. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2012, 2, 116. [Google Scholar] [CrossRef]
- Ke, B.; Fan, C.; Tu, W. The Role of Y-Box Binding Protein 1 in Kidney Injury: Friend or Foe? Cell. Physiol. Biochem. 2018, 2, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Fang, L.; Inouye, M. The CspA family in Escherichia coli: Multiple gene duplication for stress adaptation. Mol. Microbiol. 1998, 27, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Stübs, D.; Fuchs, T.M.; Schneider, B.; Bosserhoff, A.; Gross, R. Identification and regulation of cold-inducible factors of Bordetella bronchiseptica. Microbiology 2005, 151, 1895–1909. [Google Scholar] [CrossRef] [PubMed]
- Graumann, P.; Wendrich, T.M.; Weber, M.H.W.; Schröder, K.; Marahiel, M.A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 1997, 25, 741–756. [Google Scholar] [CrossRef]
- Schmid, B.; Klumpp, J.; Raimann, E.; Loessner, M.J.; Stephan, R.; Tasara, T. Role of Cold Shock Proteins in Growth of Listeria monocytogenes under Cold and Osmotic Stress Conditions. Appl. Environ. Microbiol. 2009, 75, 1621–1627. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Zhang, W. RNA-Seq-Based Analysis of Cold Shock Response in Thermoanaerobacter tengcongensis, a Bacterium Harboring a Single Cold Shock Protein Encoding Gene. PLoS ONE 2014, 9, e93289. [Google Scholar] [CrossRef]
- Trun, N.; Johnston, D. Folding Chromosomes in Bacteria: Examining the Role of Csp Proteins and Other Small Nucleic Acid-Binding Proteins. Curr. Top. Dev. Biol. 2003, 55, 173–201. [Google Scholar] [CrossRef]
- Schärer, K.; Stephan, R.; Tasara, T. Cold shock proteins contribute to the regulation of listeriolysin O production in Listeria monocytogenes. Foodborne Pathog. Dis. 2013, 10, 1023–1029. [Google Scholar] [CrossRef]
- Eshwar, A.K.; Guldimann, C.; Oevermann, A.; Tasara, T. Cold-shock domain family proteins (Csps) are involved in regulation of virulence, cellular aggregation, and flagella-based motility in Listeria monocytogenes. Front. Cell Infect. Microbiol. 2017, 7, 453. [Google Scholar] [CrossRef]
- Muchaamba, F.; Stephan, R.; Tasara, T. Listeria monocytogenes Cold Shock Proteins: Small Proteins with A Huge Impact. Microorganisms 2021, 9, 1061. [Google Scholar] [CrossRef]
- Czapski, T.R.; Trun, N. Expression of csp genes in E. coli K-12 in defined rich and defined minimal media during normal growth, and after cold-shock. Gene 2014, 547, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef]
- Cardoza, E.; Singh, H. Involvement of CspC in response to diverse environmental stressors in Escherichia coli. J. Appl. Microbiol. 2021, 132, 785–801. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Severinov, K. RNA remodeling and gene regulation by cold shock proteins. RNA Biol. 2010, 7, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Sommerville, J. Activities of cold-shock domain proteins in translation control. BioEssays 1999, 21, 319–325. [Google Scholar] [CrossRef]
- Burd, C.C.; Dreyfuss, G. Conserved structure and diversity of functions of RNA-binding proteins. Science 1994, 265, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, K.; Hill, K.; Perry, S.E.; Sentoku, N.; Long, J.A.; Karlson, D.T. Arabidopsis cold shock domain proteins: Relationships to floral and silique development. J. Exp. Bot. 2009, 60, 1047–1062. [Google Scholar] [CrossRef]
- Park, S.J.; Kwak, K.J.; Oh, T.R.; Kim, Y.O.; Kang, H. Cold Shock Domain Proteins Affect Seed Germination and Growth of Arabidopsis thaliana Under Abiotic Stress Conditions. Plant Cell Physiol. 2009, 50, 869–878. [Google Scholar] [CrossRef]
- Fusaro, A.F.; Bocca, S.N.; Ramos, R.L.B.; Barrôco, R.M.; Magioli, C.; Jorge, V.C.; Coutinho, T.C.; Rangel-Lima, C.M.; De Rycke, R.; Inzé, D.; et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 2007, 225, 1339–1351. [Google Scholar] [CrossRef]
- Caballero, C.J.; Menendez-Gil, P.; Catalan-Moreno, A.; Vergara-Irigaray, M.; García, B.; Segura, V.; Irurzun, N.; Villanueva, M.; Mozos, I.R.D.L.; Solano, C.; et al. The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus. Nucleic Acids Res. 2018, 46, 1345–1361. [Google Scholar] [CrossRef]
- Lindquist, J.A.; Brandt, S.; Bernhardt, A.; Zhu, C.; Mertens, P.R. The role of cold shock domain proteins in inflammatory diseases. J. Mol. Med. 2014, 92, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, L.; Alidousty, C.; Djudjaj, S.; Frye, B.C.; Rauen, T.; Boor, P.; Mertens, P.R.; van Roeyen, C.R.; Tacke, F.; Heymann, F.; et al. YB-1 Is an Early and Central Mediator of Bacterial and Sterile Inflammation In Vivo. J. Immunol. 2013, 191, 2604–2613. [Google Scholar] [CrossRef]
- Wang, J.; Djudjaj, S.; Gibbert, L.; Lennartz, V.; Breitkopf, D.M.; Rauen, T.; Hermert, D.; Martin, I.V.; Boor, P.; Braun, G.S.; et al. YB-1 orchestrates onset and resolution of renal inflammation via IL10 gene regulation. J. Cell Mol. Med. 2017, 21, 3494–3505. [Google Scholar] [CrossRef] [PubMed]
- Anaganti, N.; Padwal, M.K.; Dani, P.; Basu, B. Pleiotropic effects of a cold shock protein homolog PprM on the proteome of Deinococcus radiodurans. Biochim. Biophys. Acta-Proteins Proteom. 2019, 1867, 98–106. [Google Scholar] [CrossRef]
- Yamanaka, K.; Zheng, W.; Crooke, E.; Wang, Y.H.; Inouye, M. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 2001, 39, 1572–1584. [Google Scholar] [CrossRef] [PubMed]
- Shenhar, Y.; Biran, D.; Ron, E.Z. Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ. Microbiol. Rep. 2012, 4, 532–539. [Google Scholar] [CrossRef]
- Shenhar, Y.; Rasouly, A.; Biran, D.; Ron, E.Z. Adaptation of Escherichia coli to elevated temperatures involves a change in stability of heat shock gene transcripts. Environ. Microbiol. 2009, 11, 2989–2997. [Google Scholar] [CrossRef]
- Phadtare, S.; Inouye, M. Role of CspC and CspE in Regulation of Expression of RpoS and UspA, the Stress Response Proteins in Escherichia coli. J. Bacteriol. 2001, 183, 1205–1214. [Google Scholar] [CrossRef]
- Kim, J.; Ha, S.; Park, W. Expression and deletion analyses of cspE encoding cold-shock protein E in Acinetobacter oleivorans DR1. Res. Microbiol. 2018, 169, 244–253. [Google Scholar] [CrossRef]
- Vouga, M.; Greub, G. Emerging bacterial pathogens: The past and beyond. Clin. Microbiol. Infect. 2016, 22, 12–21. [Google Scholar] [CrossRef]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef]
- Pokharel, P.; Dhakal, S.; Dozois, C.M. The Diversity of Escherichia coli Pathotypes and Vaccination Strategies against This Versatile Bacterial Pathogen. Microorganisms 2023, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- Josenhans, C.; Suerbaum, S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Ramos, H.C.; Rumbo, M.; Sirard, J.C. Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004, 12, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Flint, A.; Butcher, J.; Stintzi, A. Stress responses, adaptation, and virulence of bacterial pathogens during host gastrointestinal colonization. Virulence Mech. Bact. Pathog. 2016, 4, 385–411. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, X.; Zhang, X.S.; Grigoriu, S.; Page, R.; Peti, W.; Wood, T.K. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010, 12, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wood, T.K. Toxins Hha and CspD and Small RNA Regulator Hfq Are Involved in Persister Cell Formation Through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2011, 391, 209–213. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.; Wu, Q. Cold shock protein A plays an important role in the stress adaptation and virulence of Brucella melitensis. FEMS Microbiol. Lett. 2014, 354, 27–36. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, W.; Wu, T.; Bie, P.; Wu, Q. RNA-seq reveals the critical role of CspA in regulating Brucella melitensis metabolism and virulence. Sci. China Life Sci. 2016, 59, 417–424. [Google Scholar] [CrossRef]
- Ray, S.; Da Costa, R.; Thakur, S.; Nandi, D. Salmonella typhimurium encoded cold shock protein E is essential for motility and biofilm formation. Microbiology 2020, 166, 460–473. [Google Scholar] [CrossRef]
- Michaux, C.; Holmqvist, E.; Vasicek, E.; Sharan, M.; Barquist, L.; Westermann, A.J.; Gunn, J.S.; Vogel, J. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. USA 2017, 114, 6824–6829. [Google Scholar] [CrossRef]
- Loepfe, C.; Raimann, E.; Stephan, R.; Tasara, T. Reduced host cell invasiveness and oxidative stress tolerance in double and triple csp gene family deletion mutants of Listeria monocytogenes. Foodborne Pathog. Dis. 2010, 7, 775–783. [Google Scholar] [CrossRef]
- Kragh, M.L.; Muchaamba, F.; Tasara, T.; Hansen, L.T. Cold-shock proteins affect desiccation tolerance, biofilm formation and motility in Listeria monocytogenes. Int. J. Food Microbiol. 2020, 329, 108662. [Google Scholar] [CrossRef]
- Muchaamba, F.; von Ah, U.; Stephan, R.; Stevens, M.J.A.; Tasara, T. Deciphering the global roles of Cold shock proteins in Listeria monocytogenes nutrient metabolism and stress tolerance. Front. Microbiol. 2022, 13, 1057754. [Google Scholar] [CrossRef]
- Tomlinson, B.R.; Denham, G.A.; Torres, N.J.; Brzozowski, R.S.; Allen, J.L.; Jackson, J.K.; Eswara, P.J.; Shaw, L.N. Assessing the Role of Cold-Shock Protein C: A Novel Regulator of Acinetobacter baumannii Biofilm Formation and Virulence. Infect. Immun. 2022, 90, e0037622. [Google Scholar] [CrossRef]
- Duval, B.D.; Mathew, A.; Satola, S.W.; Shafer, W.M. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents Chemother. 2010, 54, 2283–2290. [Google Scholar] [CrossRef]
- Derman, Y.; Söderholm, H.; Lindström, M.; Korkeala, H. Role of csp genes in NaCl, pH, and ethanol stress response and motility in Clostridium botulinum ATCC 3502. Food Microbiol. 2015, 46, 463–470. [Google Scholar] [CrossRef]
- Söderholm, H.; Lindström, M.; Somervuo, P.; Heap, J.; Minton, N.; Lindén, J.; Korkeala, H. CspB encodes a major cold shock protein in Clostridium botulinum ATCC 3502. Int. J. Food Microbiol. 2011, 146, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Martini, C.; Shioya, K.; Lecheheb, S.A.; Budin-verneuil, A.; Cosette, P.; Sanguinetti, M.; Hartke, A.; Verneuil, N.; Giard, J.-C. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J. Bacteriol. 2012, 194, 6900–6908. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tan, X.; Cheng, H.; Gong, J.; Zhang, Y.; Wang, D.; Ding, W. The cold shock family gene cspD3 is involved in the pathogenicity of Ralstonia solanacearum CQPS-1 to tobacco. Microb. Pathog. 2020, 142, 104091. [Google Scholar] [CrossRef]
- Wei, W.; Sawyer, T.; Burbank, L. Csp1, a Cold Shock Protein Homolog in Xylella fastidiosa Influences Cell Attachment, Pili Formation, and Gene Expression. Microbiol. Spectr. 2021, 9, e01591-21. [Google Scholar] [CrossRef]
- Burbank, L.P.; Stenger, D.C. A Temperature-Independent Cold-Shock Protein Homolog Acts as a Virulence Factor in Xylella fastidiosa. Mol. Plant Microbe Interact. 2016, 29, 335–344. [Google Scholar] [CrossRef]
- Wu, L.; Ma, L.; Li, X.; Huang, Z.; Gao, X. Contribution of the cold shock protein CspA to virulence in Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 2019, 20, 382–391. [Google Scholar] [CrossRef]
- Patwa, L.G.; Fan, T.J.; Tchaptchet, S.; Liu, Y.; Lussier, Y.A.; Sartor, R.B.; Hansen, J.J. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology 2011, 141, 1842–1851.e10. [Google Scholar] [CrossRef]
- Michaux, C.; Saavedra, L.F.R.; Reffuveille, F.; Bernay, B.; Goux, D.; Hartke, A.; Verneuil, N.; Giard, J.-C. Cold-shock RNA-binding protein CspR is also exposed to the surface of Enterococcus faecalis. Microbiology 2013, 159, 2153–2161. [Google Scholar] [CrossRef]
- Cohen-or, I.; Shenhar, Y.; Biran, D.; Ron, E.Z. CspC regulates rpoS transcript levels and complements hfq deletions. Res. Microbiol. 2010, 161, 694–700. [Google Scholar] [CrossRef]
- Duan, Q.; Zhou, M.; Zhu, L.; Zhu, G. Flagella and bacterial pathogenicity. J. Basic. Microbiol. 2013, 53, 1–8. [Google Scholar] [CrossRef]
- Sartor, R.B. Microbial Influences in Inflammatory Bowel Diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Lin, W.H.; Tseng, C.C.; Wu, A.B.; Wang, M.C.; Wu, J.J. The complex interplay among bacterial motility and virulence factors in different Escherichia coli infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2157–2162. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.K.; González Barrios, A.F.; Herzberg, M.; Lee, J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 72, 361–367. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Ray, S.; Da Costa, R.; Das, X.M.; Nandi, X.D. Interplay of cold shock protein E with an uncharacterized protein, YciF, lowers porin expression and enhances bile resistance in Salmonella typhimurium. J. Biol. Chem. 2019, 294, 9084–9099. [Google Scholar] [CrossRef]
- Yair, Y.; Michaux, C.; Biran, D.; Bernhard, J.; Vogel, J.; Barquist, L.; Ron, E.Z. Cellular RNA Targets of Cold Shock Proteins CspC and CspE and Their Importance for Serum Resistance in Septicemic Escherichia coli. mSystems 2022, 7, e00086-22. [Google Scholar] [CrossRef]
- Gould, G.W. Methods for preservation and extension of shelf life. Int. J. Food Microbiol. 1996, 33, 51–64. [Google Scholar] [CrossRef]
- Amit, S.K.; Uddin, M.M.; Rahman, R.; Islam, S.M.R.; Khan, M.S. A review on mechanisms and commercial aspects of food preservation and processing. Agric. Food Secur. 2017, 6, 51. [Google Scholar] [CrossRef]
- Leblanc, L.; Leroi, F.; Hartke, A.; Auffray, Y. Do stresses encountered during the smoked salmon process influence the survival of the spoiling bacterium Shewanella putrefaciens? Lett. Appl. Microbiol. 2000, 30, 437–442. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.A.S.; Xu, J.; Moore, J.E.; Blair, I.S.; Mcdowell, D.A. Environmental Stress and Antibiotic Resistance in Food-Related Pathogens. Appl. Environ. Microbiol. 2007, 73, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Tadigotla, V.; Shin, W.; Sengupta, A.; Severinov, K. Analysis of Escherichia coli Global Gene Expression Profiles in Response to Overexpression and Deletion of CspC and CspE. J. Bacteriol. 2006, 188, 2521–2527. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Inouye, M. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 1999, 33, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Ron, E.Z. Novel Interaction between the Major Bacterial Heat Shock Chaperone (GroESL) and an RNA Chaperone (CspC). J. Mol. Biol. 2013, 426, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Ke, H.; Inouye, M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 2001, 40, 179–188. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K. The link between bacterial radiation resistance and cold adaptation. J. Biosci. 2002, 27, 71–73. [Google Scholar] [CrossRef]
- Hossain, M.A.; Mostofa, M.G.; Fujita, M. Cross Protection by Cold-shock to Salinity and Drought Stress-induced Oxidative Stress in Mustard (Brassica campestris L.) Seedlings. Mol. Plant Breed. 2013, 4, 50–70. [Google Scholar] [CrossRef]
- García, S.; Limón, J.C.; Heredia, N.L. Cross protection by heat and cold shock to lethal temperatures in Clostridium perfringens. Braz. J. Microbiol. 2001, 32, 110–112. [Google Scholar] [CrossRef]
- Whiting, G.C.; Rowbury, R.J. Increased resistance of Escherichia coli to acrylic acid and to copper ions after cold-shock. Lett. Appl. Microbiol. 1995, 20, 240–242. [Google Scholar] [CrossRef]
- Wouters, J.A.; Rombouts, F.M.; Kuipers, O.P.; de Vos, W.M.; Abee, T. The Role of Cold-Shock Proteins in Low-Temperature Adaptation of Food-Related Bacteria. Syst. Appl. Microbiol. 2000, 23, 165–173. [Google Scholar] [CrossRef]
- Leblanc, L.; Leboeuf, C.; Leroi, F.; Hartke, A.; Auffray, Y. Comparison between NaCl tolerance response and acclimation to cold temperature in Shewanella putrefaciens. Curr. Microbiol. 2003, 46, 157–162. [Google Scholar] [CrossRef]
- Dahlsten, E.; Lindström, M.; Korkeala, H. Mechanisms of food processing and storage-related stress tolerance in Clostridium botulinum. Res. Microbiol. 2015, 166, 344–352. [Google Scholar] [CrossRef]
- Mishra, V.; Abrol, G.S.; Dubey, N. Sodium and Calcium Hypochlorite as Postharvest Disinfectants for Fruits and Vegetables. In Postharvest Disinfection of Fruits and Vegetables; Elsevier Inc.: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Feliziani, E.; Lichter, A.; Smilanick, J.L.; Ippolito, A. Disinfecting agents for controlling fruit and vegetable diseases after harvest. Postharvest Biol. Technol. 2016, 122, 53–69. [Google Scholar] [CrossRef]
- Sun, S.H.; Kim, S.J.; Kwak, S.J.; Yoon, K.S. Efficacy of sodium hypochlorite and acidified sodium chlorite in preventing browning and microbial growth on fresh-cut produce. Prev. Nutr. Food Sci. 2012, 17, 210–216. [Google Scholar] [CrossRef]
- De Corato, U. Improving the shelf-life and quality of fresh and minimally-processed fruits and vegetables for a modern food industry: A comprehensive critical review from the traditional technologies into the most promising advancements. Crit. Rev. Food Sci. Nutr. 2020, 60, 940–975. [Google Scholar] [CrossRef] [PubMed]
- Mei, G.Y.; Tang, J.; Bach, S.; Kostrzynska, M. Changes in gene transcription induced by hydrogen peroxide treatment of verotoxin-producing Escherichia coli O157:H7 and non-O157 serotypes on romaine lettuce. Front. Microbiol. 2017, 8, 477. [Google Scholar] [CrossRef] [PubMed]
- Mei, G.Y.; Tang, J.; Carey, C.; Bach, S.; Kostrzynska, M. The effect of oxidative stress on gene expression of Shiga toxin-producing Escherichia coli (STEC) O157: H7 and non-O157 serotypes. Int. J. Food Microbiol. 2015, 215, 7–15. [Google Scholar] [CrossRef]
- Al-nabulsi, A.A.; Osaili, T.M.; Shaker, R.R.; Olaimat, A.N.; Jaradat, Z.W.; Zain, N.A.; Elabedeen, N.A.Z.; Holley, R.A. Effects of osmotic pressure, acid, or cold stresses on antibiotic susceptibility of Listeria monocytogenes. Food Microbiol. 2015, 46, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Chanda, P.K.; Mondal, R.; Sau, K.; Sau, S. Antibiotics, arsenate and H2O2 induce the promoter of Staphylococcus aureus cspC gene more strongly than cold. J. Basic Microbiol. 2009, 49, 205–211. [Google Scholar] [CrossRef]
- Cardoza, E.; Singh, H. C Group-Mediated Antibiotic Stress Mimics the Cold Shock Response. Curr. Microbiol. 2021, 78, 3372–3380. [Google Scholar] [CrossRef]
- Cruz-Loya, M.; Kang, T.M.; Lozano, N.A.; Watanabe, R.; Tekin, E.; Damoiseaux, R.; Savage, V.M.; Yeh, P.J. Stressor interaction networks suggest antibiotic resistance co-opted from stress responses to temperature. ISME J. 2019, 13, 12–23. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Inouye, M. CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J. Bacteriol. 1999, 181, 1827–1830. [Google Scholar] [CrossRef]
- Jiang, W.; Jones, P.; Inouye, M. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J. Bacteriol. 1993, 175, 5824–5828. [Google Scholar] [CrossRef] [PubMed]
| Bacterium | Major Disease and Transmission | Csps * | Csp Expression | Δcsp Deletion | References |
|---|---|---|---|---|---|
| [A] Human pathogens | |||||
| Escherichia coli | UTI, pneumonia, bacteremia, abdominal and pelvic infection Part of normal microbiota. Transmission by contaminated food | 9 CspA-CspI | CspD: induction during starvation and oxidative stress; influences biofilm and persister cell formation | - | [57,58] |
| Brucella melitensis | Brucellosis, zoonosis (contaminated milk products or unpasteurized milk) | 4, CspA | Stress responses of acid, cold, oxidative | ΔcspA affected metabolism and virulence | [59,60] |
| Salmonella typhimurium | Gastroenteritis. Foodborne, or through contaminated environment | 6 CspA-E, CspH | Stress response to cold, oxidative, motility, and biofilm formation | ΔcspC and ΔcspE altered responses to stress, motility, biofilm, and virulence as well as affected host invasion and survival | [61,62] |
| Listeria monocytogenes | Meningitis and encephalitis. Transmission through contaminated food and mother-to-fetus | 3 CspA, CspB, CspD | Nutrient utilization and stress tolerance to cold, osmotic, and oxidative stress. | Deletion of csps impairs the utilization of C-sources and compromises cold, pH, and oxidative and osmotic stress tolerance. Mutants show reduced expression of virulence factors, are susceptible to antimicrobials, and are defective in motility, host invasion, and biofilm formation | [26,30,63,64,65] |
| Acinetobacter baumannii | Infection of the lung, blood, wound, and urinary tract. Person-to-person transmission | CspC | - | Hampers biofilm formation, survival, and multiplication in host | [66] |
| Staphylococcus aureus | Bacteremia, infective endocarditis, skin, and bone infections. Person-to-person transmission | 3, CspA, CspB, CspC | Stress response to cold | ΔcspA upregulated virulence and proteins related to pathogenesis. Downregulated stress response genes, including oxidative stress genes ΔcspB shows resistance and susceptibility to certain antimicrobials | [41,67] |
| Clostridium botulinum | Botulism. Transmission through dermal contact and contaminated food | 3 CspA, CspB, CspC | Stress response to cold; osmotic | ΔcspB and ΔcspC are sensitive to low pH, ethanol, and salt | [68,69] |
| Acinetobacter oleivorans DR1 | Infection of the lung, blood, wound, and urinary tract. Person-to-person transmission | 6 | CspA, CspB, CspC, CspE: cold adaptation CspE expression in antibiotic and alkane degradation and downregulation in paraquat and PMS | ΔcspE low-temperature growth defect and enhanced biofilm formation | [50] |
| Enterococcus faecalis | Endocarditis, UTI, bacteremia, intra-abdominal, and wound infection Person-to-person transmission | CspR | Cold shock response, stationary phase survival, role in virulence | ΔcspR is less virulent than the wild type | [70] |
| [B] Phytopathogens | |||||
| Ralstonia solanacearum CQPS-1 | Bacterial wilt | 4 | - | ΔcspD3 increased swimming motility and decreased virulence-associated genes and virulence potential | [71] |
| Xylella fastidiosa | Bacterial leaf scorch, phony peach disease, Pierce’s disease of grapes, citrus variegated chlorosis | Csp1 | Cold and salt stress adaptation | Δcsp1 impaired cell and surface attachment, biofilm, motility, and virulence | [72,73] |
| Xanthomonas oryzae | Bacterial leaf blight of rice | 4, CspA-D | Cold adaptation and virulence | ΔcspA affected biofilm and EPS production | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoza, E.; Singh, H. From Stress Tolerance to Virulence: Recognizing the Roles of Csps in Pathogenicity and Food Contamination. Pathogens 2024, 13, 69. https://doi.org/10.3390/pathogens13010069
Cardoza E, Singh H. From Stress Tolerance to Virulence: Recognizing the Roles of Csps in Pathogenicity and Food Contamination. Pathogens. 2024; 13(1):69. https://doi.org/10.3390/pathogens13010069
Chicago/Turabian StyleCardoza, Evieann, and Harinder Singh. 2024. "From Stress Tolerance to Virulence: Recognizing the Roles of Csps in Pathogenicity and Food Contamination" Pathogens 13, no. 1: 69. https://doi.org/10.3390/pathogens13010069
APA StyleCardoza, E., & Singh, H. (2024). From Stress Tolerance to Virulence: Recognizing the Roles of Csps in Pathogenicity and Food Contamination. Pathogens, 13(1), 69. https://doi.org/10.3390/pathogens13010069






