Abstract
Acanthamoeba act as hosts for various microorganisms and pathogens, causing Acanthamoeba Keratitis (AK). To investigate the association between endosymbionts and AK progression, we performed a metagenomics study to characterize the intracellular microbiome from five lenses associated with AK isolates and standard strains to characterize the role of ocular flora in AK progression. The used clinical isolates were axenic cultured from lenses associated with AK patients. AK isolates and standard controls such as 16S ribosomal RNA sequencing techniques were used for analysis. The microbiome compositions and relative abundance values were compared. The orders of Clostridiales and Bacteroidales presented major populations of intracellular microbes belonging to all isolates. Comparison of the different source isolates showed that most of the abundance in keratitis isolates came from Ruminococcus gnavus (121.0 folds), Eubacterium dolichum (54.15 folds), Roseburia faecis (24.51 folds), and Blautia producta (3.15 folds). Further analysis of the relative abundance data from keratitis isolates showed that Blautia producta was positively correlated with the disease course. In contrast, Bacteroides ovatus was found to be abundant in early-stage keratitis isolates. This study reveals the abundant anaerobic Gram-positive rods present in severe keratitis isolate and characterize the association between Acanthamoeba and ocular flora in AK progression.
1. Introduction
Acanthamoeba is a free-living amoebae (FLA) that can be found in moist environments, such as in tap water and human nasopharyngeal mucosa [1,2]. The major pathogenic strains include Acanthamoeba castellanii and Acanthamoeba polyphaga. Diseases in Acanthamebiasis contain Acanthamoeba keratitis (AK) and Granulomatous amoebic encephalitis (GAE). The protozoa obtain food sources from surrounding microorganisms via phagocytosis. Nevertheless, several bacteria can escape and latently inhabit amebae, including ocular surface flora.
The ocular surface harbors a variety of microbes due to continuous exposure to the environment [3]. The ocular commensals play a crucial role in defending against pathogens and maintaining ocular immunity [4]. A recent study demonstrated that the Acanthamoeba characteristics of phagocytosis alter the surrounding microorganism distribution, thereby causing complications [5,6]. Acanthamoeba can host and protect microorganisms, including pathogenic bacteria, fungi, and viruses [7]. However, some microorganisms can survive digestion and multiply within hosts, which are called endosymbionts. The association between endosymbionts and their hosts can be either temporary or steady. Since such “Trojan horses” can escape the intracellular killing of Acanthamoeba, these microbes could be protected from harmful environments, such as ocular immune killing [8]. Previous studies demonstrated that Pseudomonas aeruginosa is an important endosymbiont of Acanthamoeba and that it usually infects concurrently with AK [9]. Some pathogens such as Mimivirus can acquire genes from the ameba host [10]. These genes can be exchanged and subsequently produced by endosymbionts [11], suggesting the role of Acanthamoeba in microorganism evolution. In addition, several studies have indicated that endosymbionts can enhance the pathogenicity of Acanthamoeba [12,13]. In vitro and in vivo studies of cytopathic effect (CPE) showed significant increases in endosymbiont-infected Acanthamoeba [14,15]. Therefore, the intracellular microbiome profiling of Acanthamoeba clinical isolates might be a potential entry point for a pathogenicity differentiation between Acanthamoeba isolates.
Most AK patients who have been wearing contact lenses (CLs) over a long period have a high risk of contracting AK. According to previous studies, the long-term wearing of CLs, not only affects the eye’s health but also interferes with the ocular surface microbiota distribution through oxygen deprivation [16,17]. Recent studies have demonstrated the relationship between the dysbiosis of the ocular surface microbiome and several conditions, such as dry eyes [18] and the wearing of CLs [19]. Therefore, the use of CLs might create a sealed hunting ground for Acanthamoeba, which could lead to depletion of probiotics. Nevertheless, some studies have analyzed the microbiota in CLs users [20,21], whereas the characterization of the microbiome with Acanthamoeba involvement is less reported. As most AK isolates are related to CLs use [22,23], investigation of the association between the microbiome and clinical AK isolates would be necessary for disease diagnosis and prognosis.
In this study, bacterial 16S ribosomal RNA sequencing techniques were performed to profile the intracellular microbiome of phagocytic microbes in various CL-associated AK isolates in order to characterize the association between Acanthamoeba and the intracellular microbiome in AK progression in a hypoxic environment.
2. Results
2.1. The Isolates Were Collected from Various Conditions
To determine the intracellular microbiome in the Acanthamoeba spp., we collected five clinical and two standard isolates. All isolates were cultured until normalization. The clinical isolates belonged to Acanthamoeba castellanii strain T4 and contained axenic culture from Acanthamoeba keratitis (AK) patients, who were assessed at Cheng Kung University Hospital, Taiwan. All clinical isolates were collected from patients who had lenses as a risk factor [24]. Standard isolate ATCC-30010 was collected from environmental soil in the United State and isolate ATCC-50492 was isolated from a keratitis patient from India, as described by the official website of ATCC. The NCKUH-A, B, and C strains were isolated from patients receiving medical treatment for early keratitis. The NCKUH-D and H strain patients had received surgery for late keratitis (Table 1). The isolate profiles were used as analytic factors in this study.

Table 1.
Characteristics of Acanthamoeba castellanii isolates used in the study. Profiles for isolates ATCC-30010 and ATCC-50492 are as described on the American Type Culture Collection (ATCC®) official website. The A, B, and C strains were isolated from early keratitis patients. The D and H strains were isolated from keratitis patients treated with surgery at late progression.
2.2. Specific Bacterial 16S Primers Had Detected the Intracellular Microbes in All Isolates
Acanthamoeba spp. DNA was extracted from the axenic cultures and the bacterial specific 16S primer pairs were used to pre-examine the endosymbionts using RT-PCR. No contamination was detected in this study after the screening of isolate ATCC-50492 in cultured medium and NCKUH-B using PCR detection (Figure 1A). PCR electrophoresis showed only a single pattern belonging to the isolates (Figure 1B), which indicated that the designed primer pairs showed specificity and that the isolates harbored phagocytic intracellular microbes.
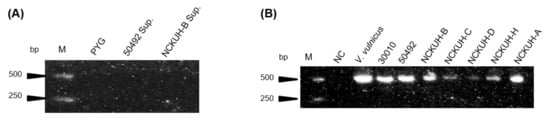
Figure 1.
Amplification of PCR cultured medium extracts and Acanthamoeba DNA samples using bacterial 16S primers. (A) Isolate ATCC-50492 and NCKUH-B cultured medium extracts were amplified by bacterial 16S primers and sterile peptone-yeast extract-glucose (PYG) medium as a negative control. (B) Acanthamoeba DNA samples amplified with bacterial 16S primers and PCR products presented at 550 bps. Vibrio vulnicus was used as positive control and distillation–distillation H2O was used as a negative control for the 16S primers. M: marker; NC: negative control
2.3. Clostridiales and Bacteroidales Are the Major Intracellular Microbes of Isolates
Regarding the 16S sequencing data from individual isolates, there were no unique differences present in all isolates. All isolates had major populations of the orders Clostridiales and Bacteroidales (Figure 2).
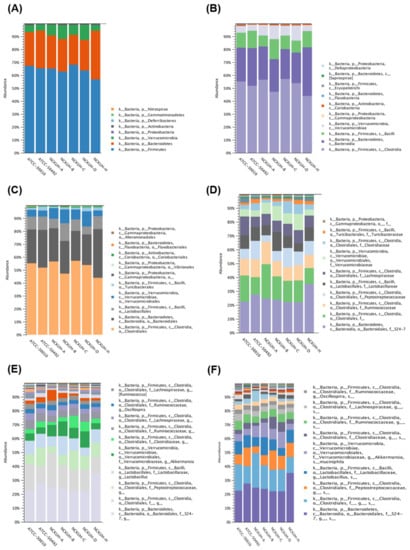
Figure 2.
16S rRNA gene sequencing of Acanthamoeba castellanii isolates. The intracellular microbes content in the individual strains of A. castellanii. The comparison data from phylum to species are shown in (A–F), respectively.
To further investigate the diversity between various Acanthamoeba spp. isolates, we compared different groups, which are described in Table 1. Comparing various isolates, either isolate ATCC-30010 from the soil source or the others from keratitis patients, all contained Clostridiales and Bacteroidales as the major populations of intracellular microbes (Figure 3). The same populations also appeared in the disease progression comparison (Figure 4). However, the two major groups could not be defined at the genus or species level owing to technical obstacles in the 16S ribosomal RNA sequencing comparison.
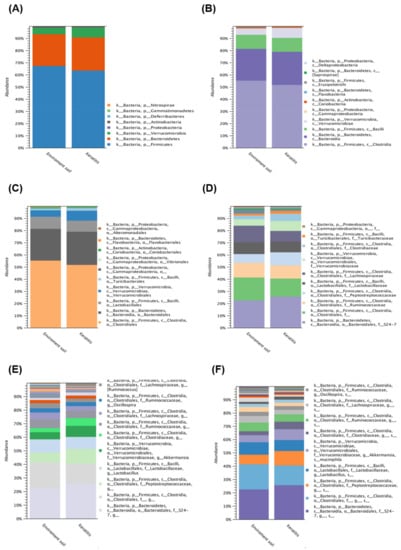
Figure 3.
16S rRNA sequencing data comparison of the isolated resource. The intracellular microbes content in the isolates source of soil and Acanthamoeba keratitis (AK) patients. The comparison data from phylum to species are shown in (A–F) respectively.
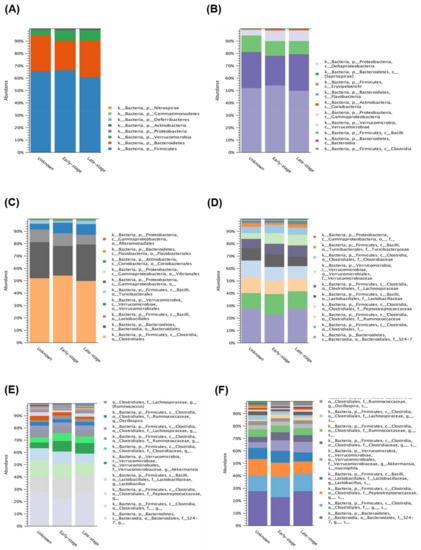
Figure 4.
16S rRNA sequencing data comparison of different progression in Acanthamoeba keratitis. The intracellular microbes content in the progression of AK isolates. The comparison data from phylum to species are shown in (A–F) respectively.
2.4. Blautia Product Showed Abundance in the Isolates of Severe Acanthamoeba Keratitis
To investigate the microbiome diversity with Acanthamoeba spp. pathogenicity, we compared the relative abundances from the different groups of gene expression databases. Interestingly, the clinical keratitis isolates showed a greater abundance of Ruminococcus gnavus (121.0 folds), Eubacterium dolichum (54.15 folds), Roseburia faecis (24.51 folds), and Blautia producta (3.15 folds) than those from soil sources. Although Akkermansia muciniphila was abundant in soil-source ameba (11.08 folds), the relative abundance was not different in the genus validation. These abundant intracellular microbes were also presented in genera of Ruminococcus (38.79 folds), Eubacterium (27.27 folds), Roseburia (19.16 folds), and Blautia (27.01 folds) during the validated comparison (Figure 5A). Furthermore, the comparison of intracellular microbe abundance in diverse keratitis progression stages showed that late-stage keratitis isolates had more Blautia producta (3.81 folds). This significant difference was also observed during the genus validation of Blautia (19.15 folds). The early-stage keratitis isolates harbored abundant microbes of Bacteroides ovatus (2.60 folds), as proven by genus validation (8.55 folds) (Figure 5B).
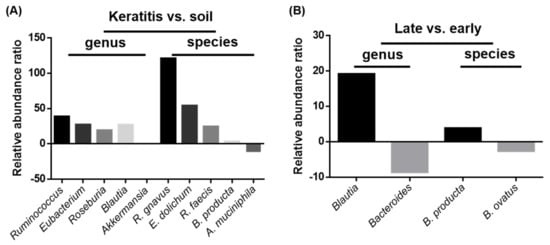
Figure 5.
Intracellular microbe relative abundance ratios of keratitis vs. soil source and late vs. early keratitis comparisons at genus and species levels. (A) Microbe relative abundance ratios of keratitis vs. soil source at genus and species levels. (B) Microbe relative abundance ratios of late vs. early keratitis patients at genus and species levels. The compared microbes are displayed on the X axis and were validated by identification in both genus and species.
3. Discussion
Free-living Acanthamoeba hosts harbor various intracellular microbes, which have been known for a long time [25]. In the present study, we analyzed five clinical isolates and two standard isolates to reveal the abundant microbes: Ruminococcus and Blautia. Of a total of seven Acanthamoeba isolates, six were obtained from lens-associated AK patients and one from a soil source which was considered as a non-disease-related strain. Of the five clinical isolates, three (NCKUH-A, B, and C) were isolated from patients who received medically successful AK treatment, whereas two (NCKUH-D and H) were from patients who finally had surgery owing to poor progression of AK. The intracellular microbiome content conversion profiles in different groups showed no obvious differences. However, the intracellular microbe relative abundance profiles from gene expression databases showed interesting diversity. The clinical isolates from lens-associated keratitis had a greater abundance of intracellular microbes of R. gnavus, E. dolichum, R. faecis, and B. producta than non-disease related strains. In contrast, the NCKUH-D and H isolate from medical failure cases had more B. producta and less B. ovatus in comparison with medical success isolates. Hence, the intracellular microbe relative abundance data suggested that the anaerobic Gram-positive bacillus Blautia producta might play roles in Acanthamoeba spp. pathogenicity in lens-wearing patients.
B. producta was described in 1941 [26] and is commonly found in human feces. It has the capacity for rapid growth at the expense of carbon monoxide (CO) through anaerobic digestion [27]. B. ovatus was first reported in 1933 [28] and provides an inherent biosafety feature in the human gut [29]. However, B. producta is Gram-positive and B. ovatus is Gram-negative. According to previous studies, human ocular microbiota was characterized to be commonly identified in Gram-positive bacteria [30], while Acanthamoeba apparently prefer Gram-negative bacteria [31]. Previous studies had demonstrated that amoebae prefer certain species of bacteria as a food source, such as P. aeruginosa and S. marcescens [32], but not S. epidermidis [33,34]. It was presumed that the gram-positive cell wall was thick and hard to be digestion for Acanthamoeba. Therefore, the early-stage keratitis isolates harbored abundant Gram-negative bacteria, with B. ovatus suspected as the early food source during initial infection. Notably, the relative abundance of B. ovatus was significantly higher in patients with atopic dermatitis as an immune disorder [35], whereas B. producta had demonstrated prevention of Vancomycin-resistant Enterococcus (VRE) colonization [36]. These results suggest the importance of Acanthamoeba ingestion of bacteria, which might lead to secondary inflammation or infection due to ocular surface dysbiosis.
Additionally, both B. producta and B. ovatus are anaerobic microbes, which were not the major populations of ocular microbiota. This was probably due to vigorous reproduction on the ocular surface, which made a hypoxic environment, leading to lens obstruction. Although several diseases can lead to ocular hypoxia, including diabetes, herpes simplex virus infection, dry eye syndrome, and limbal stem cell deficiency [37], lens use has become the common factor in induced ocular hypoxia, due to the increasing use of CLs [38]. Previous studies had demonstrated that amoebae containing endosymbionts had more high pathogenicity than endosymbiont-free amoebae. Endosymbionts may influence Acanthamoeba pathogenicity, virulence, or susceptibility to drugs [39]. However, the ingested microorganisms might play roles in the protection of the cornea. Recent studies on ocular microbiota found that common genera, such as Staphylococci and Streptococcus, can stimulate ocular immunity as a natural barrier on the corneal surface [40]. Under hypoxia and less remaining microbiota condition, AK could be predicted to poor progression and Acanthamoeba isolates were consider as high pathogenicity. Therefore, the analysis of endosymbionts in Acanthamoeba might be beneficial for AK patient prognosis.
In conclusion, these findings indicate that the types of intracellular microbes were associated with AK progression, while the abundant anaerobic bacteria in the keratitis isolates were associated with CLs wearing, suggesting hypoxia might correlate to AK induction. Further investigation of Acanthamoeba intracellular microbes will be necessary and may provide more information for the differential diagnostic and prognostic evaluation of AK.
4. Materials and Methods
4.1. Culture of Acanthamoeba Protozoa
The standard isolates ATCC-30010 and ATCC-50492 were Acanthamoeba castellanii which were isolated from soil and human cornea with keratitis and purchased from ATCC (Manassas, VA, USA). The clinical isolates were collected from the corneal ulcers of patients who were diagnosed with AK in the Cheng Kung University Hospital [24] and are characterized in Table 1. The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of National Cheng Kung University Hospital (Protocol code: A-BR-101-124; Date of approval: 9 November 2012). Acanthamoeba isolates were cultured in protease peptone-yeast extract-glucose (PYG) medium (pH 6.5) at 28 °C in cell culture flasks and maintained after Page’s modified Neff’s amoeba saline (PAS: 1.2 g NaCl, 0.04 g MgSO4×7H2O, 0.03 g CaCl2, 1.42 g Na2HPO4, 1.36 g KH2PO4 in 1 L ddH2O) washing.
4.2. Genomic DNA Extraction and Sample Preparation
To identify the endosymbionts in A. castellanii, protozoan cells were grown in axenic culture and subsequently harvested. The cell pellets were suspended in PAS for genome extraction. DNA extraction was performed using a LabPrep DNA Mini kit® (TAIGEN Bioscience Corporation, Taipei, Taiwan) according to the protocol described in the package insert.
4.3. 16S Ribosomal RNA Sequencing
The enriched DNA samples were then subjected to PCR reactions with specified forward (TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCA G) and reverse (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGAC TACHVG GGTATCTAATCC) primers [41] to amplify the V3–V4 genomics region of bacterial 16S rRNA genes. To tested axenic cultures, the unused PYG medium and the medium from cultured Acanthamoeba were analyzed by polymerase chain reaction (PCR) (10 min at 95 °C, followed by 25 cycles of 30 secs at 95 °C, 30 secs at 55 °C, and 30 secs at 72 °C). To tested intracellular symbionts presenting, all the extracted DNA of isolates were analyzed by PCR as mentioned above. With approximately 550 bp PCR product, as confirmed by gel electrophoresis, the DNA samples were further subjected to sample preparation for metagenomics sequencing. DNA samples were prepared according to the 16S Metagenomics Sequencing Library Preparation instructions (Illumina, California, USA). The prepared amplicons were sequenced on a MiSeq instrument (Illumina, California, USA) using a 600-cycle sequencing reagent and with the specified paired-end mode.
4.4. Data Analyses
The generated Next-generation sequencing (NGS) data were analyzed with the Microbial Genomics Module of CLC Genomics Workbench 9.5.4 (Qiagen, Stockach, Germany). As shown in Figure 6, the raw NGS reads were first subjected to quality trimming from the 3′ end, an optimal merging of paired reads, and fixed-length trimming, followed by operational taxonomic unit (OTU) clustering. After the OTU clustering step, the generated OTU table was further analyzed to calculate the alpha and beta diversity. Meanwhile, the generated OTU table was also subject to PERMANOVA analysis and differential abundance analysis. The parameters for each step were mainly the default values, with the specific values shown in the corresponding dialogue boxes.
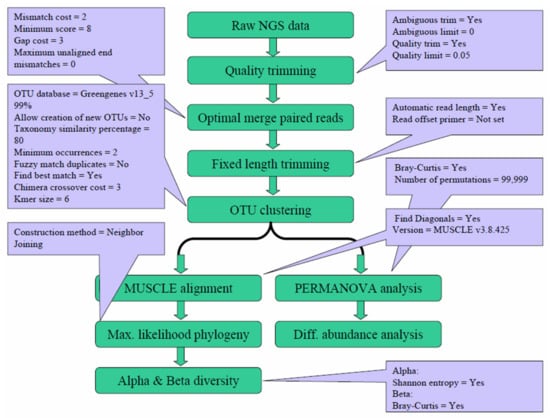
Figure 6.
Workflow and parameters used for data analysis. The generated Next-generation sequencing (NGS) reads were subjected to quality trimming, merging, and fixed-length trimming, followed by operational taxonomic unit (OTU) clustering. The generated OTU table was analyzed for alpha and beta diversity. The parameters for each step are shown in the corresponding dialogue boxes.
4.5. Gene Abundance Comparison
The comparison of intracellular bacteria gene abundance was analyzed from the NGS database. The analyzed targets were classified into keratitis, soil, early-stage AK, and late-stage AK. Keratitis includes ATCC-50492, NCKUH-A, -B, -C, -D, and –H; soil includes ATCC-30010; early-stage AK includes NCKUH-A, -B, and –C; late-stage AK includes NCKUH-D and –H. To characterize the intracellular microbiome relative abundance ratio, gene expression fold change of keratitis was divided by soil and late-stage AK was divided by early-stage AK. The significant bacteria showed consistent abundance in the comparison within family, genus, and species.
Author Contributions
Conceptualization, W.-C.L. and F.-C.H.; methodology, W.-C.L. and F.-C.H.; software, S.-C.L.; validation, Y.-J.W. and S.-C.L.; formal analysis, Y.-J.W.; investigation, Y.-J.W.; resources, F.-C.H.; data curation, Y.-J.W.; writing—original draft preparation, Y.-J.W.; writing—review and editing, W.-C.L. and F.-C.H.; visualization, W.-C.L. and F.-C.H.; supervision, W.-C.L. and F.-C.H.; project administration, W.-C.L. and F.-C.H.; funding acquisition, W.-C.L. All authors have read and agreed to the published version of the manuscript.
Funding
The Ministry of Science and Technology (MOST) to Wei-Chen Lin (grant MOST 109-2628-B-006-022).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of National Cheng Kung University Hospital (Protocol code: A-BR-101-124; Date of approval: 9 November 2012).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
Our thanks to the Department of Ophthalmology in National Cheng Kung University Hospital who provided the clinical isolates. Our thanks also to the Department of Medical Research in Kaohsiung Chang Gung Memorial Hospital for supporting the technology of 16S ribosomal RNA sequencing techniques.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Denet, E.; Coupat-Goutaland, B.; Nazaret, S.; Pélandakis, M.; Favre-Bonté, S. Diversity of free-living amoebae in soils and their associated human opportunistic bacteria. Parasitol. Res. 2017, 116, 3151–3162. [Google Scholar] [CrossRef] [PubMed]
- Pagnier, I.; Valles, C.; Raoult, D.; La Scola, B. Isolation of Vermamoeba vermiformis and associated bacteria in hospital water. Microb. Pathog. 2015, 80, 14–20. [Google Scholar] [CrossRef]
- Wen, X.; Miao, L.; Deng, Y.; Bible, P.W.; Hu, X.; Zou, Y.; Liu, Y.; Guo, S.; Liang, J.; Chen, T. The influence of age and sex on ocular surface microbiota in healthy adults. Invest. Ophthalmol. Vis. Sci. 2017, 58, 6030–6037. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, H.; Hu, M.; Ma, Y.; Chen, P.; Zhao, Z.; Li, J.; Ye, Y.; Zheng, M.; Lou, Y. Alterations in the Ocular Surface Microbiome in Traumatic Corneal Ulcer Patients. Invest. Ophthalmol. Vis. Sci. 2020, 61, 35. [Google Scholar] [CrossRef]
- Scheid, P. Relevance of free-living amoebae as hosts for phylogenetically diverse microorganisms. Parasitol. Res. 2014, 113, 2407–2414. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.; Closures, B. The Hunt for Microbial ’Trojan Horses’. Oceanus Magazine 2009, 47, 3. [Google Scholar]
- Guimaraes, A.J.; Gomes, K.X.; Cortines, J.R.; Peralta, J.M.; Peralta, R.H. Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiol. Res. 2016, 193, 30–38. [Google Scholar] [CrossRef]
- Steenbergen, J.; Shuman, H.; Casadevall, A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. 2001, 98, 15245–15250. [Google Scholar] [CrossRef]
- Willcox, M.D. Pseudomonas aeruginosa infection and inflammation during contact lens wear: A review. Optom. Vis. Sci. 2007, 84, 273–278. [Google Scholar] [CrossRef]
- Moliner, C.; Raoult, D.; Fournier, P.-E. Evidence of horizontal gene transfer between amoeba and bacteria. Clin. Microbiol. Infect. 2009, 15, 178–180. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, M. Comparative genomic analysis of acanthamoeba endosymbionts highlights the role of amoebae as a “Melting Pot” Shaping the rickettsiales evolution. Genome Biol. Evol. 2017, 9, 3214–3224. [Google Scholar] [CrossRef]
- Paterson, G.N.; Rittig, M.; Siddiqui, R.; Khan, N.A. Is Acanthamoeba pathogenicity associated with intracellular bacteria? Exp. Parasitol. 2011, 129, 207–210. [Google Scholar] [CrossRef]
- Iovieno, A.; Ledee, D.R.; Miller, D.; Alfonso, E.C. Detection of bacterial endosymbionts in clinical Acanthamoeba isolates. Ophthalmology 2010, 117, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, T.R.; Sobek, D.; Gautom, R.K. Enhancement of in vitro cytopathogenicity by Acanthamoeba spp. following acquisition of bacterial endosymbionts. FEMS Microbiol. Lett. 1998, 166, 231–236. [Google Scholar] [CrossRef]
- Badenoch, P.R.; Johnson, A.M.; Christy, P.E.; Coster, D.J. Pathogenicity of Acanthamoeba and a Corynebacterium in the rat cornea. Arch. Ophthalmol. 1990, 108, 107–112. [Google Scholar] [CrossRef]
- Shin, H.; Price, K.; Albert, L.; Dodick, J.; Park, L.; Dominguez-Bello, M.G. Changes in the eye microbiota associated with contact lens wearing. MBio 2016, 7, 16. [Google Scholar] [CrossRef]
- Boost, M.; Cho, P.; Wang, Z. Disturbing the balance: Effect of contact lens use on the ocular proteome and microbiome. Clin. Exp. Optom. 2017, 100, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.E.; Moore, J.E.; Jiru, X.; Moore, J.E.; Goodall, E.A.; Dooley, J.S.; Hayes, V.E.; Dartt, D.A.; Downes, C.S.; Moore, T.C. Ocular pathogen or commensal: A PCR-based study of surface bacterial flora in normal and dry eyes. Invest. Ophthalmol. Vis. Sci. 2007, 48, 5616–5623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, F.; Hutchinson, D.S.; Sun, W.; Ajami, N.J.; Lai, S.; Wong, M.C.; Petrosino, J.F.; Fang, J.; Jiang, J. Conjunctival microbiome changes associated with soft contact lens and orthokeratology lens wearing. Invest. Ophthalmol. Vis. Sci. 2017, 58, 128–136. [Google Scholar] [CrossRef]
- Sweeney, D.F.; Stapleton, F.; Leitch, C.; Taylor, J.; Holden, B.A.; Willcox, M.D. Microbial colonization of soft contact lenses over time. Optom. Vis. Sci. 2001, 78, 100–105. [Google Scholar] [CrossRef]
- Stapleton, F.; Naduvilath, T.; Keay, L.; Radford, C.; Dart, J.; Edwards, K.; Carnt, N.; Minassian, D.; Holden, B. Risk factors and causative organisms in microbial keratitis in daily disposable contact lens wear. PLoS ONE 2017, 12, e0181343. [Google Scholar] [CrossRef] [PubMed]
- Pacella, E.; La Torre, G.; De Giusti, M.; Brillante, C.; Lombardi, A.M.; Smaldone, G.; Lenzi, T.; Pacella, F. Results of case-control studies support the association between contact lens use and Acanthamoeba keratitis. Clin. Ophthalmol. (Auckland, NZ) 2013, 7, 991. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.M.; Tártara, L.I.; Cid, A.G.; Real, J.P.; Bermúdez, J.M.; Rajal, V.B.; Palma, S.D. Acanthamoeba in the eye, can the parasite hide even more? Latest developments on the disease. Cont. Lens Anterior Eye 2018, 41, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-L.; Shih, M.-H.; Huang, F.-C.; Tseng, S.-H.; Chen, C.-C. Treatment of early Acanthamoeba keratitis with alcohol-assisted epithelial debridement. Cornea 2012, 31, 442–446. [Google Scholar] [CrossRef]
- Hall, J.; Voelz, H. Bacterial endosymbionts of Acanthamoeba sp. J. Parasitol. 1985, 71, 89–95. [Google Scholar] [CrossRef]
- Prévot, A.; Boorsma, H. Au sujet de la fermentation du glucose parPl. Tetani. Antonie van Leeuwenhoek 1941, 7, 239–241. [Google Scholar] [CrossRef]
- Ma, K.; Wohlfarth, G.; Diekert, G. Acetate formation from CO and CO 2 by cell extracts of Peptostreptococcus productus (strain Marburg). Arch. Microbiol. 1991, 156, 75–80. [Google Scholar] [CrossRef]
- Eggerth, A.H.; Gagnon, B.H. The bacteroides of human feces. J. Bacteriol. 1933, 25, 389. [Google Scholar] [CrossRef]
- Hamady, Z.Z.; Scott, N.; Farrar, M.D.; Lodge, J.P.A.; Holland, K.T.; Whitehead, T.; Carding, S.R. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut 2010, 59, 461–469. [Google Scholar] [CrossRef]
- Ozkan, J.; Willcox, M.; Wemheuer, B.; Wilcsek, G.; Coroneo, M.; Thomas, T. Biogeography of the human ocular microbiota. Ocul. Surf. 2019, 17, 111–118. [Google Scholar] [CrossRef]
- Gorlin, A.I.; Gabriel, M.M.; Wilson, L.A.; Ahearn, D.G. Effect of adhered bacteria on the binding of Acanthamoeba to hydrogel lenses. Arch. Ophthalmol. 1996, 114, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Penland, R.L.; Wilhelmus, K.R. Comparison of axenic and monoxenic media for isolation of Acanthamoeba. J. Clin. Microbiol. 1997, 35, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J.; Madayag, R.M.; Qureshi, M.N. Acanthamoeba keratitis: Synergy between amebic and bacterial cocontaminants in contact lens care systems as a prelude to infection. J. Clin. Microbiol. 1992, 30, 2447–2450. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.; Pere, A.; Gordon, R.; Qureshi, M. Differential binding capacity and internalisation of bacterial substrates as factors in growth rate of Acanthamoeba spp. J. Med. Microbiol. 1994, 40, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Yan, F.; Wang, H.; Mo, X.; Liu, J.; Zhang, Y.; Li, H.; Chen, D. Diversity analysis of gut microbiota between healthy controls and those with atopic dermatitis in a Chinese population. J. Dermatol. 2020, 48, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Caballero, S.; Kim, S.; Carter, R.A.; Leiner, I.M.; Sušac, B.; Miller, L.; Kim, G.J.; Ling, L.; Pamer, E.G. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 2017, 21, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Onochie, O.E.; Onyejose, A.J.; Rich, C.B.; Trinkaus-Randall, V. The role of hypoxia in Corneal extracellular matrix deposition and cell motility. Anat. Rec. 2020, 303, 1703–1716. [Google Scholar] [CrossRef]
- Ladage, P.M.; Yamamoto, K.; Ren, D.H.; Li, L.; Jester, J.V.; Petroll, W.M.; Cavanagh, H.D. Effects of rigid and soft contact lens daily wear on corneal epithelium, tear lactate dehydrogenase, and bacterial binding to exfoliated epithelial cells. Ophthalmology 2001, 108, 1279–1288. [Google Scholar] [CrossRef]
- Greub, G.; Raoult, D. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 2004, 17, 413–433. [Google Scholar] [CrossRef]
- Lu, L.J.; Liu, J. Focus: Microbiome: Human microbiota and ophthalmic disease. Yale J. Biol. Med. 2016, 89, 325. [Google Scholar]
- Kato, Y.; Sato, K.; Asai, K.; Akutsu, T. Rtips: Fast and accurate tools for RNA 2D structure prediction using integer programming. Nucleic Acids Res. 2012, 40, 29–34. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).