Diagnostic Evaluation of the IS1081-Targeted Real-Time PCR for Detection of Mycobacterium bovis DNA in Bovine Milk Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of M. bovis BCG Stock
2.2. Preparation of Spiked Milk Samples, DNA Extraction, and Real-Time PCR
2.3. Clinical Milk Samples from Naturally Infected Animals
2.4. Inactivation of M. bovis in PrimeStore® MTM
2.5. Molecular Evaluation of Samples Preserved in MTM Tubes
2.6. Statistical Analysis
3. Results
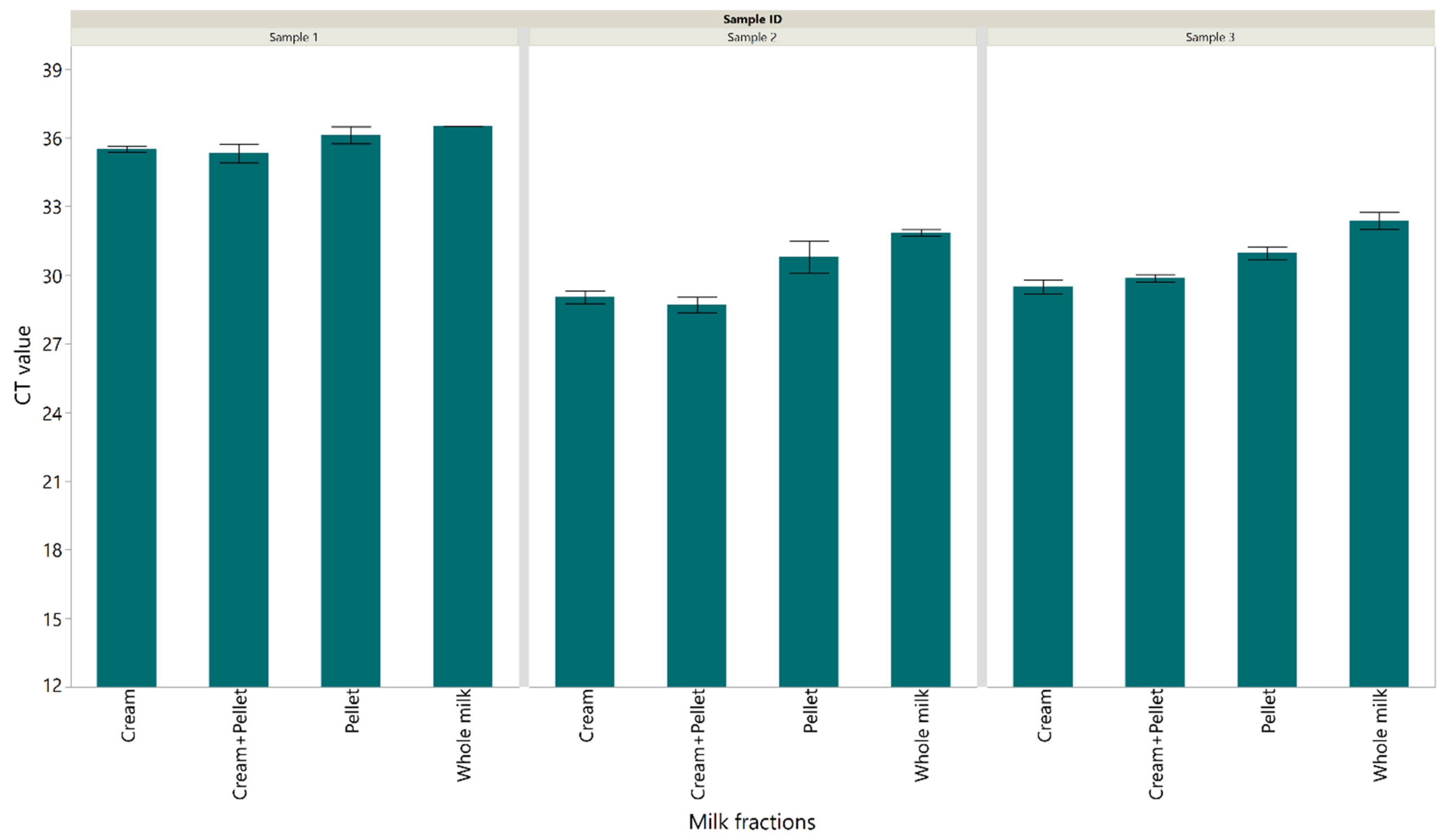
3.1. Detection of M. bovis DNA in Milk Sample Fractions
3.2. Milk Samples from Naturally Infected Animals
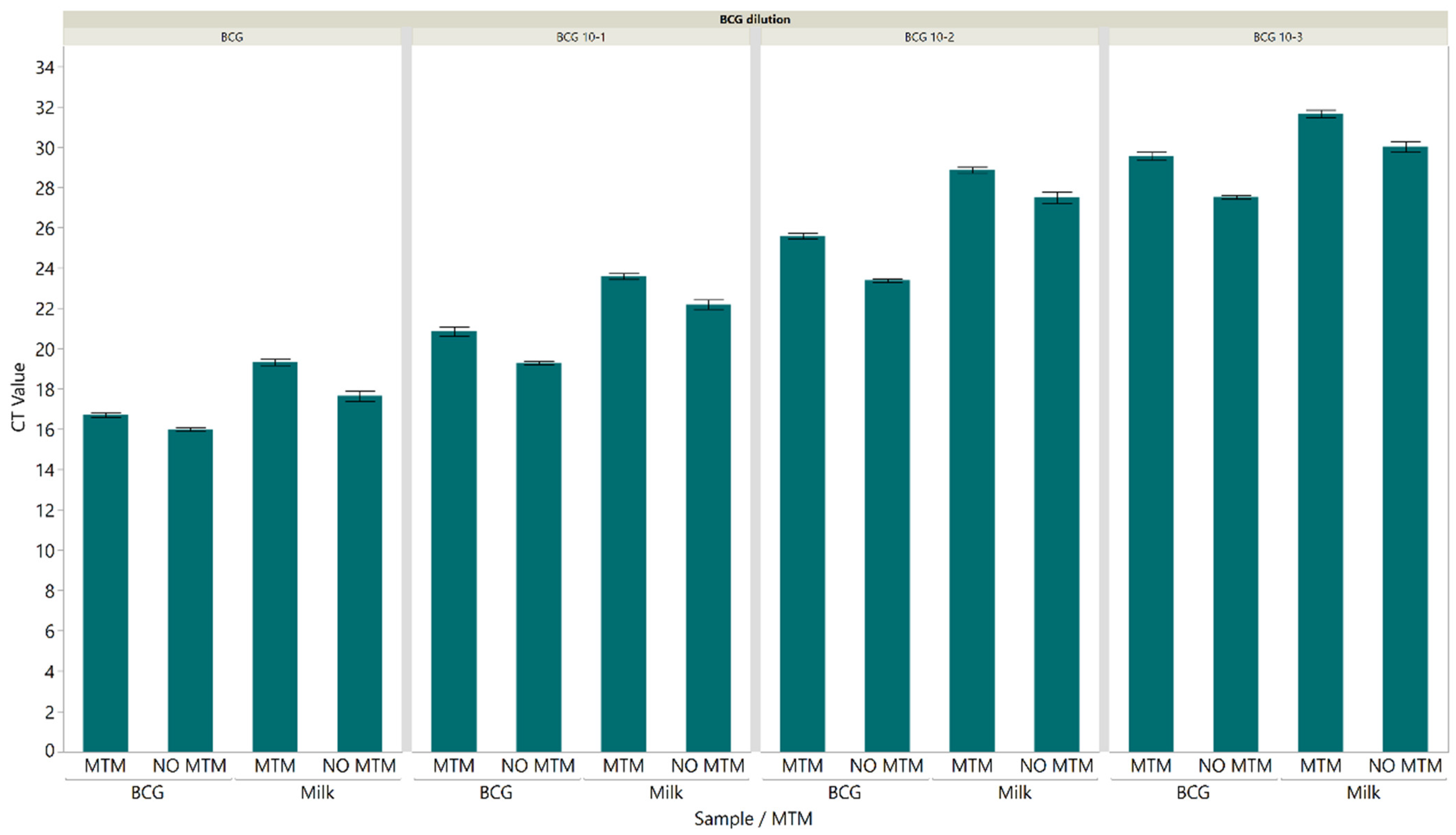
3.3. Inactivation of M. bovis BCG in PrimeStore® MTM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zinsstag, J.; Schelling, E.; Roth, F.; Kazwala, R.; Thoen, C.; Steele, J. Economics of bovine tuberculosis. In Mycobacterium bovis Infection in Animals and Humans; John Wiley & Sons: Hoboken, NJ, USA, 2006; Volume 2, pp. 68–83. [Google Scholar]
- Palmer, M. Mycobacterium bovis: Characteristics of wildlife reservoir hosts. Transbound. Emerg. Dis. 2013, 60, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guindi, S.; Ahmed, O.; Awad, W.; El-Saban, M. Incidence of bovine and human tubercle bacilli in milk and milk products. Agric. Res. Rev. 1980, 58, 75–84. [Google Scholar]
- Cornejo, B.J.; Sahagún-Ruiz, A.; Suárez-Guemes, F.; Thornton, C.G.; Ficht, T.A.; Adams, L.G. Comparison of C18-carboxypropylbetaine and glass bead DNA extraction methods for detection of Mycobacterium bovis in bovine milk samples and analysis of samples by PCR. Appl. Environ. Microb. 1998, 64, 3099–3101. [Google Scholar] [CrossRef] [PubMed]
- Kleeberg, H. Human tuberculosis of bovine origin in relation to public health. Rev. Sci. Tech. Off. Int. Epiz. 1984, 3, 11–32. [Google Scholar] [CrossRef] [PubMed]
- De la Rua-Domenech, R. Human Mycobacterium bovis infection in the United Kingdom: Incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis 2006, 86, 77–109. [Google Scholar] [CrossRef]
- Schiller, I.; Oesch, B.; Vordermeier, H.; Palmer, M.; Harris, B.; Orloski, K.; Buddle, B.; Thacker, T.; Lyashchenko, K.; Waters, W. Bovine tuberculosis: A review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound. Emerg. Dis. 2010, 57, 205–220. [Google Scholar] [CrossRef]
- Da Silva Cezar, R.D.; Lucena-Silva, N.; Batista Filho, A.F.B.; de Melo Borges, J.; de Oliveira, P.R.F.; Lúcio, É.C.; Arruda-Lima, M.; de Assis Santana, V.L.; Junior, J.W.P. Molecular detection of Mycobacterium bovis in cattle herds of the state of Pernambuco, Brazil. BMC Vet. Res. 2016, 12, 31. [Google Scholar]
- BhanuRekha, V.; Gunaseelan, L.; Pawar, G.; Nassiri, R.; Bharathy, S. Molecular detection of Mycobacterium tuberculosis from bovine milk samples. J. Adv. Vet. Anim. Res. 2015, 2, 80–83. [Google Scholar] [CrossRef]
- Kahla, I.B.; Boschiroli, M.; Souissi, F.; Cherif, N.; Benzarti, M.; Boukadida, J.; Hammami, S. Isolation and molecular characterisation of Mycobacterium bovis from raw milk in Tunisia. Afr. Health Sci. 2011, 11, 2–5. [Google Scholar]
- Bolaños, C.A.D.; Paula, C.L.d.; Guerra, S.T.; Franco, M.M.J.; Ribeiro, M.G. Diagnosis of mycobacteria in bovine milk: An overview. Rev. Do Inst. Med. Trop. São Paulo 2017, 59, e40. [Google Scholar] [CrossRef]
- Aydın, F.E.; Ulger, M.; Emekdaş, G.; Aslan, G.; Günal, S. Isolation and identification of Mycobacterium bovis and non-tuberculous mycobacteria in raw milk samples in Mersin province. Mikrobiyoloji Bul. 2012, 46, 283–289. [Google Scholar]
- Vitale, F.; Capra, G.; Maxia, L.; Reale, S.; Vesco, G.; Caracappa, S. Detection of Mycobacterium tuberculosis complex in cattle by PCR using milk, lymph node aspirates, and nasal swabs. J. Clin. Microbiol. 1998, 36, 1050–1055. [Google Scholar] [CrossRef]
- Al-Saqur, I.; Al-Thwani, A.; Al-Attar, I. Detection of Mycobacteria spp. in cows milk using conventional methods and PCR. Iraqi J. Vet. Sci. 2009, 23, 259–262. [Google Scholar]
- Zumárraga, M.J.; Soutullo, A.; García, M.I.; Marini, R.; Abdala, A.; Tarabla, H.; Echaide, S.; López, M.; Zervini, E.; Canal, A. Detection of Mycobacterium bovis–infected dairy herds using PCR in bulk tank milk samples. Foodborne Pathog. Dis. 2012, 9, 132–137. [Google Scholar] [CrossRef] [PubMed]
- de Souza Figueiredo, E.E.; Júnior, C.A.C.; Furlanetto, L.V.; Silva, F.G.S.; Duarte, R.S.; Silva, J.T.; Lilenbaum, W.; Paschoalin, V.M.F. Molecular techniques for identification of species of the Mycobacterium tuberculosis complex: The use of multiplex PCR and an adapted HPLC method for identification of Mycobacterium bovis and diagnosis of bovine tuberculosis. In Understanding Tuberculosis-Global Experiences and Innovative Approaches to the Diagnosis; IntechOpen: London, UK, 2012. [Google Scholar]
- Mishra, A.; Singhal, A.; Chauhan, D.; Katoch, V.; Srivastava, K.; Thakral, S.; Bharadwaj, S.; Sreenivas, V.; Prasad, H. Direct detection and identification of Mycobacterium tuberculosis and Mycobacterium bovis in bovine samples by a novel nested PCR assay: Correlation with conventional techniques. J. Clin. Microbiol. 2005, 43, 5670–5678. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Moreno, B.; Romero, T.; Arriaga, C.; Torres, R.; Pereira-Suárez, A.; García-Salazar, J.; Estrada-Chávez, C. High frequency of Mycobacterium bovis DNA in colostra from tuberculous cattle detected by nested PCR. Zoonoses Public Health 2008, 55, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Rådström, P.; Knutsson, R.; Wolffs, P.; Lövenklev, M.; Löfström, C. Pre-PCR processing. Mol. Biotechnol. 2004, 26, 133–146. [Google Scholar] [CrossRef]
- Barbano, D.; Lynch, J. Major advances in testing of dairy products: Milk component and dairy product attribute testing. J. Dairy Sci. 2006, 89, 1189–1194. [Google Scholar] [CrossRef]
- Sun, L.; Dicksved, J.; Priyashantha, H.; Lundh, Å.; Johansson, M. Distribution of bacteria between different milk fractions, investigated using culture-dependent methods and molecular-based and fluorescent microscopy approaches. J. Appl. Microbiol. 2019, 127, 1028–1037. [Google Scholar] [CrossRef]
- Poms, R.; Glössl, J.; Foissy, H. Increased sensitivity for detection of specific target DNA in milk by concentration in milk fat. Eur. Food Res. Technol. 2001, 213, 361–365. [Google Scholar] [CrossRef]
- Rutala, W.A.; Cole, E.C.; Wannamaker, N.S.; Weber, D.J. Inactivation of Mycobacterium tuberculosis and Mycobacterium bovis by 14 hospital disinfectants. Am. J. Med. 1991, 91, S267–S271. [Google Scholar] [CrossRef]
- Grant, I.; Ball, H.; Rowe, M. Thermal inactivation of several Mycobacterium spp. in milk by pasteurization. Lett. Appl. Microbiol. 1996, 22, 253–256. [Google Scholar] [CrossRef]
- Daum, L.; Worthy, S.; Yim, K.; Nogueras, M.; Schuman, R.; Choi, Y.; Fischer, G. A clinical specimen collection and transport medium for molecular diagnostic and genomic applications. Epidemiol. Infect. 2011, 139, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Daum, L.T.; Schuman, R.; Sei, C.; Rikhi, N.; Mesadieu, A.; Gerald, F. Real-time PCR of whole blood specimens transported in PrimeStore MTM® to detect and monitor MTB bacteremia. Int. J. Infect. Dis. 2016, 45, 392. [Google Scholar] [CrossRef][Green Version]
- Daum, L.; Choi, Y.; Worthy, S.; Rodriguez, J.; Chambers, J.; Fischer, G. A molecular transport medium for collection, inactivation, transport, and detection of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 2014, 18, 847–849. [Google Scholar] [CrossRef]
- Van Bockel, D.; Munier, C.M.L.; Turville, S.; Badman, S.G.; Walker, G.; Stella, A.O.; Aggarwal, A.; Yeang, M.; Condylios, A.; Kelleher, A.D. Evaluation of commercially available viral transport medium (VTM) for SARS-CoV-2 inactivation and use in point-of-care (POC) testing. Viruses 2020, 12, 1208. [Google Scholar] [CrossRef] [PubMed]
- Dykema, P.E.; Stokes, K.D.; Beckwith, N.R.; Mungin, J.W.; Xu, L.; Vickers, D.J.; Reising, M.M.; Bravo, D.M.; Thomsen, B.V.; Robbe-Austerman, S. Development and validation of a direct real-time PCR assay for Mycobacterium bovis and implementation into the United States national surveillance program. PeerJ PrePrints 2016, 4, e1703v1701. [Google Scholar]
- Zeineldin, M.; Camp, P.; Farrell, D.; Lehman, K.; Thacker, T. Whole genome sequencing of Mycobacterium bovis directly from clinical tissue samples without culture. Front. Microbiol. 2023, 14, 1141651. [Google Scholar] [CrossRef]
- Cosivi, O.; Grange, J.M.; Daborn, C.J.; Raviglione, M.C.; Fujikura, T.; Cousins, D.; Robinson, R.; Huchzermeyer, H.; de Kantor, I.; Meslin, F.-X. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 1998, 4, 59. [Google Scholar] [CrossRef]
- Sgarioni, S.A.; Hirata, R.D.C.; Hiroyuki Hirata, M.; Leite, C.Q.F.; Prince, K.D.; Leite, S.R.d.A.; Vedovello Filho, D.; Siqueira, V.L.D.; Caleffi-Ferracioli, K.R.; Cardoso, R.F. Occurrence of Mycobacterium bovis and non-tuberculous mycobacteria (NTM) in raw and pasteurized milk in the northwestern region of Paraná, Brazil. Braz. J. Microbiol. 2014, 45, 707–711. [Google Scholar] [CrossRef][Green Version]
- Rowe, M.T.; Donaghy, J. Mycobacterium bovis: The importance of milk and dairy products as a cause of human tuberculosis in the UK. A review of taxonomy and culture methods, with particular reference to artisanal cheeses. Int. J. Dairy Technol. 2008, 61, 317–326. [Google Scholar] [CrossRef]
- Jayasumana, M.; Galappaththi, T.; Pushpakumara, P.; Gamage, C.; Smith, N.; Jinadasa, H. Screening milk for bovine tuberculosis in dairy farms in central province, Sri Lanka. Trop. Agric. Res. 2018, 30, 12–18. [Google Scholar] [CrossRef]
- Zarden, C.F.; Marassi, C.D.; Figueiredo, E.E.; Lilenbaum, W. Mycobacterium bovis detection from milk of negative skin test cows. Vet. Rec. 2013, 172, 130. [Google Scholar] [CrossRef] [PubMed]
- Elsohaby, I.; Mahmmod, Y.S.; Mweu, M.M.; Ahmed, H.A.; El-Diasty, M.M.; Elgedawy, A.A.; Mahrous, E.; El Hofy, F.I. Accuracy of PCR, mycobacterial culture and interferon-gamma assays for detection of Mycobacterium bovis in blood and milk samples from Egyptian dairy cows using Bayesian modelling. Prev. Vet. Med. 2020, 181, 105054. [Google Scholar] [CrossRef]
- Euber, J.; Brunner, J. Reexamination of fat globule clustering and creaming in cow milk. J. Dairy Sci. 1984, 67, 2821–2832. [Google Scholar] [CrossRef]
- Brewster, J.D.; Paul, M. Improved method for centrifugal recovery of bacteria from raw milk applied to sensitive real-time quantitative PCR detection of Salmonella spp. J. Dairy Sci. 2016, 99, 3375–3379. [Google Scholar] [CrossRef]
- Caplan, Z.; Melilli, C.; Barbano, D. Gravity separation of fat, somatic cells, and bacteria in raw and pasteurized milks. J. Dairy Sci. 2013, 96, 2011–2019. [Google Scholar] [CrossRef]
- Sánchez-Juanes, F.; Alonso, J.; Zancada, L.; Hueso, P. Distribution and fatty acid content of phospholipids from bovine milk and bovine milk fat globule membranes. Int. Dairy J. 2009, 19, 273–278. [Google Scholar] [CrossRef]
- van Soolingen, D.; Hermans, P.; De Haas, P.; Van Embden, J. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: A reliable tool for recognizing Mycobacterium bovis BCG. J. Clin. Microbiol. 1992, 30, 1772–1777. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.M.; Stephens, D.M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol. Lett. 1991, 83, 11–15. [Google Scholar] [CrossRef][Green Version]
- Dziadek, J.; Sajduda, A. Specificity of insertion sequence-based PCR assays for Mycobacterium tuberculosis complex. Int. J. Tuberc. Lung Dis. 2001, 5, 569–574. [Google Scholar] [PubMed]
- Clarke, C.; Smith, K.; Goldswain, S.J.; Helm, C.; Cooper, D.V.; Kerr, T.J.; Kleynhans, L.; Van Helden, P.D.; Warren, R.M.; Miller, M.A. Novel molecular transport medium used in combination with Xpert MTB/RIF ultra provides rapid detection of Mycobacterium bovis in African buffaloes. Sci. Rep. 2021, 11, 7061. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.; Cooper, D.V.; Miller, M.A.; Goosen, W.J. Detection of Mycobacterium tuberculosis complex DNA in oronasal swabs from infected African buffaloes (Syncerus caffer). Sci. Rep. 2022, 12, 1834. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeineldin, M.M.; Lehman, K.; Camp, P.; Farrell, D.; Thacker, T.C. Diagnostic Evaluation of the IS1081-Targeted Real-Time PCR for Detection of Mycobacterium bovis DNA in Bovine Milk Samples. Pathogens 2023, 12, 972. https://doi.org/10.3390/pathogens12080972
Zeineldin MM, Lehman K, Camp P, Farrell D, Thacker TC. Diagnostic Evaluation of the IS1081-Targeted Real-Time PCR for Detection of Mycobacterium bovis DNA in Bovine Milk Samples. Pathogens. 2023; 12(8):972. https://doi.org/10.3390/pathogens12080972
Chicago/Turabian StyleZeineldin, Mohamed M., Kimberly Lehman, Patrick Camp, David Farrell, and Tyler C. Thacker. 2023. "Diagnostic Evaluation of the IS1081-Targeted Real-Time PCR for Detection of Mycobacterium bovis DNA in Bovine Milk Samples" Pathogens 12, no. 8: 972. https://doi.org/10.3390/pathogens12080972
APA StyleZeineldin, M. M., Lehman, K., Camp, P., Farrell, D., & Thacker, T. C. (2023). Diagnostic Evaluation of the IS1081-Targeted Real-Time PCR for Detection of Mycobacterium bovis DNA in Bovine Milk Samples. Pathogens, 12(8), 972. https://doi.org/10.3390/pathogens12080972





