Simple Summary
Ectropis obliqua is a destructive leaf-eating pest that is widely distributed in China’s tea gardens. This pest shows remarkable resistance against multiple insecticides. As an environmentally friendly entomopathogen, Beauveria bassiana has been widely used to prevent agricultural pest infestations. However, the molecular mechanism of B. bassiana against E. obliqua remains unclear. We firstly isolated and identified a highly virulent B. bassiana strain. Using a transcriptome, we analyzed the differences of immune gene expression levels in fat bodies and hemocytes of E. obliqua that were infected by the B. bassiana, which provide molecular insights into the insect–pathogen interaction.
Abstract
Ectropis obliqua is a destructive masticatory pest in China’s tea gardens. Beauveria bassiana as microbial insecticides can effectively control E. obliqua larvae; however, the immune response of this insect infected by B. bassiana are largely unknown. Here, after isolating a highly virulent strain of B. bassiana from E. obliqua, the changes in gene expression among different tissues, including hemocytes and fat bodies, of E. obliqua larvae infected by the entomopathogen were investigated using transcriptome sequencing. A total of 5877 co-expressed differentially expressed genes (DEGs) were identified in hemocytes and fat bodies, of which 5826 were up-regulated in hemocytes and 5784 were up-regulated in fat bodies. We identified 249 immunity-related genes, including pattern recognition receptors, immune effectors, signal modulators, and members of immune pathways. A quantitative real-time PCR analysis confirmed that several pattern recognition receptors were upregulated in hemocytes and fat bodies; however, others were downregulated. The investigated immune effectors (ATT and PPO-1) were suppressed. The results showed that there were tissue differences in the expression of immune genes. This study provides a large number of immunity-related gene sequences from E. obliqua after being infected by B. bassiana, furthering the understanding of the molecular mechanisms of E. obliqua defenses against B. bassiana.
1. Introduction
Ectropis obliqua (Lepidoptera: Geometridae) is the most harmful leaf-feeding pest of tea gardens in China, owing to its wide distribution and great destructiveness [1,2]. The larvae feed on tea leaves, consuming all the tea leaves in a tea garden during the outbreak period, badly inhibiting tea plant growth, and reducing tea production [3,4]. In recent years, insecticide-mediated chemical management methods have not only harmed human health and the environment, but they have also caused a more serious problem: insecticide resistance in pests [5,6,7]. Therefore, there is an urgent need to develop new pest control methods that are eco-friendly, sustainable, and able to overcome insecticide resistance [8].
Beauveria bassiana, an entomopathogenic fungus, has a wide host range, is environmentally friendly, and markedly improves pest control efficacies, resulting in its use as a biological control agent worldwide [9,10]. It has significant insecticidal effects on Ectropis obliqua [11], Rhynchophorous ferrugineus [12], Spodoptera frugiperda [13], Tetranychus urticae [14], and Tuta absoluta [15,16]. Generally, the infection mechanism of B. bassiana is as follows: the fungal conidia adhere to the host surface, proteases and chitinases are then secreted to hydrolyze the host cuticle, and then, the fungi invade and colonize the host hemocoel. They grow by absorbing host nutrients, and eventually the host is killed by the secreted toxins and physiological starvation [8,17,18,19,20]. The host’s natural cellular and humoral immune responses are activated after pathogenic fungi overcome the host’s physical barriers. In previous studies, a large number of immune genes have been identified in Helicoverpa armigera [21], Clanis bilineata [21], Galleria mellonella [22], Riptortus pedestris [23], Ostrinia furnacalis [24], and Hypothenemus hampei [25] infected with B. bassiana. These data provide clues and gene candidates for further exploration on the molecular mechanisms of entomopathogenicity by this fungus. However, the immune response mechanisms of E. obliqua after infection by the pathogenic fungi B. bassiana remain unclear.
In this study, we determined the sensitivity of fifth-instar E. obliqua larvae to the adherence of B. bassiana conidia, and then, the transcriptomic profiles of hemocytes and fat body tissues from E. obliqua infected by B. bassiana were analyzed using a high-throughput RNA sequencing method. We identified potential immunity-related genes through comparisons with homologous sequences of other insects. Finally, the expression levels of several key host immune genes were verified using quantitative real-time PCR (qRT-PCR). This study provides a theoretical basis for understanding the immune mechanisms of E. obliqua against B. bassiana, providing molecular insight into the complicated interaction between host and fungal pathogen.
2. Materials and Methods
2.1. Isolation, Cultivation, and Identification of Entomopathogenic Fungi
Fungal-infected Ectropis obliqua larvae was collected from tea plant leaves in the wild at Dayangzhen Tea Garden in Hefei, China (31.92° N, 117.21° E). Entomopathogenic fungi were isolated from the infected Ectropis obliqua larvae. Mycelium on the surfaces of E. obliqua were inoculated into potato dextrose agar medium and cultured in an incubator at 28 °C for 5 days. The isolated strains were purified three times. The benzyl chloride method was used to extract fungal DNA [26]. Using the genomic DNA of the strain as the template, the ITS-rDNA sequences were amplified using PCR with the universal primer pair ITS4 (5′-TCCTCCGCTTATTGATATGC-3′)/ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′). Briefly, the PCR reaction conditions were as follows: pre-denaturation at 95 °C for 5 min and 30 cycles of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min, followed by a final elongation step at 72 °C for 10 min. PCR products were detected by electrophoresis in 1% agarose gels. Later, the target ITS sequences were submitted to National Center for Biotechnology Information (NCBI) and Basic Local Alignment Search Tool (BLAST) were used for database search, and the identified homologous sequences were downloaded. A phylogenetic tree was constructed using MEGA-11 software (Home (megasoftware.net, accessed on 6 December 2021)).
2.2. Insect Rearing and Infection Bioassays
E. obliqua moths were acquired from the State Key Laboratory of Tea Plant Biology, Anhui Agricultural University, Hefei, China (31.86° N, 117.27° E). The larvae were reared on tea leaves at 23 ± 2 °C and 70–80% relative humidity with a 16 h light/8 h dark photoperiod in the insect-rearing laboratory. Tea leaves used in the experiment were inserted into flower mud for storage. A total of 120 fifth-instar larvae were randomly selected and were injected with B. bassiana conidial suspension (2 μL, 1 × 107 conidial mL−1) and Tween 80 solution (2 μL) using microliter syringes (Shanghai GaoGe Co., Shanghai, China) (n = 60). The numbers of dead larvae were recorded for 10 days. During this period, the dead larvae were collected and kept moisturized to observe whether the white hyphae could grow from the larvae.
2.3. RNA Preparation and Illumina Sequencing
At 0 h and 48 h of infection, fat body tissues and hemolymph samples of larvae were collected. Fat body tissues from ten larvae were collected under the microscope with dissecting forceps as one sample, respectively. Hemolymph collected from ten larvae was pooled into a 1.5 mL RNA-free microcentrifuge tube containing 0.1% 1-phenyl-2-thiourea and centrifuged at 500× g for 5 min at 4 °C to collect hemocytes [19]. Each sample was performed in triplicate. The samples were stored at −80 °C. Complementary DNA (cDNA) libraries were constructed using an Illumina Truseq™ Sample Prep Kit (Illumina, CA, USA). All the samples were sequenced on an Illumina Novaseq6000 platform (Illumina, San Diego, CA, USA). Original data were stored in FASTQ format.
2.4. Assembly and Annotation of Transcripts
Before assembly, to obtain satisfactory determination results, low-quality sequences (Quality < 20), sequences with nitrogen contents over 10%, and adapter sequences were removed. Trinity software was used to de novo assemble all the clean reads to produce contigs and singletons [27]. By comparing the known sequences in the public database, the functional annotation of these unigenes was obtained. The assembled unigene sequences were annotated against the NCBI non-redundant protein sequence and Swiss-prot databases using BLASTX (E-value < 10−5) [20,28]. The gene ontology (GO) annotation was obtained using the Blast2GO program [19,20]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was performed to discover the potential enriched pathways [29].
Differentially expressed genes (DEGs) were calculated using the fragments per kilobases per million mapped reads (FPKM) method, and the screening conditions (|log2fold change| ≥ 1 and p-value < 0.05) were used to identify the DEGs [30]. Through a comparison with known amino acid sequences of immunity-related genes in other insects, the E. obliqua unigenes were obtained using the tblastn method. Subsequently, the E. obliqua immunity-related genes were manually confirmed.
2.5. Tissue Differential Expression Assessed by qRT-PCR
Another sixty E. obliqua larvae were randomly selected for a test of injection with B. bassiana conidial suspension (1 × 107 conidial mL−1) and Tween 80 solution (n = 30). Ten larvae in each treatment were randomly chosen, and total RNA was extracted from fat body tissues and hemolymph. Each sample was performed in triplicate. In total, 1 μg of RNA was reverse transcribed using the StarScript II First-strand cDNA Synthesis Kit (GenStar, Beijing, China). Quantitative real-time PCR (qRT-PCR) was performed on an ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using GoTaq qPCR Master Mix (Promega, Madison, WI, USA) in a 20-µL volume. Each well plate was loaded with 1μL cDNA. The reaction steps were as follows: 95 °C for 2 min and 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The data were analyzed using the 2−ΔΔCt method [20,31]. The β-actin gene of E. obliqua was used as the reference gene. The primers were described previously (Table S1).
2.6. Data Analysis
GraphPad Prism 7.0, AI, Tbtools, IBM SPSS Statistics 25 was used for statistical analyses and to construct figures. A t-test was used to analyze the survival curve, and qPT-PCR data were analyzed using the 2−ΔΔCt method.
3. Results
3.1. Biological Identification of B. bassiana and Its Virulence against E. obliqua
On the basis of the phylogenetic tree, the gene sequences of the target strain were clustered with those of multiple Beauveria bassiana strains (Figure 1 and Figure S1); consequently, the strain Bb-1 was identified as B. bassiana. Over 97% of E. obliqua fifth-instar larvae were killed by the B. bassiana infection after 60 h, which was significantly greater than the larvae killed by the Tween 80 treatment (p < 0.01) (Figure 2A). After the injection of B. bassiana spores, all the larvae died and became stiff after 60 h treatment, and the white hyphae on the surface of infected larvae were visualized at 4 days post-infection, which then covered the whole insect body after another 3 days of infection (Figure 2B).
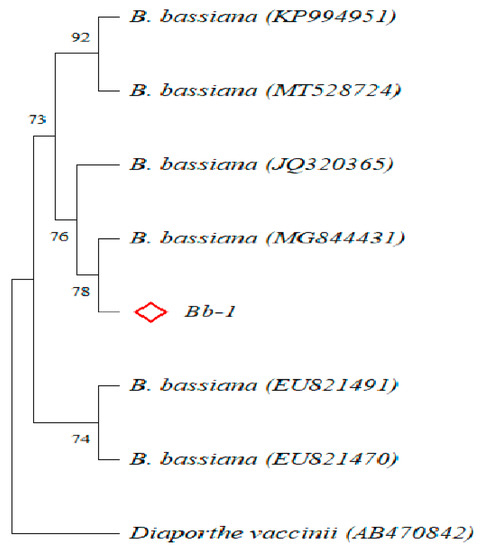
Figure 1.
Construction of phylogenetic tree (Neighbour-Joining method; ◊ represent one B. bassiana strain isolated in this study; AB470842 Diaporthe vaccinii as outgroup).
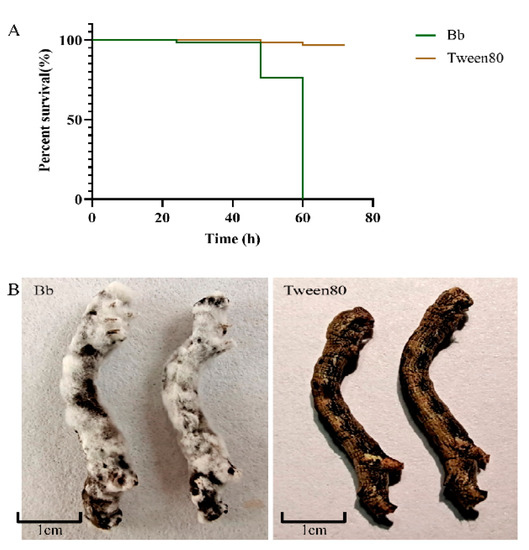
Figure 2.
(A) Effects of injection on the survival rate of E. obliqua larvae. (B) Different phenotypes of the E. oblique larvae infected with B. bassiana and Tween 80.
3.2. Transcriptome Analysis after B. bassiana Infection
To obtain a transcriptome of Ectropis obliqua, the hemocytes and fat body tissues were collected from B. bassiana-infected larvae after 48 h. A total of 56,412 unigenes were annotated using BLASTX searches. A Venn diagram analysis indicated that 5877 genes were co-expressed between hemocyte and fat body samples (5826 upregulated and 51 downregulated in hemocytes and 5784 upregulated and 93 downregulated in fat bodies) (Figure 3, Table S2). The DEGs in the hemocytes and fat bodies were enriched in the following GO terms: biological processes (cellular activity and metabolic process), molecular functions (catalytic activity and binding), and cellular components (cell part, membrane part, organelle, and protein-containing complex) (Figure 4).
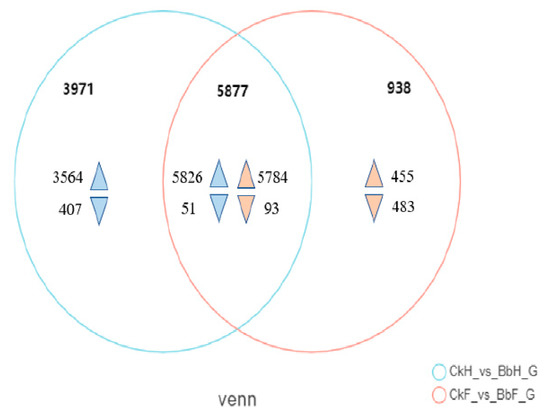
Figure 3.
Venn diagram of the statistics of differentially expressed genes (DEGs) between fat body and hemocyte in B. bassiana- and Tween 80-treated E. oblique larvae.
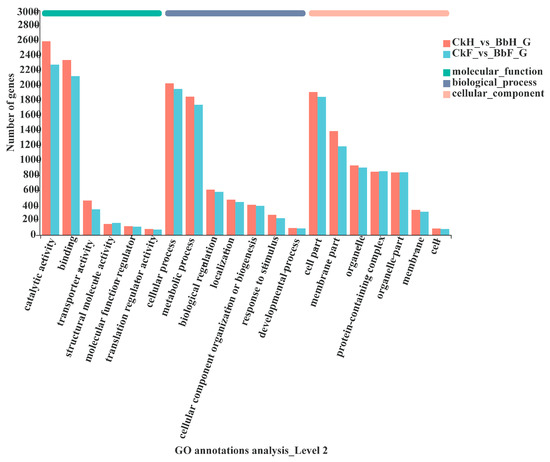
Figure 4.
Gene ontology (GO) annotation of DEGs in the E. oblique transcriptome.
To compare gene expression levels in the hemocytes and fat bodies of B. bassiana-infected larvae, a hierarchical clustering analysis of the DEGs was performed using fragments per kilobases per million mapped reads. Four major gene clusters were identified that exhibited distinct expression patterns among the different groups (Figure 5, Table S2). The 482 genes in cluster 2 and the 24 genes in cluster 3 had higher and lower expression levels, respectively, in the hemocytes and fat bodies of B. bassiana-infected larvae. The 5275 genes in cluster 1 showed significantly greater RNA levels in hemocytes from entomopathogenic fungi-infected larvae compared with those of non-infected larvae. Cluster 4 (96 genes) showed identical expression patterns in the hemocytes and fat bodies of B. bassiana- and Tween-infected larvae. Four gene clusters were categorized into thirteen functional pathways using KEGG classifications. Most of the upregulated genes in clusters 1 and 2 were involved in immune responses, energy, and carbohydrate, lipid, and amino acid metabolic processes (Figure 6).
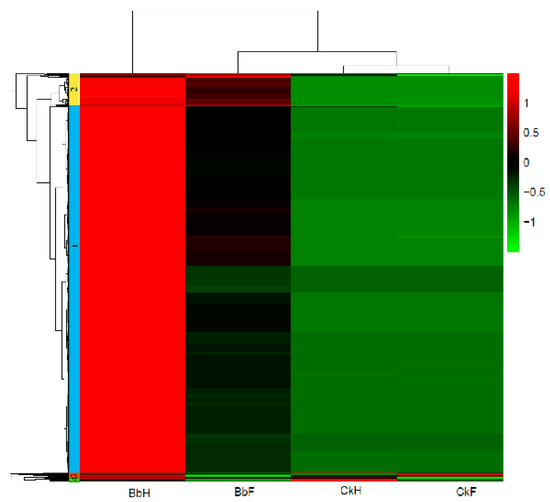
Figure 5.
Hierarchical clustering analysis of mutual DEGs in fat body and hemocytes of E. oblique larvae.
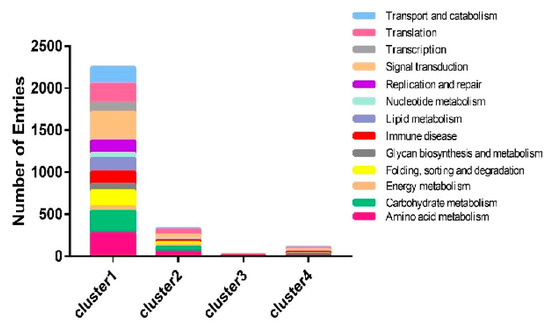
Figure 6.
Functional classification of DEGs.
Furthermore, 249 E. obliqua immunity-related genes were identified from 56,412 unigenes, and they were compared with known protein sequences in Drosophila melanogaster [32], Bombyx mori [33], and Helicoverpa armigera [28]. The E. obliqua immunity-related genes were classified as recognition molecules and immune response effectors, as well as components of extracellular signal modulation and intracellular signal transduction on the basis of their functions. Intracellular signal transduction genes included members of the Toll, IMD, JNK, and JAK/STAT pathways (Figure 7, Table S3). The differential expression analysis indicated that 16 genes (14 upregulated and 2 downregulated) were overexpressed in hemocytes and fat bodies (Figure 8).
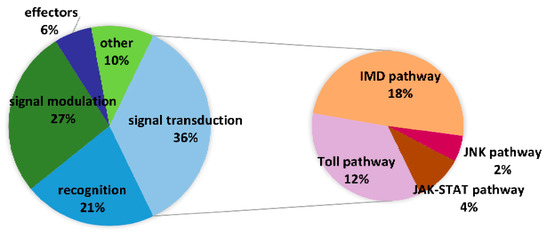
Figure 7.
Distribution of E. oblique immunity-related genes in the categories of pathogen recognition, signal modulation, signal transduction (Toll, IMD, JNK, and JAK/STAT pathways), and immune.
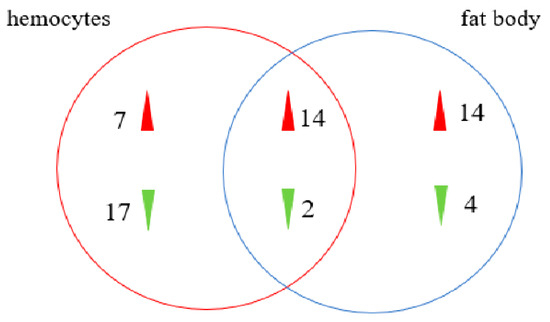
Figure 8.
The immunity-related DETs in hemocytes and fat body after B. bassiana infection.
In addition, we compared and analyzed some immunity-related genes of E. obliqua with those of other major insects, including members of Lepidoptera (Bombyx mori [33], Plutella xylostella [34,35], Helicoverpa armigera [36], Lymantria dispar [19], and Hepialus xiaojinensis [37]), Coleoptera (Tribolium castaneum [38] and Dendroctonus valens [20]), Hymenoptera (Apis mellifera [39]), and Diptera (Anopheles gambiae [40] and Drosophila melanogaster [40]). The immunity-related genes of E. obliqua (148) were similar to those of L. dispar (150). The number of immunity-related genes was highest in D. melanogaster (180) and lowest in A. mellifera (63) (Table 1)

Table 1.
Families and counts of innate immune genes from the insects. (The gene counts of B. mori, T. castaneum, A. mellifera, A. gambiae, D. melanogaster were based on genomic data.).
3.3. Expression Patterns of Immune-Related Genes Verified by qRT-PCR
In total, ten immunity-related genes with differential expression patterns in the transcriptome were selected for qRT-PCR (Figure 9). After 48 h, the B. bassiana-infected larvae’s genes involved in the recognition of C-type lectin CTL-10 were markedly upregulated, whereas the peptidoglycan recognition protein PGRP-5 and PGRP-6 genes were dramatically downregulated in hemocytes and fat bodies. The GNBP-2 gene was downregulated in hemocytes but upregulated in fat bodies. While SPH-5 was upregulated, serpin-7 was not differentially expressed in fat bodies. The immune response effector genes ATT and PPO-1 were significantly downregulated in hemocytes and fat bodies. Among the Toll pathway genes, SPZ 1–5 genes were all mostly up-regulated in hemocytes and down-regulated in fat bodies, according to our transcriptome data. SPZ-2 gene was further verified by qRT-PCR, which was consistent with the transcriptome data (Table S3).
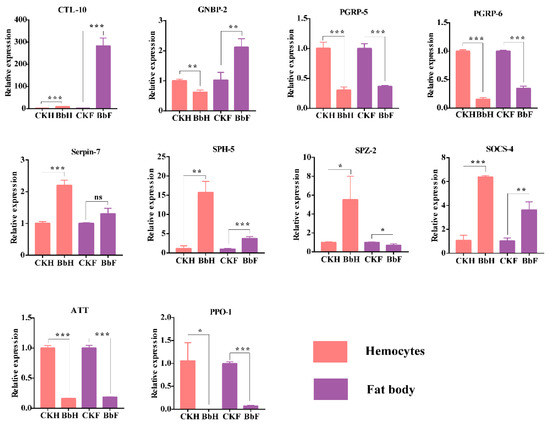
Figure 9.
Quantitative real-time PCR analysis of the E. oblique immunity-related gene expression in hemocytes and fat body after B. bassiana (48 h) injection. β-actin was used as an internal reference gene. The data is represented as the mean ± S.D. (n = 3). * p < 0.05; ** p < 0.01. *** p < 0.001. ns: no significance.
4. Discussion
We analyzed changes in gene expression levels in different tissues, including hemocytes and fat body tissues, of E. obliqua infected with B. bassiana using a transcriptome. We identified 5877 DEGs in hemocytes and fat body tissues of E. obliqua larvae. This indicated that the pathogenic fungal invasion can profoundly provoke the immune system of E. obliqua. These results were consistent with the gene expression in L. dispar infected by B. bassiana [19]. The infection of E. obliqua larvae by B. bassiana activated a series of molecular responses; besides, B. bassiana may also consume carbohydrates and lipids, according to the GO and KEGG annotations. In total, we identified 249 immunity-related genes, which was more than identified previously in other insects. In total, 190, 233, 232, 149, and 244 immunity-related genes have been identified in O. furnacalis [41], H. armigera [28], M. sexta [42], P. xylostella [34], and L. dispar [19], respectively. These immunity-related genes are involved in recognition, signal modulation, and signal transduction, and some act as effectors. However, the Toll signaling pathway does not contain the plasmosin pelle, and the IMD signaling pathway does not contain the TAK-binding protein TAB in this insect. This result needs to be clarified in the genome data of E. oblogua in the future. Similarly, the Hop signal molecules that activate the JAK/STAT signaling pathway have not been identified in the B. mori genome [33]. Plasmosin MyD88, which activates NF-kB transcription factors in the Toll signaling pathway, has not been identified in the P. xylostella transcriptome [34].
Insects perceive the presence of pathogens through intracorporal pattern recognition receptors in combination with pathogen-associated molecular patterns, which activate immune responses. Insect pattern recognition receptors mainly include CTLs, β-1,3-glucan recognition proteins, and PGRPs. CTLs recognize pathogens, enhance host phenol oxidase activity levels, and activate encapsulation and melanization reactions [43,44]. The injection of gram-negative bacteria into M. sexta larvae induces the synthesis of CTLs and activates PO [45]. In this study, the CTL-10 gene was upregulated in both hemocytes and fat body tissues, indicating that it plays important roles in the immune responses of E. obliqua. The β-1,3-glucan recognition proteins, known as GNBPs, recognize the glucan component of fungal cell walls and bind to gram-negative bacteria. For example, GNBPs of D. melanogaster are recognized by fungi to activate the Toll pathway [46]. The GNBP-2 gene in this study was upregulated in fat body tissues and downregulated in hemocytes, and this pattern may result from the use of different signaling pathways in different tissues. PGRP-LC and PGRP-LE in D. melanogaster recognize the meso-diaminopimelic acid-type PGN in gram-negative cell walls, thereby activating the IMD signal transduction pathway [32]. As shown in the transcriptome data (Table S2), all PGRP1-9 were down-regulated in hemocytes and fat bodies after B. bassiana infection, and the expression of PGRP-5 and PGRP-6 were further confirmed by qPT-PCR results. Moreover, effector genes (such as ATT and PPO) were downregulated in all the samples infected by B. bassiana. We speculate that the 48 h sample time point allowed a sufficient amount of time for the release of entomopathogenic toxins.
After insects recognize pathogens, the protease activity signals of serine proteases (SPs) and their homologs (SP homologs, SPHs) are amplified step by step and finally transmitted to the Toll pathway’s extracellular ligands SPZ or PPO. The enzyme-linked reactions of SPs are usually regulated by SP inhibitors (serpins). These proteases play crucial roles in the reduction or amplification of immune signals [40]. In this experiment, SPH-5, SPZ-2, and serpin-7 all showed upregulated trends in the immune responses of E. obliqua. Signal transduction molecules are the most common type of immunity-related genes, and those in the Toll and IMD signaling pathways mostly showed upregulated trends. In addition, in this transcriptome data, the JNK pathway is activated after pathogen recognition, leading to the activation of the transcription factor to turn on expression of antimicrobial peptides (AMP), such as cecropin and gloverin. Both gloverin and cecropin of antimicrobial peptides were down-regulated in the hemocytes after B. bassiana infection.
In this study, we chose only one time point (48 h post-infection) to profile and characterize the immune reaction of E. obliqua against B. bassiana, which is insufficient to fully understand when the insect immune is initially responding and how immune genes are temporally expressed to counter the invasion of B. bassiana. Furthermore, we aimed at identifying a highly virulent B. bassiana strain, profiling the immune system of E. obliqua, and partly revealing the immune reaction of the pest against the entomopathogen. The difference of the immune reaction in E. obliqua against high and low virulence strains is an interesting question and deserves investigation. In summary, our study provided new insights into the expression profiles of host defense genes in non-model insects exposed to pathogenic fungi, allowing us to further investigate the functions of antifungal immunity-related genes and to improve our understanding of host–pathogen interactions.
5. Conclusions
After the isolation of a highly virulent strain of B. bassiana from E. obliqua, we identified 249 immunity-related genes from E. obliqua infected by the B. bassiana using a transcriptome and compared their expression with those of homologous genes in other insects. Then, we confirmed the expression patterns of certain genes using qRT-PCR. Our study provides a molecular basis for the function studies of antifungal immunity-related genes and advances the understanding of host–pathogen interactions of E. obliqua.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13030225/s1, Figure S1. Morphological characteristics of strain and optical microscopic observation, Table S1. Primers for qRT-PCR, Table S2. Annotation and expression profile of all differently expressed genes in four treatments, Table S3. Immunity-related genes identified in Ectropis obliqua transcriptome.
Author Contributions
Conceptualization, T.G. and Q.S.; methodology, T.G. and X.L.; software, Q.S.; validation, T.G., S.L. and Y.Z.; formal analysis, Y.L.; investigation, T.G., Q.S. and S.L.; resources, Y.Y.; data curation, Y.L., T.G. and L.Z.; writing—original draft preparation, Y.L. and T.G.; writing—review and editing, Y.L., T.G. and L.X.; visualization, T.G. and Q.S.; supervision, L.X.; project administration, Y.Y.; funding acquisition, Y.L. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 32072421 and 31870635; National Key Research and Development Programs, grant number 2021YFD1601105; Anhui Key Research and Development Program, grant number 202004e11020006.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the Supplementary Materials section.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pan, Y.; Fang, G.; Wang, Z.; Cao, Y.; Liu, Y.; Li, G.; Liu, X.; Xiao, Q.; Zhan, S. Chromosome-level genome reference and genome editing of the tea geometrid. Mol. Ecol. Resour. 2021, 21, 2034–2049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Feng, B.W.; Zhang, J.; Titus, I.; Chen, L.L. Research progress on green control techniques of Ectropis grisescens Warren and Ectropis obliqua Prout. J. Environ. Entomol. 2020, 42, 1121–1138. (In Chinese) [Google Scholar]
- Jiang, N.; Liu, S.X.; Xue, D.Y.; Tang, M.J.; Xiao, Q.; Han, H.X. External morphology and molecular identification of two tea Geometrid moth from southern China. Chin. J. Appl. Entomol. 2014, 51, 987–1002. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, T.Z.; Deng, J.; Wu, Z.; Yang, Y.; Long, Y. Effect of feed on gut bacterial diversity and larval survival of Ectropis obliqua (Lepidoptera: Geometridae). Acta Microbiol. Sinica 2019, 59, 881–890. (In Chinese) [Google Scholar] [CrossRef]
- Dang, K.; Doggett, S.L.; Singham, G.V.; Lee, C.-Y. Insecticide resistance and resistance mechanisms in bed bugs, Cimex spp. (Hemiptera: Cimicidae). Parasites Vectors 2017, 10, 318. [Google Scholar] [CrossRef] [Green Version]
- Liu, N. Insecticide Resistance in Mosquitoes: Impact, Mechanisms, and Research Directions. Annu. Rev. Èntomol. 2015, 60, 537–559. [Google Scholar] [CrossRef]
- Naqqash, M.N.; Gökçe, A.; Bakhsh, A.; Salim, M. Insecticide resistance and its molecular basis in urban insect pests. Parasitol. Res. 2016, 115, 1363–1373. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, C.; Wang, G.; Li, Y.; Wang, S. Insects defend against fungal infection by employing microRNAs to silence virulence-related genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2023802118. [Google Scholar] [CrossRef]
- Guo, D.; Ren, G.; Cheng, J.; Liu, Y.; An, D. Advance of Classification of Genus Beauveria. Acta Agric. Boreali-Occident. Sin. 2019, 28, 497–509. (In Chinese) [Google Scholar]
- Mascarin, G.M.; Jaronski, S.T. The production and uses of Beauveria bassiana as a microbial insecticide. World J. Microbiol. Biotechnol. 2016, 32, 177. [Google Scholar] [CrossRef]
- Xie, X.; Jiang, X.; Pang, B.; Wang, X. Field trial of several new pesticides on control of Ectropis obliqua Newsletter of Sericulture and Tea. Newsl. Seric. Tea. 2015, 2, 24–25. [Google Scholar]
- Sutanto, K.D.; Husain, M.; Rasool, K.G.; Al-Qahtani, W.H.; Aldawood, A.S. Pathogenicity of local and exotic entomopathogenic fungi isolates against different life stages of red palm weevil (Rhynchophorus ferrugineus). PLoS ONE 2021, 16, e0255029. [Google Scholar] [CrossRef] [PubMed]
- Kuzhuppillymyal-Prabhakarankutty, L.; Ferrara-Rivero, F.; Tamez-Guerra, P.; Gomez-Flores, R.; Rodríguez-Padilla, M.; Ek-Ramos, M. Effect of Beauveria bassiana-Seed Treatment on Zea mays L. Response against Spodoptera frugiperda. Appl. Sci. 2021, 11, 2887. [Google Scholar] [CrossRef]
- Kheradmand, K.; Heidari, M.; Sedaratian-Jahromi, A.; Talaei-Hassanloui, R.; Havasi, M. Biological responses of Tetranychus urticae (Acari: Tetranychidae) to sub-lethal concentrations of the entomopathogenic fungus Beauveria bassiana. Bull. Èntomol. Res. 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Aynalem, B.; Muleta, D.; Venegas, J.; Assefa, F. Molecular phylogeny and pathogenicity of indigenous Beauveria bassiana against the tomato leafminer, Tuta absoluta Meyrick 1917 (Lepidoptera: Gelechiidae), in Ethiopia. J. Genet. Eng. Biotechnol. 2021, 19, 1–15. [Google Scholar] [CrossRef]
- Erasmus, R.; Berg, J.V.D.; du Plessis, H. Susceptibility of Tuta absoluta (Lepidoptera: Gelechiidae) Pupae to Soil Applied Entomopathogenic Fungal Biopesticides. Insects 2021, 12, 515. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S. Insect Pathogenic Fungi: Genomics, Molecular Interactions, and Genetic Improvements. Annu. Rev. Èntomol. 2017, 62, 73–90. [Google Scholar] [CrossRef]
- Wraight, S.; Ramos, M.; Avery, P.; Jaronski, S.; Vandenberg, J. Comparative virulence of Beauveria bassiana isolates against lepidopteran pests of vegetable crops. J. Invertebr. Pathol. 2010, 103, 186–199. [Google Scholar] [CrossRef]
- Bai, J.; Xu, Z.; Li, L.; Ma, W.; Xu, L.; Ma, L. Temporospatial modulation of Lymantria dispar immune system against an entomopathogenic fungal infection. Pest Manag. Sci. 2020, 76, 3982–3989. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Zhang, S.; Deng, J.; Lu, M.; Zhang, L.; Zhang, J. Comparative analysis of the immune system of an invasive bark beetle, Dendroctonus valens, infected by an entomopathogenic fungus. Dev. Comp. Immunol. 2018, 88, 65–69. [Google Scholar] [CrossRef]
- Liu, J.; Ling, Z.; Wang, J.; Xiang, T.; Xu, L.; Gu, C.; Liu, R.; Xu, J.; Xu, C.; Zhou, W.; et al. In vitro transcriptomes analysis identifies some special genes involved in pathogenicity difference of the Beauveria bassiana against different insect hosts. Microb. Pathog. 2021, 154, 104824. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Wang, Y.; Shao, Y.; Zhou, Q.; Chen, S.; Wu, Y.; Chen, H.; Liu, E. Genes involved in Beauveria bassiana infection to Galleria mellonella. Arch. Microbiol. 2018, 200, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-T.; Lee, S.J.; Nai, Y.-S.; Kim, S.; Kim, J.S. Up-regulation of carbon metabolism-related glyoxylate cycle and toxin production in Beauveria bassiana JEF-007 during infection of bean bug, Riptortus pedestris (Hemiptera: Alydidae). Fungal Biol. 2016, 120, 1236–1248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, Y.; Xu, W.; Du, Q.; Sui, L.; Zhao, Y.; Li, Q. RNA sequencing analysis of Beauveria bassiana isolated from Ostrinia furnacalis identifies the pathogenic genes. Microb. Pathog. 2019, 130, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Mantilla, J.G.; Galeano, N.F.; Gaitan, A.L.; Cristancho, M.A.; Keyhani, N.O.; Góngora, C.E. Transcriptome analysis of the entomopathogenic fungus Beauveria bassiana grown on cuticular extracts of the coffee berry borer (Hypothenemus hampei). Microbiology 2012, 158, 1826–1842. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Shi, J.; Wang, Y. Isolation and molecular biological analyses of genomic DNAs from fungi using benzyl chloride. J. Dalian Inst. Light Ind. 2003, 19, 36–39. (In Chinese) [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Xiong, G.-H.; Xing, L.-S.; Lin, Z.; Saha, T.T.; Wang, C.; Jiang, H.; Zou, Z. High throughput profiling of the cotton bollworm Helicoverpa armigera immunotranscriptome during the fungal and bacterial infections. BMC Genom. 2015, 16, 321. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, M.; Kanehisa, M. Using the KEGG Database Resource. Curr. Protoc. Bioinform. 2012, 38, 1.12.1–1.12.43. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yu, L.; Xue, M.; Yu, X.; Vasseur, L.; Gurr, G.; Baxter, S.W.; Lin, H.; Lin, J.; You, M. Genome-wide characterization and expression profiling of immune genes in the diamondback moth, Plutella xylostella (L.). Sci. Rep. 2015, 5, 9877. [Google Scholar] [CrossRef]
- Xu, J.; Xu, X.; Shakeel, M.; Li, S.; Wang, S.; Zhou, X.; Yu, J.; Xu, X.; Yu, X.; Jin, F. The entomopathogenic fungi Isaria fumosorosea plays a vital role in suppressing the immune system of Plutella xylostella: RNA-Seq and DGE analysis of immunity-related genes. Front. Microbiol. 2017, 8, 1421. [Google Scholar] [CrossRef] [Green Version]
- Xiong, G.-H. Immunotranscriptome Profiling of the Helicoverpa armigera Regulated by Entomopathogenic Fungus. Ph.D. Thesis, University of Chinese Academy of Sciences, Beijing, China, 2015. (In Chinese). [Google Scholar]
- Meng, Q.; Yu, H.-Y.; Zhang, H.; Zhu, W.; Wang, M.-L.; Zhang, J.-H.; Zhou, G.-L.; Li, X.; Qin, Q.-L.; Hu, S.-N.; et al. Transcriptomic insight into the immune defenses in the ghost moth, Hepialus xiaojinensis, during an Ophiocordyceps sinensis fungal infection. Insect Biochem. Mol. Biol. 2015, 64, 1–15. [Google Scholar] [CrossRef]
- Zou, Z.; Evans, J.D.; Lu, Z.; Zhao, P.; Williams, M.; Sumathipala, N.; Hetru, C.; Hultmark, D.; Jiang, H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007, 8, R177. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Shen, D.; Zhou, F.; Wang, G.; An, C. Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-Seq analysis. PLoS ONE 2014, 9, e86436. [Google Scholar] [CrossRef] [Green Version]
- Gunaratna, R.; Jiang, H. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev. Comp. Immunol. 2013, 39, 388–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.-Q.; Kanost, M.R. Binding of hemolin to bacterial lipopolysaccharide and lipoteichoic acid—An immunoglobulin superfamily member from insects as a pattern-recognition receptor. JBIC J. Biol. Inorg. Chem. 2002, 269, 1827–1834. [Google Scholar] [CrossRef]
- Kanost, M.R.; Jiang, H.; Yu, X.-Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Q.; Kanost, M. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 2000, 275, 37373–37381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottar, M.; Gobert, V.; Matskevich, A.A.; Reichhart, J.M.; Wang, C.; Butt, T.M.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual detection of fungal infections in Drosophila through recognition of microbial structures and sensing of virulence factors. Cell 2006, 127, 1425–1437. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).