Abstract
Onchocerciasis is a neglected tropical disease targeted for elimination using ivermectin mass administration. Ivermectin kills the microfilariae and temporarily arrests microfilariae production by the macrofilariae. We genotyped 436 microfilariae from 10 people each in Ituri, Democratic Republic of the Congo (DRC), and Maridi County, South Sudan, collected before and 4–5 months after ivermectin treatment. Population genetic analyses identified 52 and 103 mitochondrial DNA haplotypes among the microfilariae from DRC and South Sudan, respectively, with few haplotypes shared between people. The percentage of genotype-based correct assignment to person within DRC was ~88% and within South Sudan ~64%. Rarefaction and extrapolation analysis showed that the genetic diversity in DRC, and even more so in South Sudan, was captured incompletely. The results indicate that the per-person adult worm burden is likely higher in South Sudan than DRC. Analyses of haplotype data from a subsample (n = 4) did not discriminate genetically between pre- and post-treatment microfilariae, confirming that post-treatment microfilariae are not the result of new infections. With appropriate sampling, mitochondrial haplotype analysis could help monitor changes in the number of macrofilariae in a population as a result of treatment, identify cases of potential treatment failure, and detect new infections as an indicator of continuing transmission.
1. Introduction
Over 120 million people are at risk of contracting onchocerciasis, a neglected tropical disease (NTD) caused by the filarial nematode Onchocerca volvulus and transmitted by blackflies in the genus Simulium, primarily in sub-Saharan Africa [1]. Onchocerciasis pathology includes skin depigmentation and lesions, ocular damage that can lead to blindness, and neurological–endocrine disorders such as epilepsy and Nakalanga syndrome [2,3,4]. The World Health Organization (WHO), in conjunction with government and non-government agencies, are targeting the elimination of O. volvulus transmission in endemic countries [5]. The strategy for interrupting transmission is the mass drug administration of ivermectin (MDAi). Ivermectin kills the larval stages (microfilariae) found in the skin of infected persons (microfilaricidal effect), and temporarily stops female adult worm reproduction (embryostatic effect). It is distributed to affected communities annually, biannually, or even quarterly. With each subsequent exposure to ivermectin, the fertility of female worms may be reduced, although the estimated degree of this cumulative effect ranges across studies [6,7,8,9]. The effects of long-term MDAi on skin microfilariae density and macrofilariae fertility have resulted in the WHO-verified elimination of transmission in most endemic areas in the Americas and may have also resulted in elimination in several endemic areas in sub-Saharan Africa [10,11,12,13,14]. These successes motivated shifting the goal from control as a public health problem to the elimination of onchocerciasis transmission [5,15].
One challenge facing the elimination of the transmission of O. volvulus with MDAi in sub-Saharan Africa is the variable response of adult worms to the embryostatic effect of ivermectin, with increases in skin microfilariae density post treatment beginning as early as a few weeks or as late as >1 year [16,17,18,19,20,21,22,23,24,25]. Repopulation of the skin with microfilariae 80 days post treatment, or the observation of stretched microfilariae in the uteri of adult female worms 80–90 days post treatment, has been referred to as a sub-optimal response (SOR) to ivermectin [16,17,18,19,20,21,22,23,24,26]. In addition, a sub-optimal microfilariae response (SOMR) has been described, in which the rate or extent of the initial reduction in skin microfilariae following ivermectin treatment is reduced [24,25,27]. The microfilaricidal and embryostatic effect of ivermectin may differ between geographic areas [20,24]. Thus, tracking community-specific changes in microfilaridermia and worm fertility is useful for the quantitative assessment of the effect of MDAi and/or for parameterizing models used to predict the optimal timing and duration of MDA for the successful elimination of transmission [28,29].
Current parasitological methods for assessing the effect of MDAi on an individual or community level [16,17,21,22] do not allow the estimation of the number of fertile adult worms in a host or in a community. Palpable subcutaneous nodules containing adult worms can be excised for analysis, but an unknown (and significant) number of worms are inaccessible deep in the body [30,31,32,33,34], and the number of individuals who can be sampled is limited by the fact that minor surgery is required. Estimates of the number of live female adult worms per person living in the West African savannah based on the statistical analyses of macrofilariae in excised nodules indicate high variability among hosts within the same community, ranging from approximately 4 to 177 [35,36] worms per person.
Our goal is to develop a tool that can quantitively measure progress towards reducing the number of fertile worms during MDAi (and complementary interventions). Because maternal sibling microfilariae sampled from the same person will have identical mitochondrial DNA sequences (or haplotypes) due to the strictly maternal nature of mitochondrial inheritance, we can estimate how many adult females (macrofilariae) are reproductively active by counting the number of unique mitochondrial haplotypes identified in the microfilariae sampled from the skin, assuming sister nematodes rarely infect the same person. Because a particular skin snip may not contain microfilariae offspring from all female worms in the person, the number of haplotypes detected will represent the minimum number of reproductively active female worms. However, assuming that the subset represents a random sample of the microfilariae in the host, changes over time in the number of haplotypes estimated should indicate whether MDAi is effectively reducing the number of reproductively active worms. More explicitly, the number of haplotypes present amongst the microfilariae should decline, as MDAi reduces transmission (and thus new infections) and adult females reach the end of their reproductive life span, thus providing a measure of progress towards the endpoint of zero fertile adult females.
We opportunistically utilized microfilariae collected in two studies whose focus was on onchocerciasis-associated epilepsy, one in the Logo Health zone in the Ituri province of the Democratic Republic of the Congo (DRC) and the other in Maridi county of South Sudan. People enrolled in the studies had epilepsy (potentially onchocerciasis-associated epilepsy) and were ivermectin-naïve in the Logo Health zone [37]. To estimate how many microfilariae per person would need to be sequenced to determine the minimum number of reproductively active adult female worms, we performed rarefaction and extrapolation analysis of mitochondrial haplotype diversity. We compared these estimates between sampling locations to explore differences in baseline infection intensity. We compared haplotype diversity between the samples from the two countries and between microfilariae obtained pre- and post-treatment for four people from whom both pre- and post-ivermectin microfilariae were available. Finally, we placed mitochondrial haplotype diversity identified in the microfilariae from DRC and South Sudan in the context of sequence data from parasites from other endemic areas in sub-Saharan Africa.
2. Materials and Methods
2.1. Origin of Microfilariae
Skin snips were collected from people with epilepsy in Maridi County in South Sudan and in the Logo Health Zone, Ituri Province, DRC in 2018 during epidemiological studies to investigate the association between onchocerciasis and epilepsy. Two snips were obtained from each participant at two time points: immediately before ivermectin treatment (“pre-treatment”) and 4 months (DRC) or 5 months (South Sudan) later (“post-treatment”). After the 24-h incubation of the snips in isotonic saline, the microfilaridermia was quantified microscopically [38,39,40] and samples were transferred to 80% ethanol and sent to La Trobe University, Victoria, Australia. In Maridi County, annual MDAi had been instituted since the early 2000s, but was interrupted for several years because of insecurity, and only reintroduced in 2017 with very low coverage (40.8%) [41]. At the time the skin snip was taken, each participant had either never taken ivermectin or had taken it only once. In the Logo Health zone, DRC, MDAi had never been instituted. A recent study concluded that today, Simulium dentulosum appears to be the main vector of human onchocerciasis in the area, and that Simulium vorax may be a secondary vector [42]. Ov16 IgG4 antibody positivity, determined with the Ov16 IgG4 Bioline rapid diagnostic test [43,44,45,46], between 2016 and 2018 was 0% (0/55) among 6-year-old and 7.1% (13/182) among 7–10-year-old children in the Logo Health zone [47]. In contrast, the Ov16 seroprevalence, determined with the same rapid test in an area in Maridi close to the blackfly breeding site, was 40% among children 3–6 years old and 66.7% among children 7–9 years old [48]. This suggests much higher ongoing O. volvulus transmission in Maridi compared to the Logo Health zone.
2.2. Origin of Adult Worms and Adult Worm Sequences
In 2016–2017, 27 adult female worms from the Centre Region and 12 from the Littoral Region in Cameroon were excised from nodules, as described in [20], and the heads (i.e., without uterine tissue) were placed in RNAlater (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).
Short-read data from adult worms were downloaded from the NCBI database from Benin, Côte d’Ivoire, Ghana, Guinea, Liberia, Mali, Sierra Leone, Liberia, Mali, and Uganda, project numbers PRJNA997216, PRJNA289926 [49], and PRJNA560089 [50].
2.3. DNA Extraction, Amplification and Sequencing
To minimize the transfer of host skin cell debris during DNA extraction, the microfilariae-containing ethanol solution was washed with Milli-Q® water (Merck, KGaA, Darmstadt, Germany) by carefully transferring the ethanol solution containing the microfilariae, but without skin snip, into a 15 mL falcon tube, filling to 15 mL with Milli-Q water, and centrifuging at 500× g for 5 min. Approximately 14 mL was aspirated carefully from the tube, leaving the microfilarial pellet in approximately 1ml of now diluted ethanol solution. This was transferred into a clean reusable glass petri dish (glass because microfilariae are less adhesive to glass compared to plastic). Each microfilaria was picked by aspiration using a pipette (0.1–2 µL) under a dissecting microscope into 20 µL lysis buffer (10mM Tris–HCl, pH 8.0; 1mM EDTA, pH 8.0; add 1% Tween®20 (Sigma-Aldrich, Burlington, MA, USA) and Proteinase K 300 µg/mL (recombinant, PCR Grade, Sigma-Aldrich) added just prior to use). The transfer of any visible cell debris was avoided while picking microfilariae. Tubes each containing a microfilaria in lysis buffer were incubated at 55 °C for 2 or 3 h, followed by heat inactivating the solution at 80 °C or 85 °C for 20 min. DNA from adult worms from Cameroon was extracted using the Isolate II Genomic DNA kit (Bioline, London, UK) as per the manufacturer’s instructions.
Quantitative PCR (qPCR) was performed on the microfilarial lysates to confirm the presence of mitochondrial DNA prior to whole genome amplification. qPCR reactions were performed using 2 µL of 1:5 diluted microfilarial lysates, 5 µL of SsoAdvanced Universal SYBR Green master mix (Bio-Rad Laboratories, Hercules, CA, USA), 2 µL of nuclease-free water, and 0.5 µL each of 10 µM forward (SP-Ov-mt-10062: 5′-attggtgaccaataaccttca-3′) and reverse (ASP-Ov-mt-10062: 5′-ttgattcaatatcagggacgta-3′) primers. A synthesized 68-bp oligonucleotide (ttg att caa tat cag gga cgt ata ttt cgt caa tct gag ttg act ttg aag gtt att ggt cac caa t) was used as a positive control and standard and HPLC water as a negative control for all reactions. Oligonucleotides were synthesized by Integrated DNA Technologies (Redwood City, CA, USA). qPCR assays were run on a CFX Real-Time System (Bio-Rad Laboratories), with an initial denaturing step of 3 min at 98 °C followed by 40 cycles (of 98 °C 10 s, 54 °C 15 s, 72 °C 15 s, read plate) including melt curve analysis at 65 °C to 95 °C for 5 s with an increment of 0.2 °C for 5 s. Each of the diluted lysates was assayed in duplicate, and the standards in triplicate. The overall statistics for the qPCR runs were assessed using the CFX Maestro Software (Bio-Rad Laboratories). The cycle threshold (Cq) values for each sample were determined as positive if the Cq < 30, and negative if Cq > 30 [51,52].
Microfilariae are relatively small in size (250–360 × 5–9 µm [29]), with low yield of DNA from a single microfilaria. A minimum amount of starting DNA is prerequisite to the successful generation of Illumina sequencing libraries. Whole genome amplification (WGA) was used as an intermediate step on microfilarial DNA lysates to generate high yields of amplified DNA as required for library construction. To generate a sufficient quantity of microfilarial DNA for Illumina library construction, high-fidelity, multiple displacement whole genome amplification (WGA) was performed on each microfilaria using 2 µL lysate as the starting material and processed according to the manufacturer’s instructions (REPLI-g, QIAGEN GmbH, Hilden, Germany). The WGA reactions were performed at 30 °C for 16 h and heat-inactivated at 65 °C for 10 min. Concentrations of the WGA DNA were determined using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
Sequencing libraries were constructed on samples whose DNA concentrations were above 1.0 ng/µL, using the Nextera DNA Flex/Illumina DNA prep library construction kits and barcoded using unique dual indexes, according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). An Agilent TapeStation analysis was performed on libraries using D1000 ScreenTapes (Agilent Technologies, Santa Clara, CA, USA) to ensure that the libraries were within the selected size range of 400–650 bp. The Qubit fluorometer was used to quantify the libraries for pooling. The final 4nM pooled library was spiked with 1% PhiX control (Illumina) and run on a NovaSeq SP, 300 cycles (i.e., 150 bp paired reads) at the Australian Genomic Research Facility (Melbourne, Australia) or on a NextSeq 500, 300 cycles, at the La Trobe Genomics Platform (Bundoora, Australia).
2.4. Variant Calling
Raw reads were trimmed using trimmomatic v.0.32 [53]. Trimmed reads were competitively mapped to the O. volvulus nuclear and mitochondrial genomes v.4 [54], the Wolbachia bacterial endosymbiont (GenBank accession number NZ_HG810405.1), and Homo sapiens (genome GRCh38.p13) using bwa v.0.7.17 [55,56]. Reads that mapped to H. sapiens were counted for quality control assessment (Supplementary Table S1) and discarded from the data using Unix command line programs grep and awk. Samtools v.1.9 [57] was used to remove secondary and supplementary reads, and only reads that mapped uniquely to O. volvulus were retained. If a microfilaria was sequenced in more than one experiment, the mapped reads were combined. Depth was assessed using bedtools v.2.26.0 [58] and coverage for each O. volvulus chromosome at a depth of at least 20× was estimated using a custom Perl script (Supplementary Table S1). Samples that did not meet a minimum coverage of 85% at a depth of 20× were not included in the downstream analyses. RepeatScout v1.0.5 [59] was used to identify repetitive regions across O. volvulus chromosomes.
For sequences from adult worms, variants were called on the mitochondrial genomes using GATK v.4.0.11 HaplotypeCaller using a minimum read quality filter of 30. For sequences from microfilariae, GATK v.4.2.6.1 was used, with de bruijn graph on. Variants for each sample were combined and GATK GenotypeGVCFs was used for final variant calling. These data were then filtered using GATK VariantFiltration for quality and depth (-filter “QD < 2.0” --filter-name “QD2” -filter “QUAL < 30.0” --filter-name “QUAL30” -filter “SOR > 3.0” --filter-name “SOR3” -filter “FS > 60.0” --filter-name “FS60” -filter “MQ < 40.0” --filter-name “MQ40” -filter “MQRankSum < −12.5” --filter-name “MQRankSum-12.5” -filter “ReadPosRankSum < −8.0” --filter-name “ReadPosRankSum-8” -filter “DP > 20” --filter-name “DP20”).
Variants were also called using freebayes v.1.0.2 [60], and filtered for quality and depth using vcftools v.0.1.13 [61]. Variant calls were normalized using bcftools v.1.2 [62] and haplotypes were simplified using the vcfallelicprimitives function in vcflib [63]. Bcftools was then used to find the intersection between the calls made by GATK and those made by freebayes. Finally, vcftools was used to further filter the dataset to remove individuals with less than 99% of variants called, to remove sites with missing data, and to identify and remove indels, singletons, and repeat regions.
2.5. Data Analysis
The R package adegenet v.2.1.5 was used to produce principal component analysis (PCA) plots of the genetic diversity within people at each of the different collection time points and to cluster the worms [64,65]. Discriminant analysis of principal components (DAPC [66]), which maximizes the differences between predefined groups, was used to estimate the probability that a microfilaria would be correctly identified to person or to date of collection based on genotype. The number of PCs to use in DAPC was estimated using cross-validation.
Rarefaction curves using microfilarial haplotypes from people from whom more than 10 microfilariae were successfully sequenced were produced using the R package iNEXT v.3.0.0 [67]. Vegan v.2.5–5 [65,68] was used to produce extrapolated estimates of total haplotype richness and associated standard error based on the Chao and abundance-based accumulated estimators, adjusted for sample bias [69,70,71].
SNPEff v.4.1l [72] was used to assess whether variants were in coding or noncoding regions and to calculate their effects on known genes (e.g., amino acid changes).
PGDSpider v.2.1.1.3 for Windows [73] was used to convert vcf to nexus format, which was then edited to add a “trait” block indicating person or country of origin. PopArt v.1.7 [74] was used to generate haplotype networks using the TCS approach [75], calculate diversity statistics, and to perform an analysis of molecular variance (AMOVA; [76]).
3. Results
3.1. Sequencing Results
Out of 804 sequenced in total, 225 microfilariae from 10 people from the DRC and 211 microfilariae from 10 people from South Sudan successfully passed the filtering criteria and were included in the analyzed data (Table 1). Of the successfully sequenced microfilariae from the DRC, 207 were collected before treatment (D0); 15 and 3 microfilariae collected four months post treatment, were, respectively, from two persons from whom we do not have pre-treatment mtDNA sequences. The dataset from South Sudan includes 163 microfilariae collected before treatment and 48 collected around five months post treatment, with four people having parasites successfully sequenced from both pre- and post-treatment microfilariae samples.
3.2. Genetic Variation of Microfilariae within People
After merging and filtering, 439 variants (places in the genome where the base differs from the reference sequence) in the 13,744 bp mitochondrial genome were called, of which 119 were singletons (i.e., found in only one worm). Because singletons could be due to experimental error (e.g., the introduction of variation during whole genome amplification) rather than biological variation, we excluded these when determining the number of mitochondrial haplotypes represented in each sample, leaving 320 variants.
The number of mitochondrial haplotypes identified for the microfilariae from a person represents the minimum number of reproductively active female worms in that person (Table 1), since there may be unidentified haplotypes from microfilariae either not present in the skin snip, lost during transfer of the skin snip to ethanol, or not successfully sequenced.

Table 1.
Number of Onchocerca volvulus mitochondrial haplotypes estimated per person in the study areas in the Democratic Republic of Congo (DRC) and South Sudan. The mean counts of microfilariae from two skin snips, the number of microfilariae successfully sequenced and in the final dataset, and the number of haplotypes before and after treatment are given. The total number of parasite haplotypes per person are also indicated, as this number will be different from the sum of the pre-treatment and post-treatment haplotypes when haplotypes are found in both samples. Totals for each country represent the total number of unique pre- and post-treatment haplotypes used in the analysis; the total number in parentheses indicates the number of unique haplotypes across all worms, as some haplotypes were found in more than one person.
Table 1.
Number of Onchocerca volvulus mitochondrial haplotypes estimated per person in the study areas in the Democratic Republic of Congo (DRC) and South Sudan. The mean counts of microfilariae from two skin snips, the number of microfilariae successfully sequenced and in the final dataset, and the number of haplotypes before and after treatment are given. The total number of parasite haplotypes per person are also indicated, as this number will be different from the sum of the pre-treatment and post-treatment haplotypes when haplotypes are found in both samples. Totals for each country represent the total number of unique pre- and post-treatment haplotypes used in the analysis; the total number in parentheses indicates the number of unique haplotypes across all worms, as some haplotypes were found in more than one person.
| Country | Person | Mean Number of Microfilariae Pre/Post [37] | Number of Microfilariae in Dataset Pre/Post | Number of Haplotypes Pre/Post (Total) |
|---|---|---|---|---|
| DRC | OAE015 | 48.5/3 | 0/15 | 0/4 (4) |
| OAE073 | 191/0 | 55/0 | 13/0 (13) | |
| OAE121 | 295.5/0 | 55/0 | 13/0 (13) | |
| OAE185 | 18.5/29 | 2/0 | 1/0 (1) | |
| OAE195 | 12.5/0 | 2/0 | 2/0 (2) | |
| OAE203 | 28/0 | 8/0 | 2/0 (2) | |
| OAE209 | 370.5/0 | 60/0 | 6/0 (6) | |
| OAE228 | 40/0 | 9/0 | 6/0 (6) | |
| OAE304 | 9/4 | 3/0 | 2/0 (2) | |
| OAE369 | 52.5/0 | 16/0 | 7/0 (7) | |
| All DRC | 210/15 | 52/4 (52) | ||
| South Sudan | K014 | 16.5/20.5 | 0/1 | 0/1 (1) |
| K028 | 78.5/20 | 27/0 | 16/0 (16) | |
| K029 | 57/143 | 0/1 | 0/1 (1) | |
| K038 | 23.5/24 | 3/0 | 3/0 (3) | |
| K096 | 108.5/1.5 | 9/0 | 8/0 (8) | |
| M204 | 59.5/46.5 | 27/10 | 13/8 (17) | |
| M206 | 105.5/62 | 33/21 | 19/ 16 (28) | |
| M219 | 45.5/64 | 13/0 | 8/0 (8) | |
| M224 | 23.5/51 | 28/9 | 15/7 (20) | |
| M238 | 23/55 | 23/6 | 14/5 (18) | |
| All South Sudan | 163/48 | 96/38 (103) | ||
| Totals | 373/63 | 148/42 (155) |
A rarefaction curve indicates how the number of haplotypes identified changes given the number of successfully sequenced microfilariae, and the shape of the curve indicates the extent to which the number of microfilariae sequenced effectively captures the genetic diversity within the population investigated, i.e., the microfilariae within an individual person (host). The rarefaction curves for samples from 12 individuals (those with microfilariae counts ≥10; Figure 1) suggest that, overall, the number of microfilariae successfully sequenced captured the genetic diversity in the parasite population in the DRC study area (green) to a larger extent than that in the South Sudan study area (purple). Extrapolated estimates for the total number of haplotypes predicted per person further suggest that the predicted number of reproductively active female worms (observed+unobserved) is greater in hosts from the South Sudan study area (average across people based on Chao estimate: 29.4 haplotypes per person; based on abundance-based accumulation: 36.4) when compared to the DRC (9.18 and 13.4, respectively), with highly variable associated standard errors (Table 2).
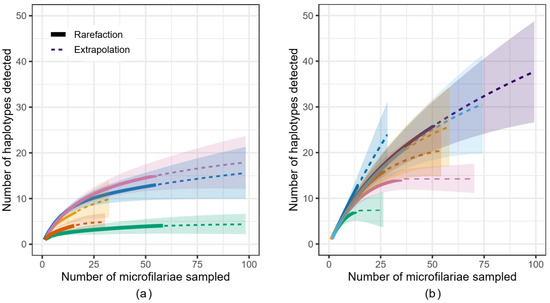
Figure 1.
Rarefaction curve indicating how the number of Onchocerca volvulus microfilariae sampled from (a) five individual hosts from the DRC and (b) seven hosts from the South Sudan study areas affects the number of haplotypes likely to be observed. Solid line: rarefaction; dotted line: extrapolation; shaded area: confidence interval.

Table 2.
Extrapolated mitochondrial haplotype richness estimates with standard error (SE) for Onchocerca volvulus microfilariae sampled from participants in the DRC and South Sudan. Table indicating the sample size (number of microfilariae sequenced), observed number of haplotypes, extrapolated number of haplotypes based on the Chao [69] and the abundance-based coverage estimator (ACE), adjusted for sample bias [70,71].
We used several different methods for assessing and comparing mtDNA genetic diversity in the microfilariae from the DRC and South Sudan locations. Because the sampling of the parasite population was non-random (i.e., the repeated sampling of sibling microfilariae in each host is non-representative of the population of parasites across all hosts), we estimated genetic diversity statistics using the set of unique maternal haplotypes rather than individual microfilariae sequences. Nucleotide diversity (π: the average number of pairwise nucleotide differences per site) of haplotypes from the DRC was 0.0198, and that of haplotypes from South Sudan was similar at 0.0212. Tajima’s D (the difference between diversity estimates derived from the mean number of pairwise differences and the number of variant sites) was negative, but not significantly so, in both populations (DRC: D = −2.591, p > 0.999; South Sudan: D = −2.609, p > 0.999). A simple, non-nested AMOVA calculated a fixation index of ΦST = 0.094 (p = 0.011) in the DRC and ΦST = 0.0351 (p = 0.1) in South Sudan. This indicates that in the DRC, most of this variation was partitioned within individual people (90.59%), with minor variation among people (9.41%). In South Sudan, this pattern of variation was even stronger (within individuals: 96.73%; between individuals 3.27%). In other words, the genetic differentiation between the microfilariae present in different people was marginally higher in the DRC than in South Sudan, but in both populations, nearly all of the genetic diversity was within the individual hosts (as in other parasites, such as the filarial nematode that causes lymphatic filariasis, Wuchereria bancrofti [77,78]).
We visualized the relationships among the microfilariae using haplotype networks, which indicate how similar specific haplotypes are to each other. There are fewer haplotypes observed across the 225 microfilariae from 10 people from the DRC (Figure 2a) than observed across the 211 microfilariae from 10 people in South Sudan (Figure 2b), and the network is correspondingly simpler. There are two haplotypes that form central nodes in the South Sudan haplotype network that are shared by microfilariae from multiple people (Figure 2b).
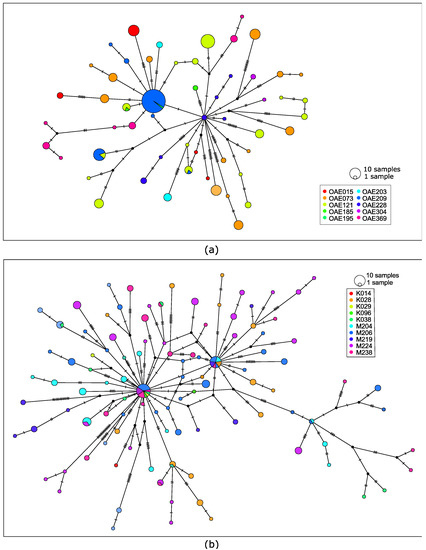
Figure 2.
(a) Haplotype network using 143 mitochondrial single-nucleotide polymorphic variants from 225 microfilariae collected from 10 people from the DRC. (b) Haplotype network based on 228 genetic variants from 211 microfilariae collected from 10 people from South Sudan. Each circle represents a haplotype and is colored based on person; circle size indicates the number of microfilariae with that haplotype. Hatch marks along connecting lines indicate the number of sequence differences between haplotypes.
We also explored whether genetic diversity was clustered by infrapopulation (the microfilariae within an individual host). The PCA of the samples from DRC suggests that there is considerable overlap in genetic diversity across microfilariae from different people (Figure 3a), but with only a few shared haplotypes (Figure 2a). DAPC is an analytical approach that maximizes differentiation among groups. A DAPC of the mitochondrial variants from DRC (using the first 80 PCs as determined by cross-validation) indicated that worms sampled from different people could be genetically differentiated (Figure 3b). While there are a few shared haplotypes, the proportion of individual microfilariae that could be correctly assigned to their host, based on their mitochondrial genotype, is 0.8756. The assignment proportion does not necessarily serve as a metric for similarity among worms within a person; rather, it indicates how different the worms sampled are between people. The higher the assignment proportion, the more distinct the parasite genotypes present in different people.
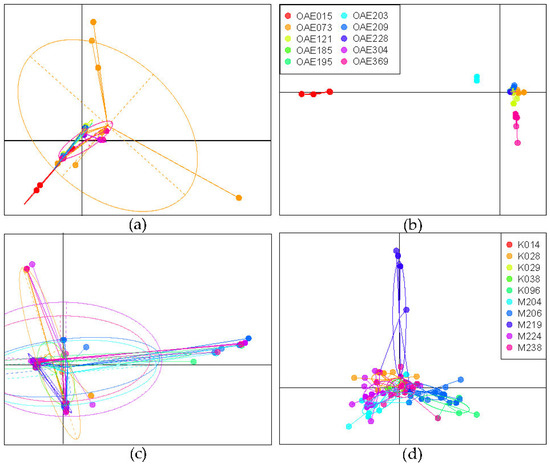
Figure 3.
Analysis of genetic differentiation based on 320 mitochondrial DNA variant sites sequences of microfilariae collected from people in the DRC and South Sudan. (a) Principal components analysis (PCA) of microfilariae genotyped from 10 people from the DRC, colored by host (as in (b)); (b) discriminant analysis of principal components (DAPC) for microfilariae from DRC, maximizing differentiation between hosts; (c) PCA of microfilariae from 10 people from South Sudan, colored by host (as in (d)); (d) DAPC of microfilariae from South Sudan, maximizing differentiation between hosts.
The PCA of mitochondrial genetic variation in the South Sudan microfilariae similarly suggests overlap in genetic diversity across the microfilariae in different hosts (Figure 3c), with a couple of haplotypes found in several people (Figure 2b). DAPC (using the first 60 PCs) was less able to discriminate among the parasites from different people compared to the DAPC performed on the DRC samples (Figure 3d), and the assignment proportion for individual microfilariae to the infected person from which they came was only 0.641, indicating that discriminating infrapopulations based on maternal mitochondrial haplotype was not feasible.
We focused further analysis on the 98 microfilarial haplotypes in the four people from South Sudan from whom we had both pre- and post-ivermectin treatment samples. It was not possible to discriminate haplotypes based on whether they were collected prior to or following treatment: haplotypes found before treatment (gray) were also found post-treatment (black) and some of the haplotypes were found in more than one person. Figure 4a visually demonstrates this genetic overlap between microfilariae haplotypes, and Figure 4b indicates the inability of the discriminant function to differentiate between pre- and post-treatment haplotypes: each haplotype identified (columns) has a similar probability of being assigned to either the pre-treatment (gray) or post-treatment (black) category based on their genotype. When discriminating based on human host and the day sampled, the post-treatment haplotypes largely overlapped with the pre-treatment haplotypes sampled from the same individual, although haplotypes from different individuals were moderately differentiated (Figure 4c,d).
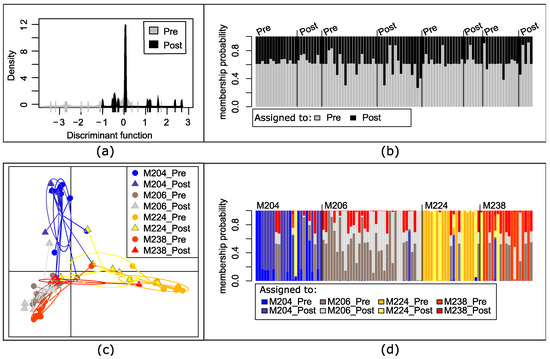
Figure 4.
Analysis of genetic differentiation based on the mitochondrial genotypes of microfilariae collected from four people in South Sudan with both pre- and post-treatment samples. (a) Discriminate analysis of principal components (DAPC) by time collected: pre-treatment (pre) or 5 months post-treatment (post). (b) Each column represents a mitochondrial haplotype found within a person and the shading the probability that haplotype would be assigned to time of collection based on the DAPC, with the actual day collected indicated above and columns arranged by individual person, as in (d); (c) DAPC of haplotypes maximizing differentiation between each individual host and by day collected; (d) each column represents a haplotype within a person (M204, M206, M224, or M238) and the shading the probability that it would be assigned to the pre- or post-treatment sample of one of the four individuals.
3.3. Genetic Variation in the African Context
Maternal haplotype sequences from the DRC and South Sudan mf data (i.e., retaining only one copy of a haplotype if it was found in multiple people and/or at multiple time points) were combined with sequences from Guinea (n = 4), Sierra Leone (n = 3), Liberia (n = 1), Mali (n = 10), Côte d’Ivoire (n = 13), Ghana (n = 189), Benin (n = 1), Cameroon (n = 39), and Uganda (n = 2) [49,50] for a total of 438 O. volvulus. After filtering, there were 851 single-nucleotide polymorphic variants called in total; of these, 427 were singletons. For downstream analyses, the singletons were removed.
Of the 424 polymorphic variants in the 13,744 bp mitochondrial genome, 139 variants were nonsynonymous (i.e., resulting in a change in the amino acid in the gene product), 208 were synonymous (i.e., a nonsynonymous:synonymous ratio across all genes of 0.668), 4 were nonsense mutations that would truncate the predicted gene product, and the remaining were in non-coding regions. The overall nucleotide diversity was 0.0111. The reduction in nucleotide diversity compared to that estimated from microfilariae from the DRC and South Sudan was largely driven by low nucleotide diversity among the 221 West African worms (π = 0.0064). Most of the variance in this diversity was within countries rather than between them: a simple non-nested AMOVA resulted in a fixation index of ΦST = 0.0255 (p = 0.009; significance based on 1000 permutations), where only 2.55% variance was among countries and 97.45% was within country.
Based on PCA, the haplotype diversity observed in South Sudan and DRC contained some sequence variation that overlapped with mitochondrial sequences obtained from adult female worms from West and Central Africa, and some sequence variation that was quite distinct (Figure 5a; consistent with the haplotype network presented in Figure S1). The results of a DAPC including only parasites from those countries where at least 10 parasites were sampled (Cameroon, Côte d’Ivoire, DRC, Ghana, Mali, and South Sudan), using the first 70 PCAs as determined through cross-validation, was visually consistent with the results of the PCA (Figure 5b). Assignment proportions (the proportion of parasites correctly assigned to their country of origin) were higher for parasites from Cameroon (0.90), DRC (0.94), and South Sudan (0.81) than parasites from Ghana (0.73), Côte d’Ivoire (0.08), and Mali (0.40) (Figure 5c). The very low proportions for Côte d’Ivoire and Mali could be due to fewer haplotypes being available compared to the number of sequences from countries with higher assignment proportions (i.e., an analytical bias). Consideration has to be given to the possibility that parasites in countries in West Africa are more closely related overall (due to the migration of hosts or vectors and/or the more recent divergence of those populations in evolutionary history) and thus less able to be discriminated using mitochondrial data alone.
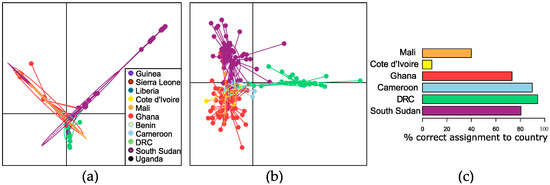
Figure 5.
(a) Principal component analysis of Onchocerca volvulus worms from Benin, Cameroon, Côte d’Ivoire, Democratic Republic of Congo (DRC), Ghana, Guinea, Liberia, Mali, Sierra Leone, South Sudan, and Uganda based on mitochondrial genome sequencing. (b) Discriminant analysis of principal components (DAPC) of worms from Cameroon, Côte d’Ivoire, DRC, Mali, and South Sudan. (c) Percentage of worms from each country that were correctly assigned to their country of origin based on DAPC.
4. Discussion
Our results showed that the O. volvulus mitochondrial DNA haplotype diversity varied between people and that the number of haplotypes in a single person was in the range of 2–26. The number of haplotypes indicates the minimum number of adult female worms that produced the microfilariae in our analyses, and is within the range of previous estimates of the number of female adult worms per person in West African populations [35,36]. Furthermore, our analyses showed that: (1) the number of haplotypes identified increased with the number of microfilariae successfully sequenced (Figure 1), indicating that data from a higher number of microfilariae than was available to us is required to estimate the number of haplotypes within one host as well as within the human population to which the host belongs; (2) the total number of microfilariae that need to be successfully sequenced to capture the genomic diversity of their parents will vary depending on the number of adult parasites within a person and geographic location (Figure 1, Table 2); and, thus, that (3) the number of haplotypes we identified underestimates the number of reproductively active female worms in most people, more so in those from South Sudan than those from DRC. If a rarefaction curve begins to asymptote, then the microfilariae genotyped is more likely to be representative of the genetic diversity within that person. In South Sudan, the number of microfilariae per person that would need to be successfully sequenced to capture the genetic diversity in that parasite population appears to be higher than for people in the DRC (Figure 1). These differences were observed despite similar numbers of microfilariae sequenced from a similar number of hosts, which further suggests that the intensity of infection, or worm burden per person, is higher in the area sampled in South Sudan than in DRC. The differences in infection intensity may be related to differences in prevalence; in the cohorts studied here, 36.5% of people recruited for the study based in Ituri compared to 84.9% of participants from South Sudan were microfilaridermic [38,40]. However, because the participants did not represent a random sampling of onchocerciasis-infected people in either location, sampling parasites from additional people would be required to confirm what would be a significant epidemiological difference between the two areas. Differences in onchocerciasis prevalence are driven by differences in annual biting rates, which are, in turn, driven by ecological conditions favorable for blackfly breeding, the amount of time people spend in areas with different biting rates, and the presence and effectiveness of interventions [79,80,81].
The statistically significant fixation index (ΦST from AMOVA) of haplotypes from the DRC and South Sudan demonstrates that each individual infected person carries a genetically variable population of worms. In population genetic terms, each infected person carries their own infrapopulation that is drawn from a much larger pool of parasites present in the population as a whole (the metapopulation). Most of the genetic variation is partitioned within people, which means that the differentiation between infrapopulations is moderate, and stronger in the DRC than South Sudan. The difference between the DRC and South Sudan infrapopulation differentiation is driven by fewer haplotypes (i.e., fewer reproductively active females) detected per host from the DRC, and thus that each sampled host contained a smaller proportion of the total parasite population’s genetic diversity than hosts in South Sudan. Regardless, the higher within- than between-infrapopulation diversity supports the hypothesis that the parasite populations in each transmission zone—the geographic area over which parasites are transmitted and thus able to interbreed—are large, and the probability of parasites that have identical-by-descent mitochondrial haplotypes being transmitted to one person is low.
The O. volvulus populations sampled in DRC and South Sudan harbor population-associated genetic variants that can be used to discriminate many of the parasites collected from these locations from parasites found elsewhere in sub-Saharan Africa. As shown in similar analyses of mitochondrial DNA data from O. volvulus [49,78], there is a widespread haplotype found in nearly all countries, which causes the overlap in genetic diversity observed (Figure 5; central node in Figure S1). This is consistent with large population sizes maintaining ancestral variation in mitochondrial genomes, which are under selective constraint to retain function.
For the four hosts from South Sudan from whom we had mitochondrial DNA sequences from microfilariae collected before as well as five months post-ivermectin treatment, we found that the haplotypes of the microfilariae collected post treatment were not distinct from those taken before treatment (Figure 4b). This shared genetic diversity indicates that the microfilariae present before and five months after ivermectin treatment are likely the offspring of those mothers that recovered fertility relatively rapidly. However, some haplotypes identified post treatment were not found in the pre-treatment sample. New post-treatment infections can be discounted as a source of microfilariae in the skin only five months post treatment, given that it takes 12–18 months for L3 larvae transmitted to mature into a reproductive adult worm and that it might take up to 3 years for the microfilariae of that adult to be sufficiently numerous to be detectable in skin snips [32,82,83,84]. An alternative source for post-treatment mitochondrial haplotypes not detected in a pre-treatment sample could be the random assortment of unique haplotypes into offspring from a heteroplasmic mother (i.e., a female worm with more than one mitochondrial haplotype). A likely interpretation is that sampling was not sufficient to detect all of the genetic diversity actually present within the host: some genotypes are missing from the pre-treatment sample because of under-sampling rather than true absence. We tested this hypothesis by producing a rarefaction curve, which indicates how the detection of unique mitochondrial sequences (or haplotypes) found in each person in both the DRC and South Sudan microfilariae increases with additional sequenced microfilariae (Figure 1). Under the assumption that post-treatment microfilariae would be a genetic subset of the pre-treatment worms, the results are consistent with the hypothesis that there was insufficient sampling of microfilariae from people in the South Sudan cohort to capture the genetic diversity present at the time of ivermectin treatment. Extrapolation may be useful for guiding researchers in determining when additional sequencing is required; while the extrapolated number of haplotypes varies depending on the assumptions made about detection of rare haplotypes [70,71], we found that in as many as three people (M206, M224, M238), only about half of the mitochondrial diversity was predicted to have been sampled (Table 2).
The analysis of parasite mitochondrial data to track changes in the number of reproductively active female worms could be useful for elimination programs and researchers working where there has been persistent transmission despite MDAi with high compliance and community coverage. High vector-biting rates can sustain transmission, even when skin microfilariae load is low. However, ongoing transmission might also be driven by parasites transmitted via vector or human movement from other onchocerciasis endemic areas or by SOR to ivermectin (reviewed in [85]). Changes in the estimated number of reproductively active worms over time could serve as an indicator of whether years of programmatic MDAi are successfully reducing the worm burden, or whether new imported infections (represented as new haplotypes) are identified. In this latter case, the genotyping of worms from areas with ongoing transmission that are likely to be connected by movement of people (as in [86,87]) or vectors (including long-distance, wind-assisted migration [88,89,90,91]) could help determine whether transmission among endemic areas might be contributing to persistent prevalence (see, e.g., [92,93] for modeling that explores the impact of movement of people or vectors on prevalence). We have shown here that mitochondrial haplotypes can discriminate among parasites from different countries, and, thus, may be informative where cross-border transmission might occur. Nuclear genotypes derived from the whole-genome sequencing of adult worms were able to discriminate O. volvulus collected in forest vs. savannah bioclimes within West Africa [49]; thus, the further development of cost-effective approaches for the nuclear genotyping of microfilariae would be useful for identifying transmission occurring between endemic areas.
In areas where SOR is suspected or has been demonstrated, monitoring changes in the number of reproductive SOR macrofilariae can help inform decisions about whether to deploy certain alternative and/or complementary interventions. SOR macrofilariae resume reproduction as soon as a few weeks after ivermectin treatment. Assuming there is no difference in the growth, survival, and probability of transmission of microfilariae that are the offspring of SOR and non-SOR macrofilariae, and assuming that the early resumption of reproduction is a heritable trait [20], the prevalence of reproducing SOR parasites will increase over time. This would jeopardize onchocerciasis control and elimination efforts and would require alternative intervention strategies. To date, the methods for identifying SOR parasites involve counting the microfilariae in skin snips and/or evaluating developmental stages in the uteri of macrofilariae excised from palpable subcutaneous nodules (where a fraction of the macrofilariae reside) soon (typically around 3 months) after ivermectin treatment [16,17,18,19,20,21,22,23,24,26]. Neither method can identify the number or percentage of SOR macrofilariae. A genetic approach applied to longitudinal, post-treatment samples of skin microfilariae or parasites in the vectors could indicate whether or not there is an increase in the number of SOR adult female worms producing those microfilariae repopulating the skin early, and, thus, whether alternative intervention strategies should be considered.
A major challenge for developing a genetics-based tool is the expense and sampling effort required to sufficiently capture the genetic diversity of reproducing parasites. A cost-effective approach would minimize the amount of effort needed to obtain microfilariae from skin snips, be able to sequence DNA from parasite pools, and ideally be suitable for use with blackfly pools. Long-read sequencing technologies (such as Oxford Nanopore) applied to amplified targets in the mitochondrial or nuclear genome would have the advantage of sequencing haplotypes even from DNA pools. Since national NTD programs are currently not set up for sequencing and population genetic analysis, collaboration with a research institution in the country, or building the required infrastructure and personnel capacity within the programs, would be needed.
Finally, the methods we present here may be useful, and easiest to implement, in studies evaluating the efficacy of new drugs, when it is important to know whether microfilariae appearing in the skin are due to macrofilariae reproductively active before treatment and that were not sterilized or killed by the new drug or whether they are due to new infections acquired after treatment [94]. In drug efficacy trials, four skin snips are usually taken before treatment and at varying time points after treatment. The mitochondrial haplotypes of the microfilariae sampled before and after treatment (timing dependent on the prior knowledge about the effect of the drug on the parasites) can be compared to estimate the effect of a drug on the burden of reproductively active female worms and on individual female worm fertility. Depending on the extent to which sampling captures haplotype diversity, samples taken >1 year after treatment could provide insight into the probability that the haplotypes only identified in post-treatment samples are due to new infections rather than due to parasites not affected by or having recovered from the effect of the drug. This is particularly important when the studies are conducted in areas with high transmission that could result in post-treatment infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12070971/s1, Figure S1: Haplotype network of mitochondrial haplotypes of Onchocerca volvulus collected in Benin, Cameroon, Côte d’Ivoire, Democratic Republic of Congo, Ghana, Guinea, Liberia, Mali, Sierra Leone, South Sudan, and Uganda; Table S1: Sequencing and mapping statistics for Onchocerca volvulus collected in sub-Saharan Africa.
Author Contributions
Conceptualization, S.M.H., Y.-J.C., A.C.K., M.M. (Makedonka Mitreva) and W.N.G.; validation, S.M.H. and Y.-J.C.; formal analysis, S.M.H.; investigation, S.M.H., A.K., G.C.C., W.N.G. and N.S.; resources, S.R.J., A.H., M.M. (Michel Mandro), J.N.S.F., R.C., R.A.A., G.N.A. and S.W.; data curation, S.M.H.; writing—original draft preparation, S.M.H.; writing—review and editing, all authors; visualization, S.M.H.; supervision, M.M. (Makedonka Mitreva) and W.N.G.; project administration, S.M.H., M.M. (Makedonka Mitreva) and W.N.G.; funding acquisition, W.N.G. and M.M. (Makedonka Mitreva). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (WHO/TDR), grant numbers B80149 and B80153, and the National Institute of Allergy and Infectious Diseases (NIAID), grant number R01AI144161. The epidemiological studies during which the samples were collected in DRC and South Sudan were funded by the European Research Council (ERC 671055) and in Cameroon by WHO/TDR (B40134). NIH and ERC had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. WHO/TDR, via co-author A.C.K., had a role in the conceptualization of the research, the writing of the manuscript, and in the decision to publish the results.
Data Availability Statement
The data presented in this study are openly available from the National Center for Biotechnology Information Short Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under study accession number PRJNA981587.
Acknowledgments
The authors wish to thank the participants and the teams in DRC and South Sudan who conducted the study that provided the microfilariae we examined, as well as Wilson Sebit at the National Public Health Laboratory, Republic of South Sudan, for supervising the reading of the skin snips. We additionally thank Kirsten Grant for picking so many microfilariae in the laboratory, and for helpful discussions, we thank Tim Anderson at the Texas Biomedical Research Institute and Himal Shrestha and Brooke Whitelaw at La Trobe University. We also thank Amber Hadermann from the Global Health Institute, University of Antwerp, for comments on the draft manuscript. The authors alone are responsible for the views expressed, which do not necessarily represent the views, decisions, or policies of the institutions with which the authors are affiliated.
Conflicts of Interest
A.C.K was a staff member of WHO/TDR at the time this work was conducted. The authors declare no other conflict of interest.
Ethics Committee Approval
Ethical approval was obtained from the Ethics Committee of the School of Public Health in Kinshasa (Logo: January 2017, ESP/CE/006/2017), the ethical committee of the Ministry of Health of South Sudan (September 2018, MOH/ERB 3/2018), and the Ethics Committee of the Antwerp University Hospital (24 May 2017, B300201733011); informed consent was obtained from all study participants. Cameroon ethical approval was obtained from the National Ethics Committee for Human Health Research (Ref: N° 2015/09/641/CE/CNERSH/SP). Administrative authorization was obtained from the Ministry of Public Health (Ref: No 48/L/MINSANTE/SG/DLMEP/PNLO). At the health system level and in the study communities, details on the benefits and potential risk of the study were explained to study volunteers, and, as applicable, community leaders (Quarter-heads or Chiefs), community drug distributors, front-line health personnel (FLHPs), and Regional Onchocerciasis Coordinators (ROC) using information sheets. A written consent form was signed or thumb-printed by the adult participants and by the parents or guardians of children (<18 years of age) who enrolled in the study.
References
- World Health Organization. Elimination of human onchocerciasis: Progress report, 2020. Wkly. Epidemiol. Rec. 2021, 46, 557–567. [Google Scholar]
- Chesnais, C.B.; Nana-Djeunga, H.C.; Njamnshi, A.K.; Lenou-Nanga, C.G.; Boulle, C.; Bissek, A.Z.; Kamgno, J.; Colebunders, R.; Boussinesq, M. The temporal relationship between onchocerciasis and epilepsy: A population-based cohort study. Lancet Infect. Dis. 2018, 18, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Colebunders, R.; Njamnshi, A.K.; van Oijen, M.; Mukendi, D.; Kashama, J.M.; Mandro, M.; Gumisiriza, N.; Preux, P.M.; Suykerbuyk, P.; Idro, R. Onchocerciasis-associated epilepsy: From recent epidemiological and clinical findings to policy implications. Epilepsia Open 2017, 2, 145–152. [Google Scholar] [CrossRef]
- Colebunders, R.; Njamnshi, A.K.; Menon, S.; Newton, C.R.; Hotterbeekx, A.; Preux, P.M.; Hopkins, A.; Vaillant, M.; Siewe Fodjo, J.N. Onchocerca volvulus and epilepsy: A comprehensive review using the Bradford Hill criteria for causation. PLoS Negl. Trop. Dis. 2021, 15, e0008965. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ending the Neglect to Attain the Sustainable Development Goals: A Road Map for Neglected Tropical Diseases 2021–2030; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Plaisier, A.P.; Alley, E.S.; Boatin, B.A.; Van Oortmarssen, G.J.; Remme, H.; De Vlas, S.J.; Bonneux, L.; Habbema, J.D. Irreversible effects of ivermectin on adult parasites in onchocerciasis patients in the Onchocerciasis Control Programme in West Africa. J. Infect. Dis. 1995, 172, 204–210. [Google Scholar] [CrossRef]
- Basáñez, M.-G.; Pion, S.D.; Boakes, E.; Filipe, J.A.; Churcher, T.S.; Boussinesq, M. Effect of single-dose ivermectin on Onchocerca volvulus: A systematic review and meta-analysis. Lancet Infect. Dis. 2008, 8, 310–322. [Google Scholar] [CrossRef]
- Bottomley, C.; Isham, V.; Collins, R.C.; Basáñez, M.G. Rates of microfilarial production by Onchocerca volvulus are not cumulatively reduced by multiple ivermectin treatments. Parasitology 2008, 135, 1571–1581. [Google Scholar] [CrossRef]
- Turner, H.C.; Walker, M.; Churcher, T.S.; Basáñez, M.G. Modelling the impact of ivermectin on River blindness and its burden of morbidity and mortality in African savannah: EpiOncho projections. Parasit. Vectors 2014, 7, 241. [Google Scholar] [CrossRef][Green Version]
- Sauerbrey, M.; Rakers, L.J.; Richards, F.O. Progress toward elimination of onchocerciasis in the Americas. Int. Health 2018, 10, i71–i78. [Google Scholar] [CrossRef]
- World Health Organization. Elimination of human onchocerciasis: Progress report, 2021—Élimination de l’onchocercose humaine: Rapport de situation, 2021. Wkly. Epidemiol. Rec. 2022, 96, 591–598. [Google Scholar]
- Tekle, A.H.; Zouré, H.G.; Noma, M.; Boussinesq, M.; Coffeng, L.E.; Stolk, W.A.; Remme, J.H. Progress towards onchocerciasis elimination in the participating countries of the African Programme for Onchocerciasis Control: Epidemiological evaluation results. Infect. Dis. Poverty 2016, 5, 66. [Google Scholar] [CrossRef]
- Diawara, L.; Traore, M.O.; Badji, A.; Bissan, Y.; Doumbia, K.; Goita, S.F.; Konate, L.; Mounkoro, K.; Sarr, M.D.; Seck, A.F.; et al. Feasibility of onchocerciasis elimination with ivermectin treatment in endemic foci in Africa: First evidence from studies in Mali and Senegal. PLoS Negl. Trop. Dis. 2009, 3, e497. [Google Scholar] [CrossRef] [PubMed]
- Traore, M.O.; Sarr, M.D.; Badji, A.; Bissan, Y.; Diawara, L.; Doumbia, K.; Goita, S.F.; Konate, L.; Mounkoro, K.; Seck, A.F.; et al. Proof-of-principle of onchocerciasis elimination with ivermectin treatment in endemic foci in Africa: Final results of a study in Mali and Senegal. PLoS Negl. Trop. Dis. 2012, 6, e1825. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization; African Programme for Onchocerciasis Control. Report of the Consultative Meetings on Strategic Options and Alternative Treatment Strategies for Accelerating Onchocerciasis Elimination in Africa. World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Awadzi, K.; Attah, S.K.; Addy, E.T.; Opoku, N.O.; Quartey, B.T.; Lazdins-Helds, J.K.; Ahmed, K.; Boatin, B.A.; Boakye, D.A.; Edwards, G. Thirty-month follow-up of sub-optimal responders to multiple treatments with ivermectin, in two onchocerciasis-endemic foci in Ghana. Ann. Trop. Med. Parasitol. 2004, 98, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Awadzi, K.; Boakye, D.A.; Edwards, G.; Opoku, N.O.; Attah, S.K.; Osei-Atweneboana, M.Y.; Lazdins-Helds, J.K.; Ardrey, A.E.; Addy, E.T.; Quartey, B.T.; et al. An investigation of persistent microfilaridermias despite multiple treatments with ivermectin, in two onchocerciasis-endemic foci in Ghana. Ann. Trop. Med. Parasitol. 2004, 98, 231–249. [Google Scholar] [CrossRef] [PubMed]
- Churcher, T.S.; Pion, S.D.; Osei-Atweneboana, M.Y.; Prichard, R.K.; Awadzi, K.; Boussinesq, M.; Collins, R.C.; Whitworth, J.A.; Basáñez, M.G. Identifying sub-optimal responses to ivermectin in the treatment of River Blindness. Proc. Natl. Acad. Sci. USA 2009, 106, 16716–16721. [Google Scholar] [CrossRef]
- Osei-Atweneboana, M.Y.; Eng, J.K.; Boakye, D.A.; Gyapong, J.O.; Prichard, R.K. Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: A two-phase epidemiological study. Lancet 2007, 369, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.R.; Bourguinat, C.; Nana-Djeunga, H.C.; Kengne-Ouafo, J.A.; Pion, S.D.S.; Bopda, J.; Kamgno, J.; Wanji, S.; Che, H.; Kuesel, A.C.; et al. Genome-wide analysis of ivermectin response by Onchocerca volvulus reveals that genetic drift and soft selective sweeps contribute to loss of drug sensitivity. PLoS Negl. Trop. Dis. 2017, 11, e0005816. [Google Scholar] [CrossRef]
- Osei-Atweneboana, M.Y.; Awadzi, K.; Attah, S.K.; Boakye, D.A.; Gyapong, J.O.; Prichard, R.K. Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl. Trop. Dis. 2011, 5, e998. [Google Scholar] [CrossRef]
- Nana-Djeunga, H.C.; Bourguinat, C.; Pion, S.D.; Bopda, J.; Kengne-Ouafo, J.A.; Njiokou, F.; Prichard, R.K.; Wanji, S.; Kamgno, J.; Boussinesq, M. Reproductive status of Onchocerca volvulus after ivermectin treatment in an ivermectin-naive and a frequently treated population from Cameroon. PLoS Negl. Trop. Dis. 2014, 8, e2824. [Google Scholar] [CrossRef]
- Opoku, N.O.; Bakajika, D.K.; Kanza, E.M.; Howard, H.; Mambandu, G.L.; Nyathirombo, A.; Nigo, M.M.; Kasonia, K.; Masembe, S.L.; Mumbere, M.; et al. Single dose moxidectin versus ivermectin for Onchocerca volvulus infection in Ghana, Liberia, and the Democratic Republic of the Congo: A randomised, controlled, double-blind phase 3 trial. Lancet 2018, 392, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Bakajika, D.; Kanza, E.M.; Opoku, N.O.; Howard, H.M.; Mambandu, G.L.; Nyathirombo, A.; Nigo, M.M.; Kennedy, K.K.; Masembe, S.L.; Mumbere, M.; et al. Effect of a single dose of 8 mg moxidectin or 150 μg/kg ivermectin on O. volvulus skin microfilariae in a randomized trial: Differences between areas in the Democratic Republic of the Congo, Liberia and Ghana and impact of intensity of infection. PLoS Negl. Trop. Dis. 2022, 16, e0010079. [Google Scholar] [CrossRef]
- Awadzi, K.; Opoku, N.O.; Attah, S.K.; Lazdins-Helds, J.; Kuesel, A.C. A randomized, single-ascending-dose, ivermectin-controlled, double-blind study of moxidectin in Onchocerca volvulus infection. PLoS Negl. Trop. Dis. 2014, 8, e2953. [Google Scholar] [CrossRef]
- Frempong, K.K.; Walker, M.; Cheke, R.A.; Tetevi, E.J.; Gyan, E.T.; Owusu, E.O.; Wilson, M.D.; Boakye, D.A.; Taylor, M.J.; Biritwum, N.K.; et al. Does increasing treatment frequency address suboptimal responses to ivermectin for the control and elimination of River blindness? Clin. Infect. Dis. 2016, 62, 1338–1347. [Google Scholar] [CrossRef]
- Abong, R.A.; Amambo, G.N.; Chounna Ndongmo, P.W.; Njouendou, A.J.; Ritter, M.; Beng, A.A.; Esum, M.E.; Deribe, K.; Fru-Cho, J.; Fombad, F.F.; et al. Differential susceptibility of Onchocerca volvulus microfilaria to ivermectin in two areas of contrasting history of mass drug administration in Cameroon: Relevance of microscopy and molecular techniques for the monitoring of skin microfilarial repopulation within six months of direct observed treatment. BMC Infect. Dis. 2020, 20, 726. [Google Scholar] [CrossRef]
- Stolk, W.A.; Walker, M.; Coffeng, L.E.; Basáñez, M.G.; de Vlas, S.J. Required duration of mass ivermectin treatment for onchocerciasis elimination in Africa: A comparative modelling analysis. Parasit. Vectors 2015, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Basáñez, M.G.; Walker, M.; Turner, H.C.; Coffeng, L.E.; de Vlas, S.J.; Stolk, W.A. River blindness: Mathematical models for control and elimination. Adv. Parasitol. 2016, 94, 247–341. [Google Scholar] [CrossRef]
- Duke, B.O. Onchocerciasis; deep worm bundles close to hip joints. Trans. R. Soc. Trop. Med. Hyg. 1970, 64, 791–792. [Google Scholar] [CrossRef] [PubMed]
- Meyers, W.M.; Neafie, R.C.; Connor, D.H. Onchocerciasis: Invasion of deep organs by Onchocerca volvulus. Am. J. Trop. Med. Hyg. 1977, 26, 650–657. [Google Scholar] [CrossRef]
- Duke, B.O. The population dynamics of Onchocerca volvulus in the human host. Trop. Med. Parasitol. 1993, 44, 61–68. [Google Scholar] [PubMed]
- Duerr, H.P.; Dietz, K.; Schulz-Key, H.; Buttner, D.W.; Eichner, M. The relationships between the burden of adult parasites, host age and the microfilarial density in human onchocerciasis. Int. J. Parasitol. 2004, 34, 463–473. [Google Scholar] [CrossRef]
- Albiez, E.J. Studies on nodules and adult Onchocerca volvulus during a nodulectomy trial in hyperendemic villages in Liberia and Upper Volta. I. Palpable and impalpable onchocercomata. Tropenmed. Parasitol. 1983, 34, 54–60. [Google Scholar]
- Basáñez, M.G.; Boussinesq, M. Population biology of human onchocerciasis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 809–826. [Google Scholar] [CrossRef]
- Schulz-Key, H.; Albiez, E.J. Worm burden of Onchocerca volvulus in a hyperendemic village of the rain-forest in West Africa. Tropenmedizin Parasitol. 1977, 28, 431–438. [Google Scholar]
- Dusabimana, A.; Bhwana, D.; Raimon, S.; Mmbando, B.P.; Hotterbeekx, A.; Tepage, F.; Mandro, M.; Siewe Fodjo, J.N.; Abrams, S.; Colebunders, R. Ivermectin treatment response in Onchocerca volvulus infected persons with epilepsy: A three-country short cohort study. Pathogens 2020, 9, 617. [Google Scholar] [CrossRef]
- Fodjo, J.N.S.; Mandro, M.; Mukendi, D.; Tepage, F.; Menon, S.; Nakato, S.; Nyisi, F.; Abhafule, G.; Wonya’rossi, D.; Anyolito, A.; et al. Onchocerciasis-associated epilepsy in the Democratic Republic of Congo: Clinical description and relationship with microfilarial density. PLoS Negl. Trop. Dis. 2019, 13, e0007300. [Google Scholar] [CrossRef]
- Mandro, M.; Siewe Fodjo, J.N.; Dusabimana, A.; Mukendi, D.; Haesendonckx, S.; Lokonda, R.; Nakato, S.; Nyisi, F.; Abhafule, G.; Wonya’rossi, D.; et al. Single versus multiple dose ivermectin regimen in onchocerciasis-infected persons with epilepsy treated with phenobarbital: A randomized clinical trial in the Democratic Republic of Congo. Pathogens 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elfarag, G.; Carter, J.Y.; Raimon, S.; Sebit, W.; Suliman, A.; Fodjo, J.N.S.; Olore, P.C.; Biel, K.P.; Ojok, M.; Logora, M.Y.; et al. Persons with onchocerciasis-associated epilepsy and nodding seizures have a more severe form of epilepsy with more cognitive impairment and higher levels of Onchocerca volvulus infection. Epileptic Disord. 2020, 22, 301–308. [Google Scholar] [CrossRef]
- Colebunders, R.; Carter, J.Y.; Olore, P.C.; Puok, K.; Bhattacharyya, S.; Menon, S.; Abd-Elfarag, G.; Ojok, M.; Ensoy-Musoro, C.; Lako, R.; et al. High prevalence of onchocerciasis-associated epilepsy in villages in Maridi County, Republic of South Sudan: A community-based survey. Seizure 2018, 63, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Post, R.J.; Laudisoit, A.; Mandro, M.; Lakwo, T.; Laemmer, C.; Pfarr, K.; Hoerauf, A.; Tortosa, P.; Gomard, Y.; Ukety, T.; et al. Identification of the onchocerciasis vector in the Kakoi-Koda focus of the Democratic Republic of Congo. PLoS Negl. Trop. Dis. 2022, 16, e0010684. [Google Scholar] [CrossRef] [PubMed]
- Nana-Djeunga, H.C.; Sicard, C.M.; Mogoung-Wafo, A.E.; Chesnais, C.B.; Deleglise, H.; Touka-Nounkeu, R.; Domche, A.; Golden, A.; Klion, A.D.; Nutman, T.B.; et al. Changes in onchocerciasis Ov16 IgG4 rapid diagnostic test results over one-month follow-up: Lessons for reading timeframe and decision-making. Am. J. Trop. Med. Hyg. 2022, 107, 658–661. [Google Scholar] [CrossRef]
- Hotterbeekx, A.; Perneel, J.; Mandro, M.; Abhafule, G.; Siewe Fodjo, J.N.; Dusabimana, A.; Abrams, S.; Kumar-Singh, S.; Colebunders, R. Comparison of diagnostic tests for Onchocerca volvulus in the Democratic Republic of Congo. Pathogens 2020, 9, 435. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Report of the Proceedings of the Fifth Meeting of the Onchocerciasis Technical Advisory Subgroup, Virtual Meeting, 9–10 December 2021; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- World Health Organization. Report on the Fourth Meeting of the WHO Onchocerciasis Technical Advisory Subgroup: Virtual Meeting, 28–29 October 2020; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Dusabimana, A.; Siewe Fodjo, J.N.; Ndahura, M.M.; Mmbando, B.P.; Jada, S.R.; Boven, A.; De Smet, E.; Ukety, T.; Njamnshi, A.K.; Laudisoit, A.; et al. Surveillance for onchocerciasis-associated epilepsy and OV16 IgG4 testing of children 6–10 years old should be used to identify areas where onchocerciasis elimination programs need strengthening. Pathogens 2022, 11, 281. [Google Scholar] [CrossRef]
- Lakwo, T.L.; Raimon, S.; Tionga, M.; Siewe Fodjo, J.N.; Alinda, P.; Sebit, W.J.; Carter, J.Y.; Colebunders, R. The role of the Maridi Dam in causing an onchocerciasis-associated epilepsy epidemic in Maridi, South Sudan: An epidemiological, sociological, and entomological study. Pathogens 2020, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Tyagi, R.; McNulty, S.N.; Rosa, B.A.; Ozersky, P.; Martin, J.; Hallsworth-Pepin, K.; Unnasch, T.R.; Norice, C.T.; Nutman, T.B.; et al. Genomic diversity in Onchocerca volvulus and its Wolbachia endosymbiont. Nat. Microbiol. 2016, 2, 16207. [Google Scholar] [CrossRef]
- Crawford, K.E.; Hedtke, S.M.; Kuesel, A.C.; Doyle, S.D.; Armoo, S.; Osei-Atweneboana, M.; Grant, W.N. Genome-based tools for onchocerciasis elimination: Utility of the mitochondrial genome for delineating Onchocerca volvulus transmission zones. bioRxiv 2019, 732446. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Burns, M.; Valdivia, H. Modelling the limit of detection in real-time quantitative PCR. Eur. Food Res. Technol. 2008, 226, 1513–1524. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Cotton, J.A.; Bennuru, S.; Grote, A.; Harsha, B.; Tracey, A.; Beech, R.; Doyle, S.R.; Dunn, M.; Hotopp, J.C.; Holroyd, N.; et al. The genome of Onchocerca volvulus, agent of river blindness. Nat. Microbiol. 2016, 2, 16216. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinforma. 2014, 47, 11–12. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21 (Suppl. 1), i351–i358. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Kronenberg, Z.N.; Dawson, E.T.; Pedersen, B.S.; Prins, P. Vcflib and tools for processing the VCF variant call format. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- R Development Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008.
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Chao, A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 1987, 43, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Wang, Y.T.; Walther, B.A.; Chao, A. An improved nonparametric lower bound of species richness via a modified good-turing frequency formula. Biometrics 2014, 70, 671–682. [Google Scholar] [CrossRef]
- O’Hara, R.B. Species richness estimators: How many species can dance on the head of a pin? J. Anim. Ecol. 2005, 74, 375–386. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Lischer, H.E.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Churcher, T.S.; Schwab, A.E.; Prichard, R.K.; Basanez, M.G. An analysis of genetic diversity and inbreeding in Wuchereria bancrofti: Implications for the spread and detection of drug resistance. PLoS Negl. Trop. Dis. 2008, 2, e211. [Google Scholar] [CrossRef]
- Hedtke, S.M.; Zendejas-Heredia, P.A.; Graves, P.M.; Sheridan, S.; Sheel, M.; Fuimaono, S.; Lau, C.L.; Grant, W.N. Genetic epidemiology of lymphatic filariasis in American Samoa after mass drug administration. Int. J. Parasitol. 2021, 15, 137–147. [Google Scholar] [CrossRef]
- World Health Organization; Onchocerciasis Control Programme in the Volta River Basin Area; Rolland, A.; Balay, G. Onchocerciasis Focus in the Bissa Country; Onchocercisis Control Programme in the Volta River Basin Area: Ouagadougou, Burkino Faso, 1985. [Google Scholar]
- Ngoumou, P.; Walsh, J.F. ; WHO Programme for the Prevention of Blindness. UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases. A Manual for Rapid Epidemiological Mapping of Onchocerciasis; Ngoumou, P., Walsh, J.F., Eds.; World Health Organization: Geneva, Switzerland, 1993. [Google Scholar]
- Remme, J.H.F.; Boatin, B.; Boussinesq, M. Helminthic diseases: Onchocerciasis and loiasis. In The International Encyclopedia of Public Health, 2nd ed.; Quah, S.R., Cockerham, W.C., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 3, pp. 576–587. [Google Scholar]
- Duke, B.O. Observations and reflections on the immature stages of Onchocerca volvulus in the human host. Ann. Trop. Med. Parasitol. 1991, 85, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.S. Human onchocerciasis: Notes on the history, the parasite and the life cycle. Ann. Trop. Med. Parasitol. 1991, 85, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Prost, A. Latence parasitaire dans l’onchocercose. Bull. World Health Organ. 1980, 58, 923–925. [Google Scholar]
- Hedtke, S.M.; Kuesel, A.C.; Crawford, K.E.; Graves, P.M.; Boussinesq, M.; Lau, C.L.; Boakye, D.A.; Grant, W.N. Genomic epidemiology in filarial nematodes: Transforming the basis for elimination program decisions. Front. Genet. 2020, 10, 1282. [Google Scholar] [CrossRef]
- Koala, L.; Nikiema, A.; Post, R.J.; Paré, A.B.; Kafando, C.M.; Drabo, F.; Traoré, S. Recrudescence of onchocerciasis in the Comoé valley in Southwest Burkina Faso. Acta Trop. 2017, 166, 96–105. [Google Scholar] [CrossRef]
- Koala, L.; Nikiema, A.S.; Pare, A.B.; Drabo, F.; Toe, L.D.; Belem, A.M.G.; Boakye, D.A.; Traore, S.; Dabire, R.K. Entomological assessment of the transmission following recrudescence of onchocerciasis in the Comoé Valley, Burkina Faso. Parasit. Vectors 2019, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Garms, R. The reinvasion of the onchocerciasis control programme area in the Volta River Basin by Simulium damnosum s.l., the involvement of the different cytospecies and epidemiological implications. Ann. Soc. Belg. Med. Trop. 1981, 61, 193–198. [Google Scholar]
- Garms, R.; Walsh, J.F.; Davies, J.B. Studies on the reinvasion of the Onchocerciasis Control Programme in the Volta River Basin by Simulium damnosum s.I. with emphasis on the south-western areas. Tropenmed Parasitol. 1979, 30, 345–362. [Google Scholar]
- Magor, J.I.; Rosenberg, L.J. Studies of winds and weather during migrations of Simulium damnosum Theobald (Diptera: Simuliidae), the vector of onchocerciasis in West Africa. Bull. Entomol. Res. 1980, 70, 693–716. [Google Scholar] [CrossRef]
- Baker, R.H.; Guillet, P.; Seketeli, A.; Poudiougo, P.; Boakye, D.; Wilson, M.D.; Bissan, Y.; Garms, R.; Cheke, R.A.; Sachs, R.; et al. Progress in controlling the reinvasion of windborne vectors into the western area of the Onchocerciasis Control Programme in West Africa [and discussion]. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1990, 328, 731–747. [Google Scholar] [CrossRef]
- McCulloch, K.; Hedtke, S.M.; McCaw, J.; McVernon, J.; Basáñez, M.-G.; Walker, M.; Kuesel, A.C.; Grant, W.N. Impact of human movement between hypo- and hyperendemic areas on sustainability of elimination of Onchocerca volvulus transmission. medRxiv 2023. [Google Scholar] [CrossRef]
- de Vos, A.S.; Stolk, W.A.; de Vlas, S.J.; Coffeng, L.E. The effect of assortative mixing on stability of low helminth transmission levels and on the impact of mass drug administration: Model explorations for onchocerciasis. PLoS Negl. Trop. Dis. 2018, 12, e0006624. [Google Scholar] [CrossRef] [PubMed]
- Specht, S.; Hoerauf, A.; Adjei, O.; Debrah, A.; Buttner, D.W. Newly acquired Onchocerca volvulus filariae after doxycycline treatment. Parasitol. Res. 2009, 106, 23–31. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).