Nutrigenomics of Obesity: Integrating Genomics, Epigenetics, and Diet–Microbiome Interactions for Precision Nutrition
Abstract
1. Introduction
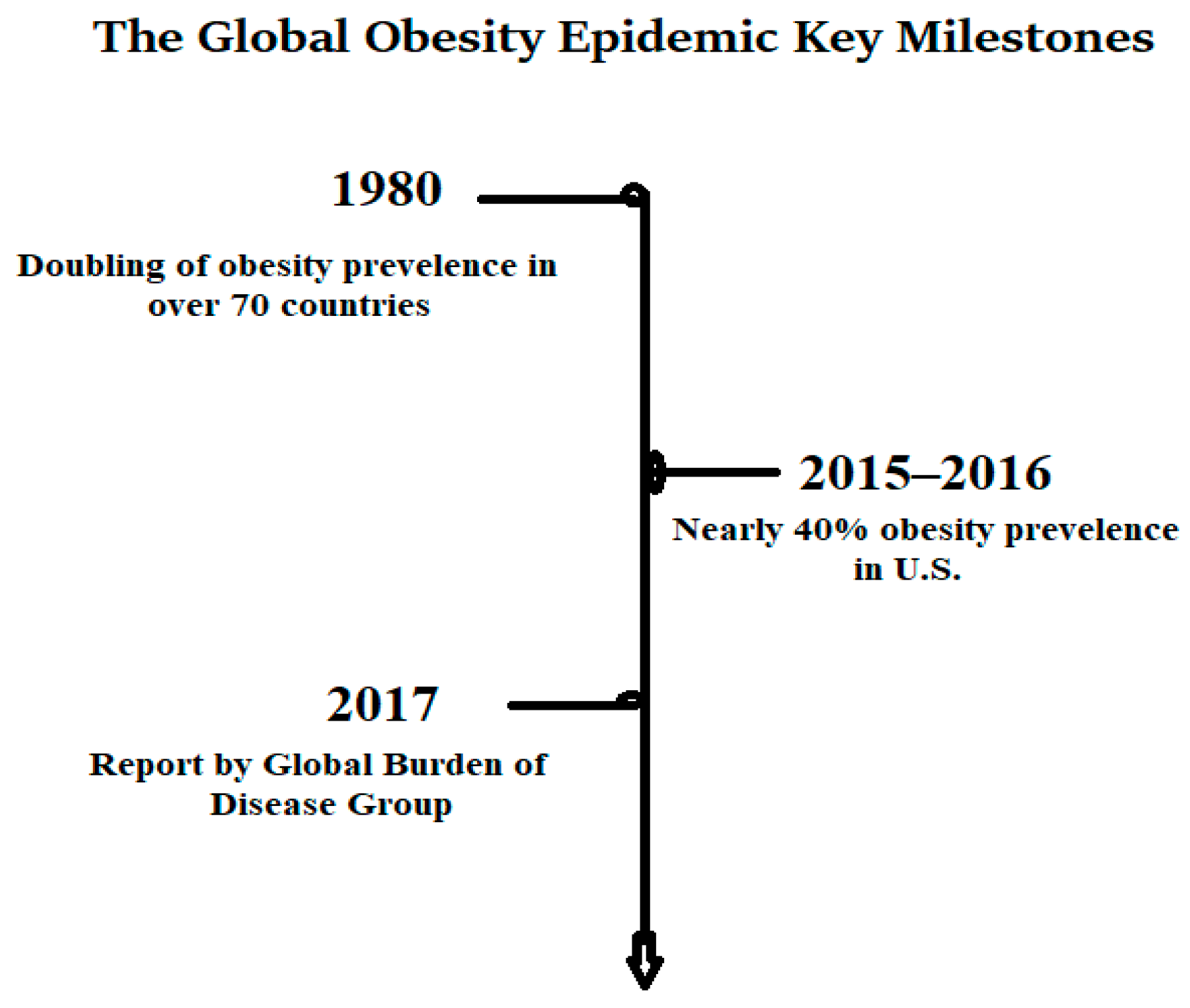
1.1. Global Epidemiological Landscape
- Most genetic discoveries lack functional annotation, limiting clinical translation;
- Population-specific obesity loci remain underexplored, especially in Asian and low-income cohorts;
- The interaction between genetics, diet, and gut microbiota composition requires deeper integrative analysis.
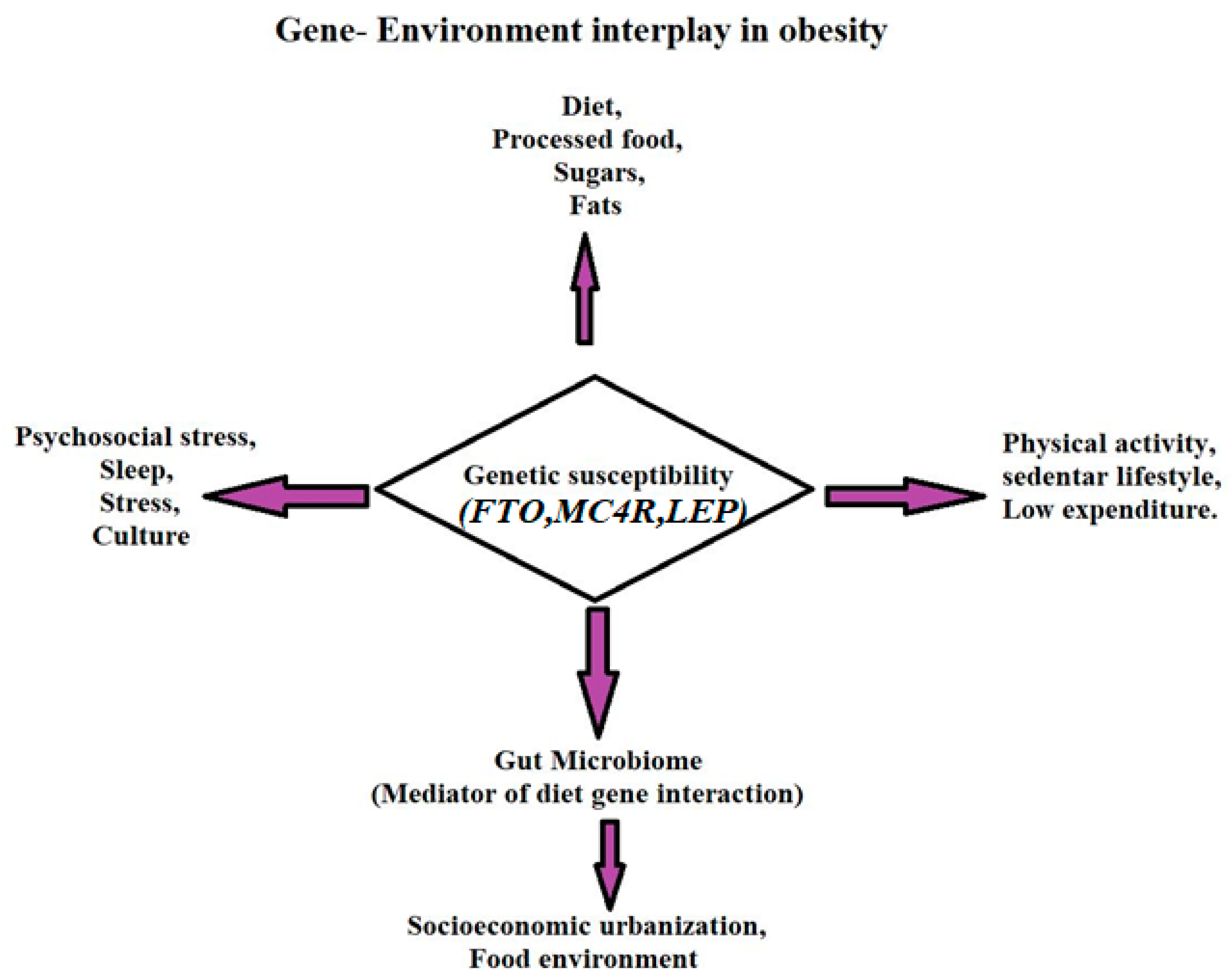
1.2. Gene-Environment Framing
1.3. Objective
- Summarize key mechanisms underlying obesity susceptibility across monogenic, polygenic, and nutrigenetic frameworks.
- Explore diet–gene–microbiome interactions shaping adiposity and metabolic plasticity;
- Highlight translational strategies for precision nutrition and individualized obesity prevention.
2. Methodology
2.1. Literature Search Strategy
2.2. Databases and Keywords
2.3. Eligibility Criteria
- Investigated genetic, epigenetic, nutrigenomic, or microbiome-driven factors influencing obesity susceptibility;
- Reported findings from large-scale GWAS, epigenomic profiling, nutrigenetic studies, or gut microbiome analyses;
- Explored diet–gene interactions or nutrient-responsive molecular pathways;
- Published in peer-reviewed, high-quality journals.
- Non-original works are excluded unless they provide meta-analytical insights;
- Animal-only studies without human translational relevance;
- Grey literature and preprints are not peer-reviewed.
2.4. Study Selection Process
- Title & Abstract Screening: Two independent reviewers screened all retrieved studies based on predefined inclusion criteria;
- Full-Text Evaluation: Articles meeting the initial screening were reviewed in detail to extract key findings related to obesity-associated genes, nutrigenomic pathways, and microbiome-modulated mechanisms.
2.5. Data Extraction and Synthesis
- Study design, sample size, and demographic details;
- Genetic loci, gene variants, and pathway-level findings;
- Nutrigenomic outcomes, microbiome diversity indices, and epigenomic signatures.
- Reported diet–gene–microbiome interactions.
- Functional pathway mapping to identify molecular convergence;
- Gene-environment response profiling to analyze nutrigenomic interactions;
- Cross-ancestry meta-analysis insights where available.
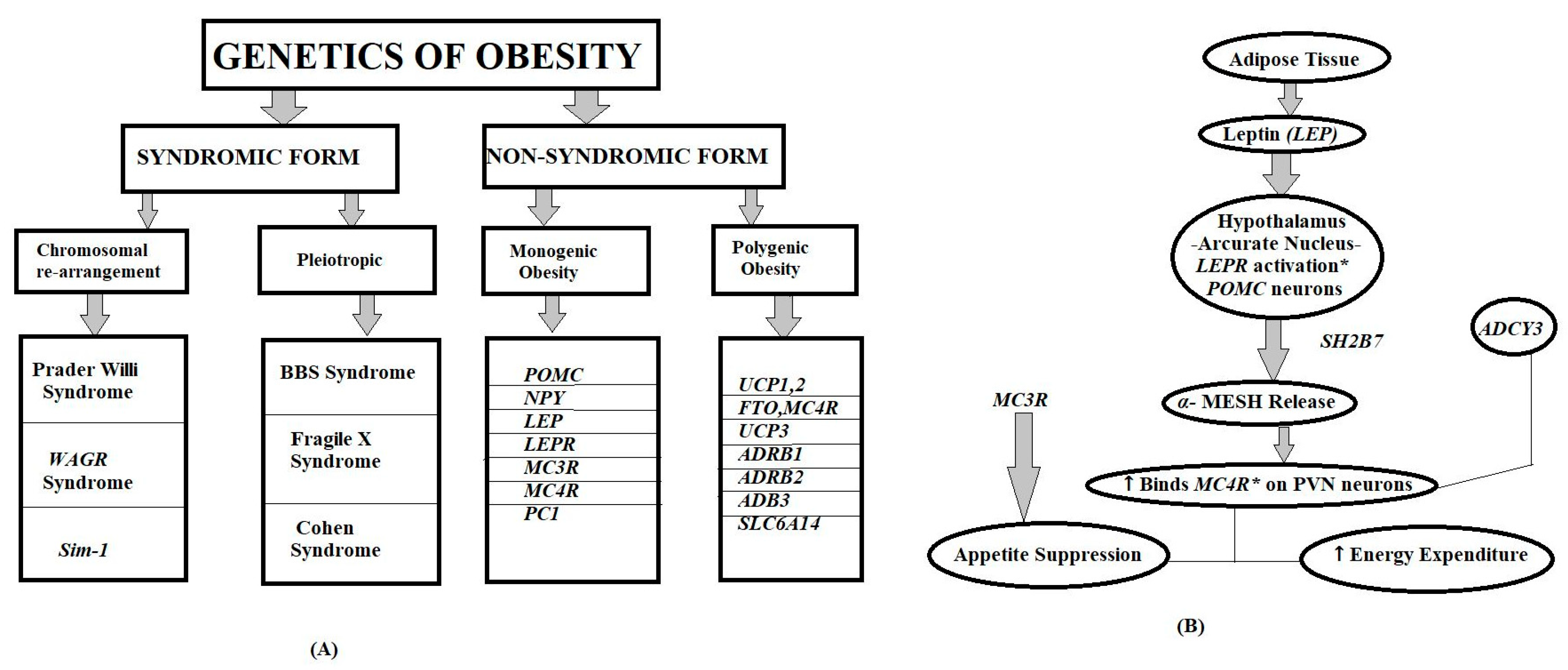
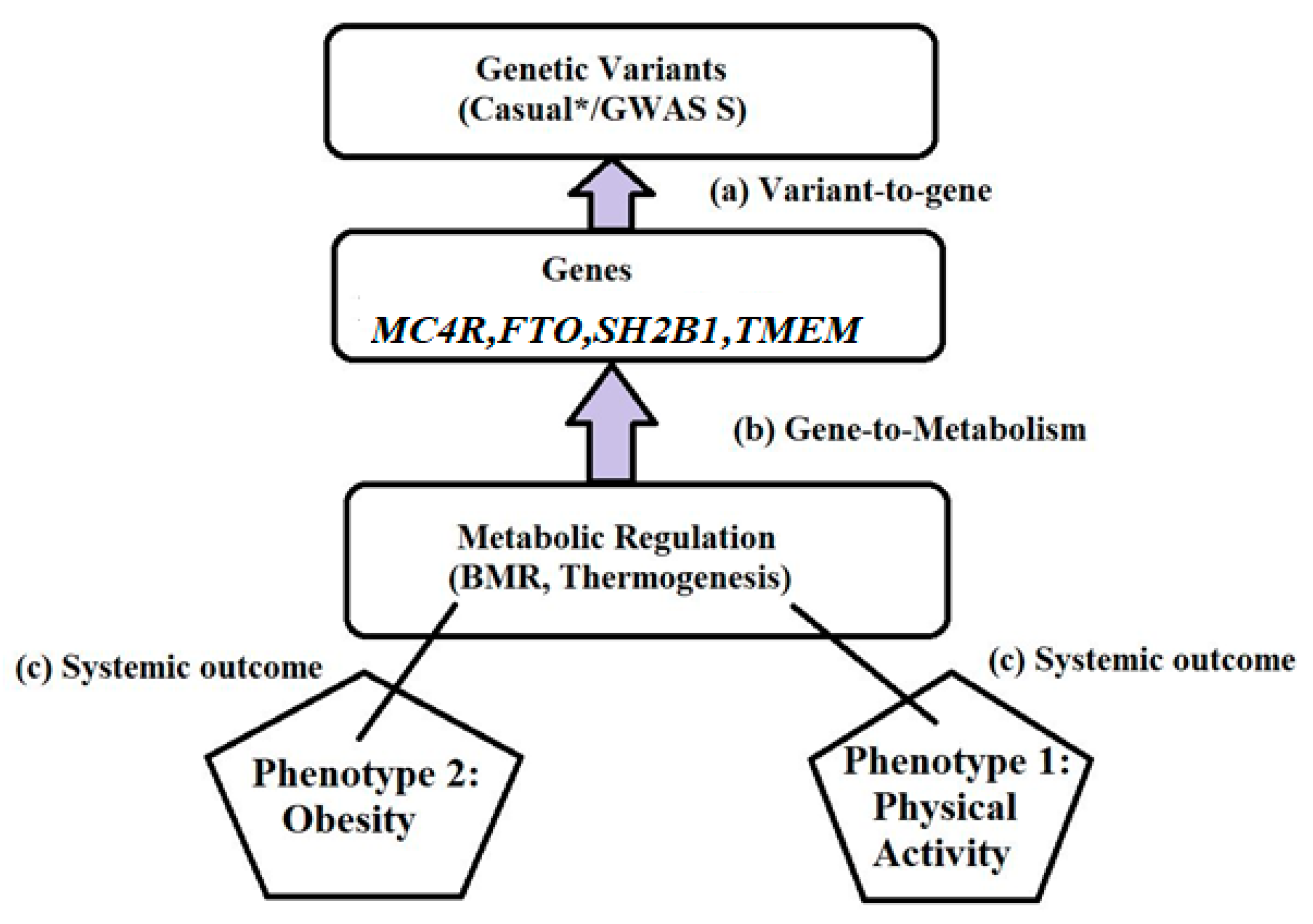
3. Genetic Basis of Obesity
3.1. Monogenic Obesity Syndromes
3.1.1. Discovery and Historical Context
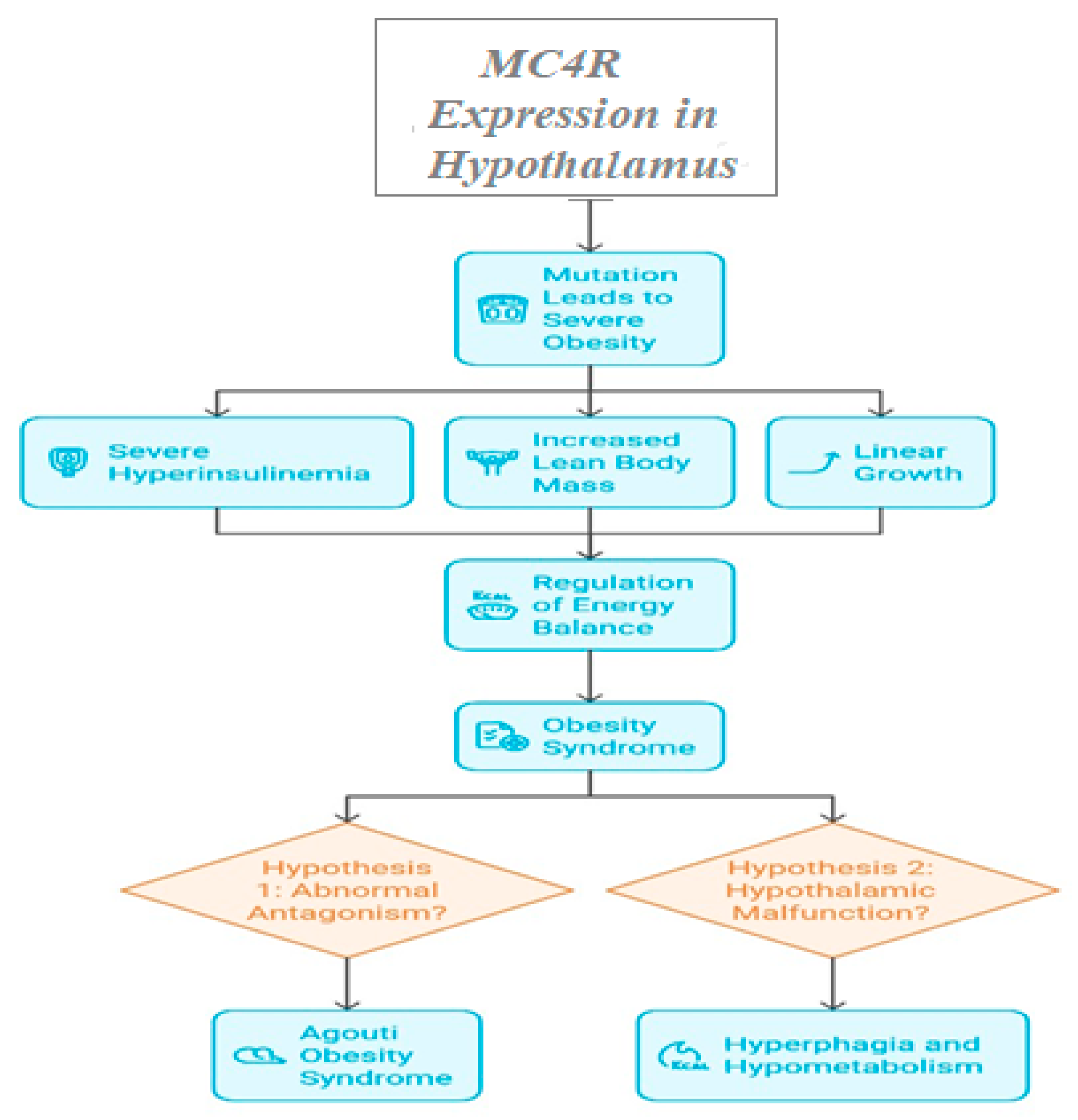
3.1.2. Key Genes Implicated and Clinical Outcomes
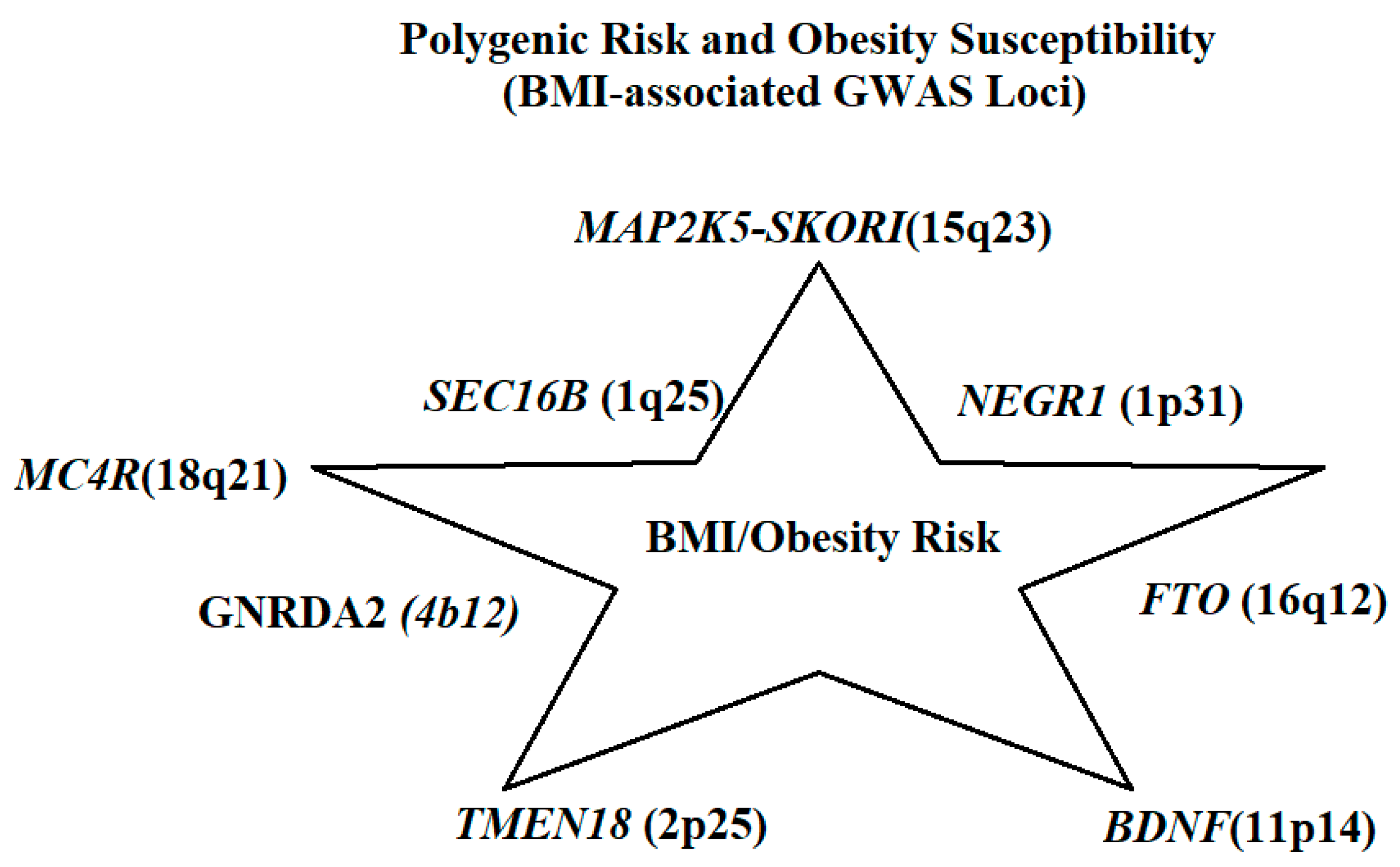
3.2. Polygenic Obesity and GWAS Findings
4. Epigenetic Modifications in Obesity
5. Genetic Mechanisms Underlying Obesity
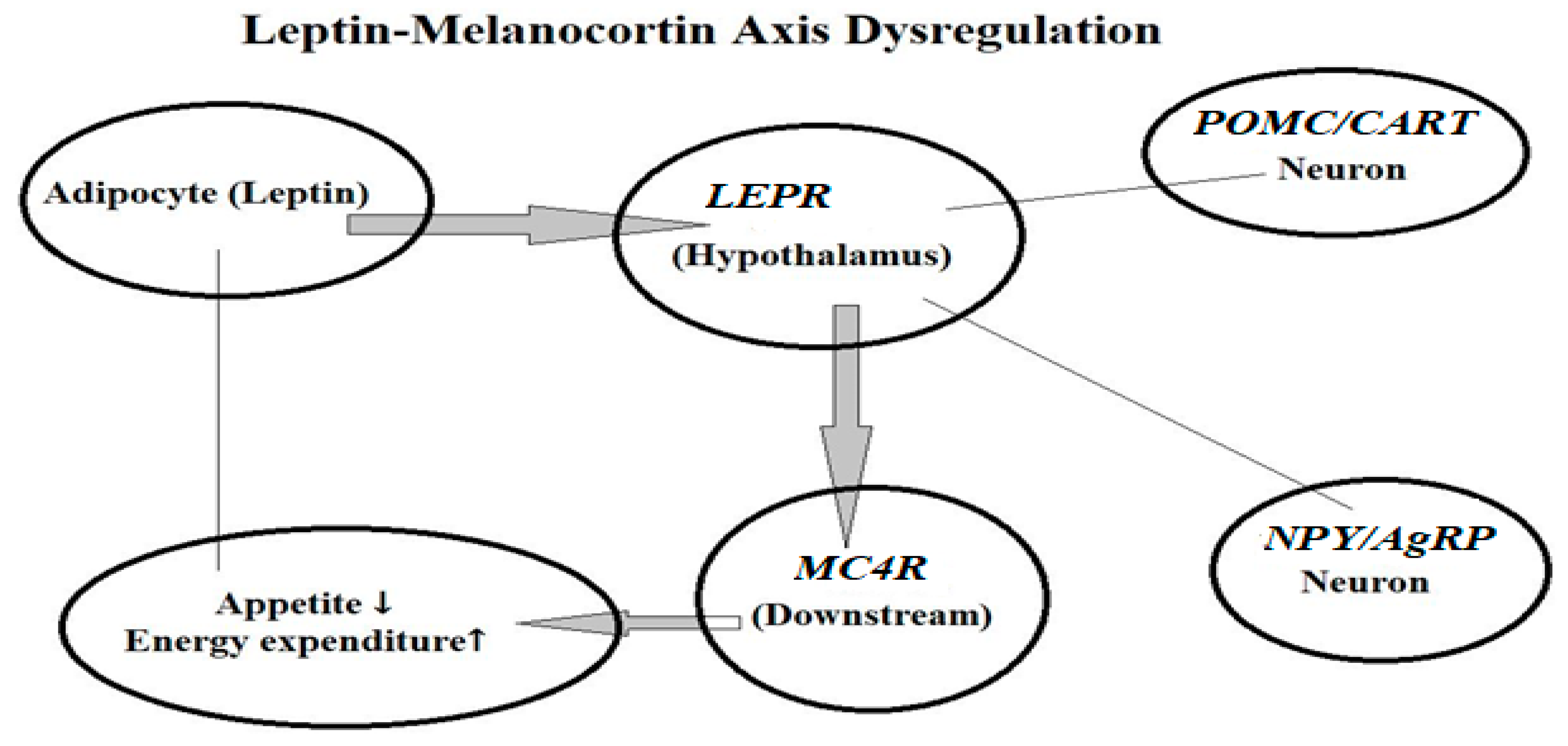
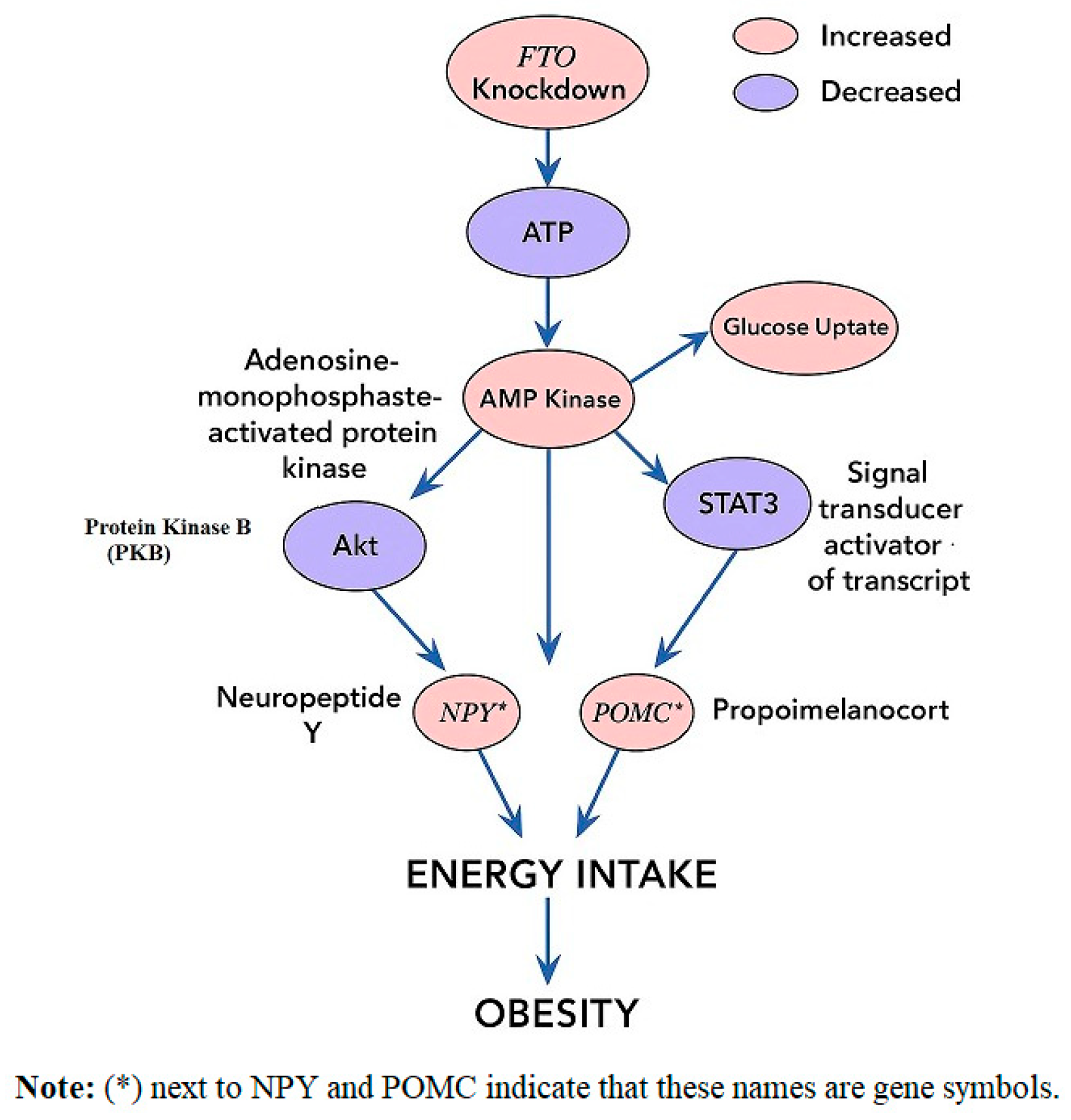
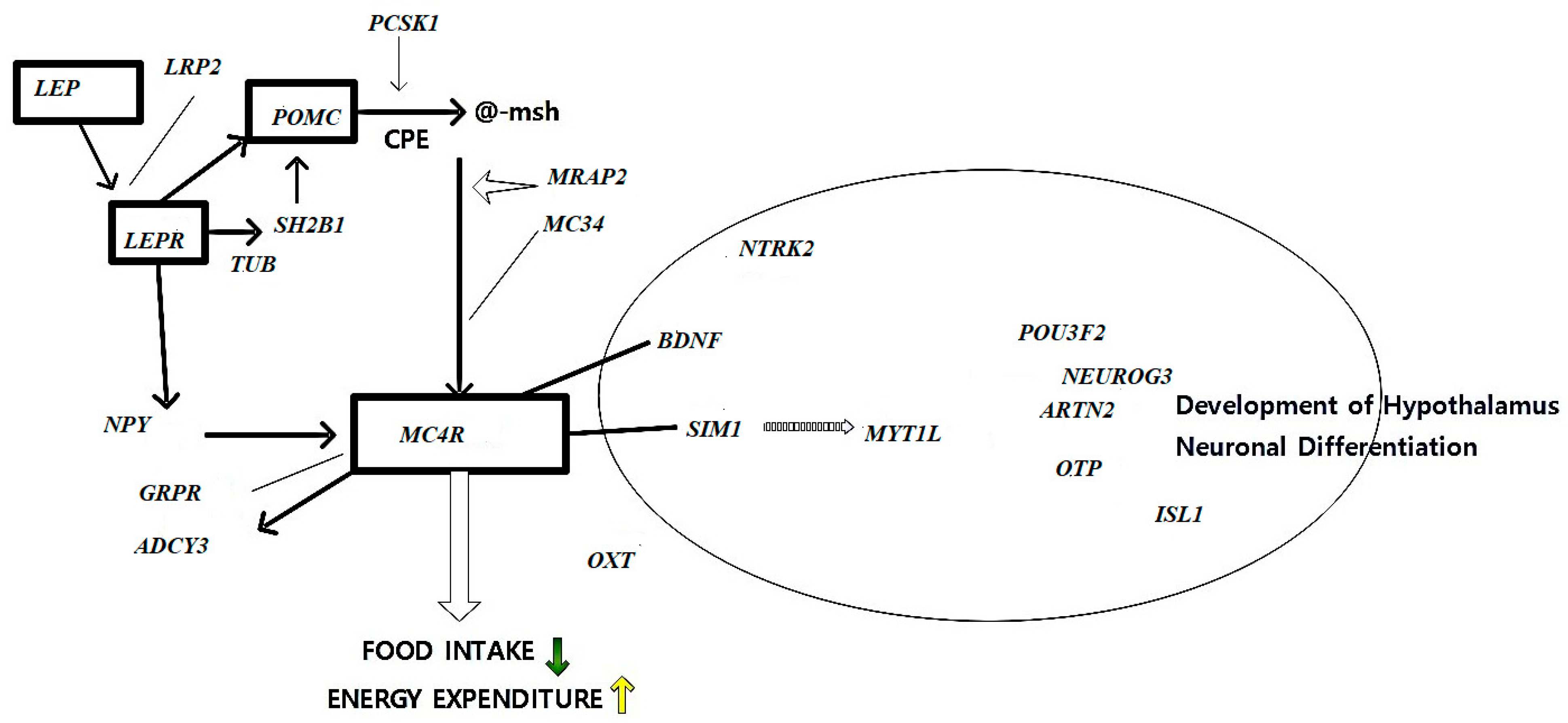
5.1. Appetite Regulation Genes
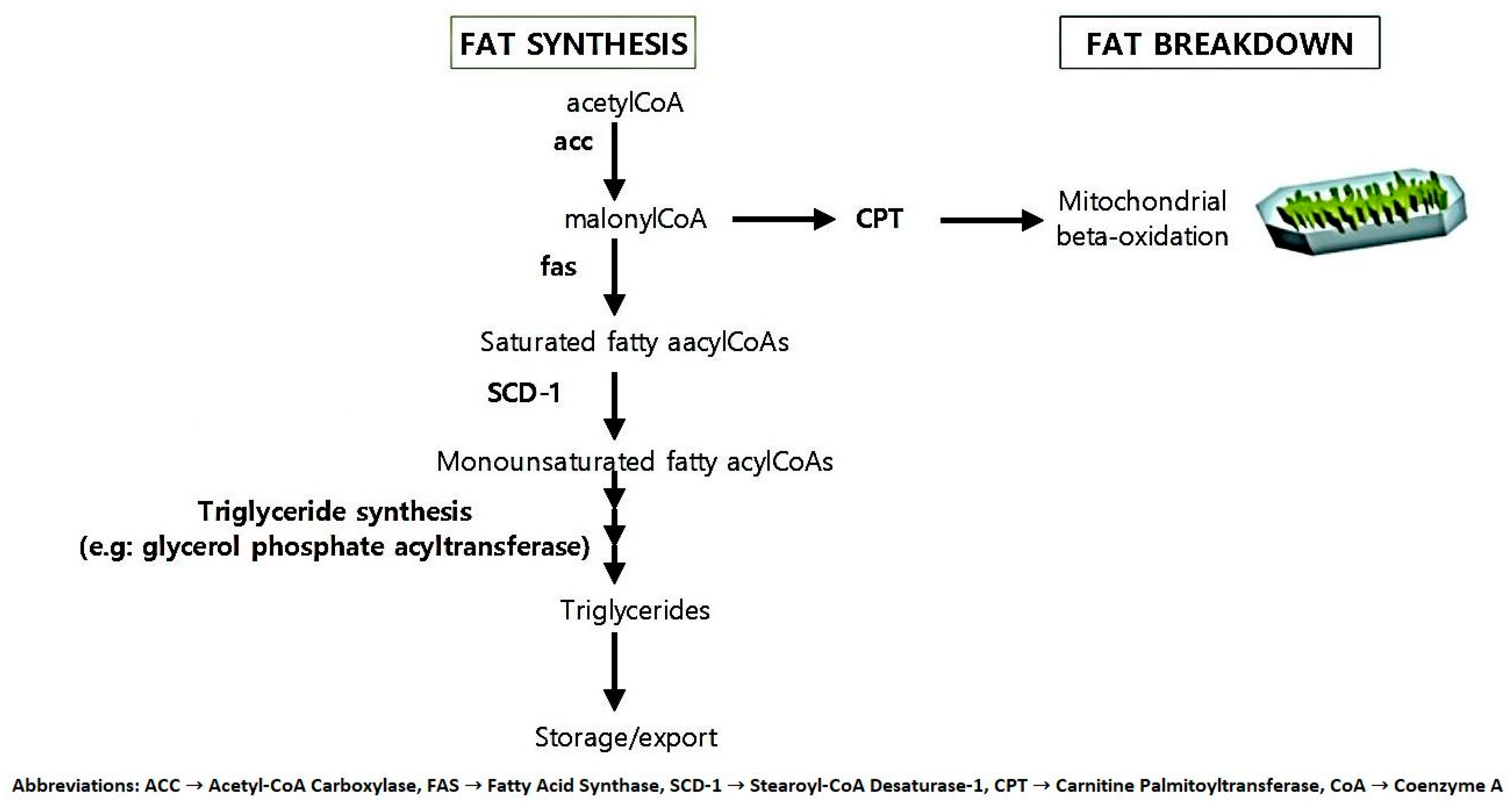
5.2. Fat Metabolism and Storage Genes
5.3. Energy Expenditure Genes
5.3.1. Gene Influencing Basal Metabolic Rate
5.3.2. Role of Physical Activity-Related Genes in Obesity Risk
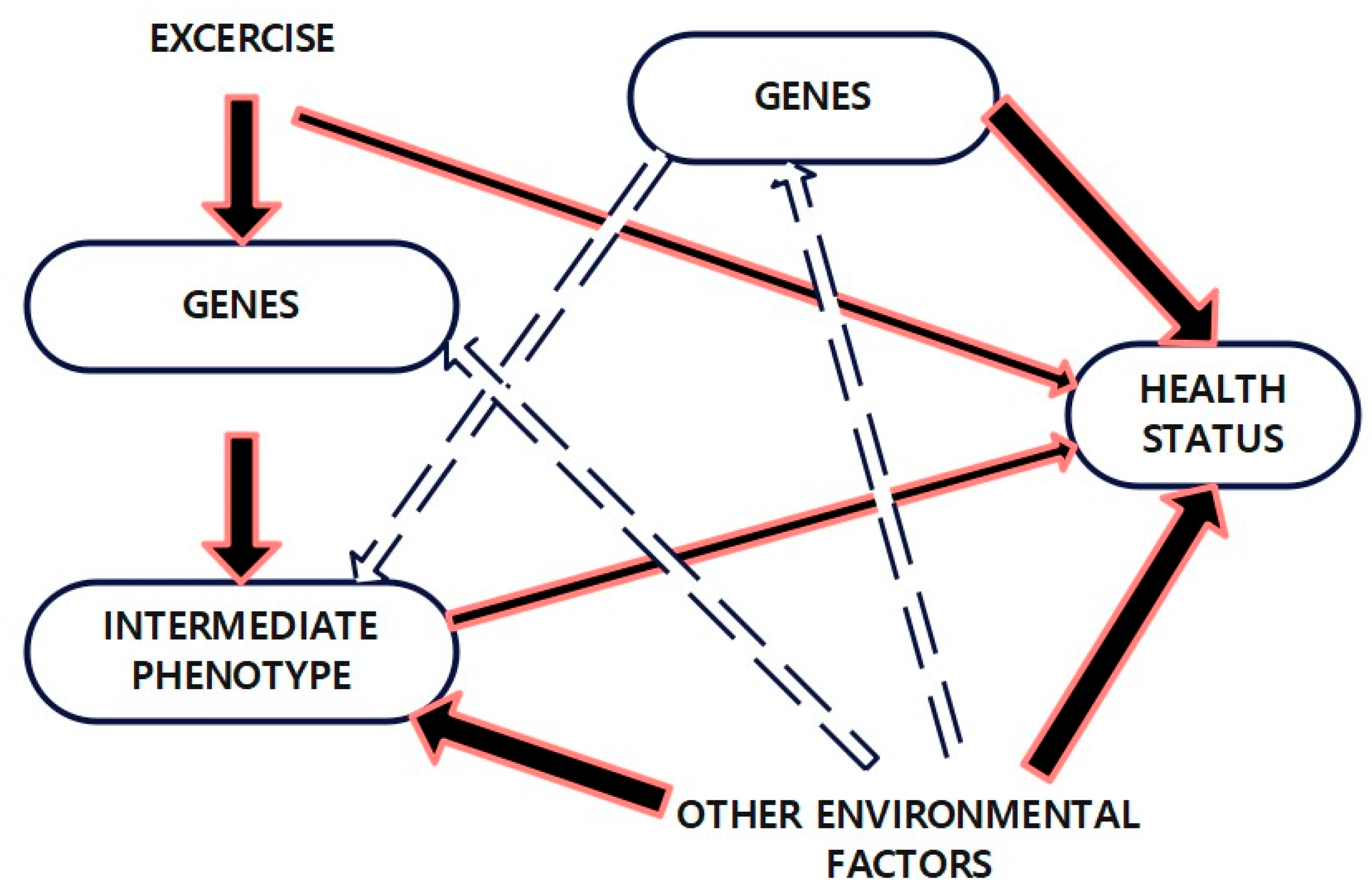
6. Gene–Environment Interaction in Obesity
Interaction Between Genetic Predispositions and Environmental Factors
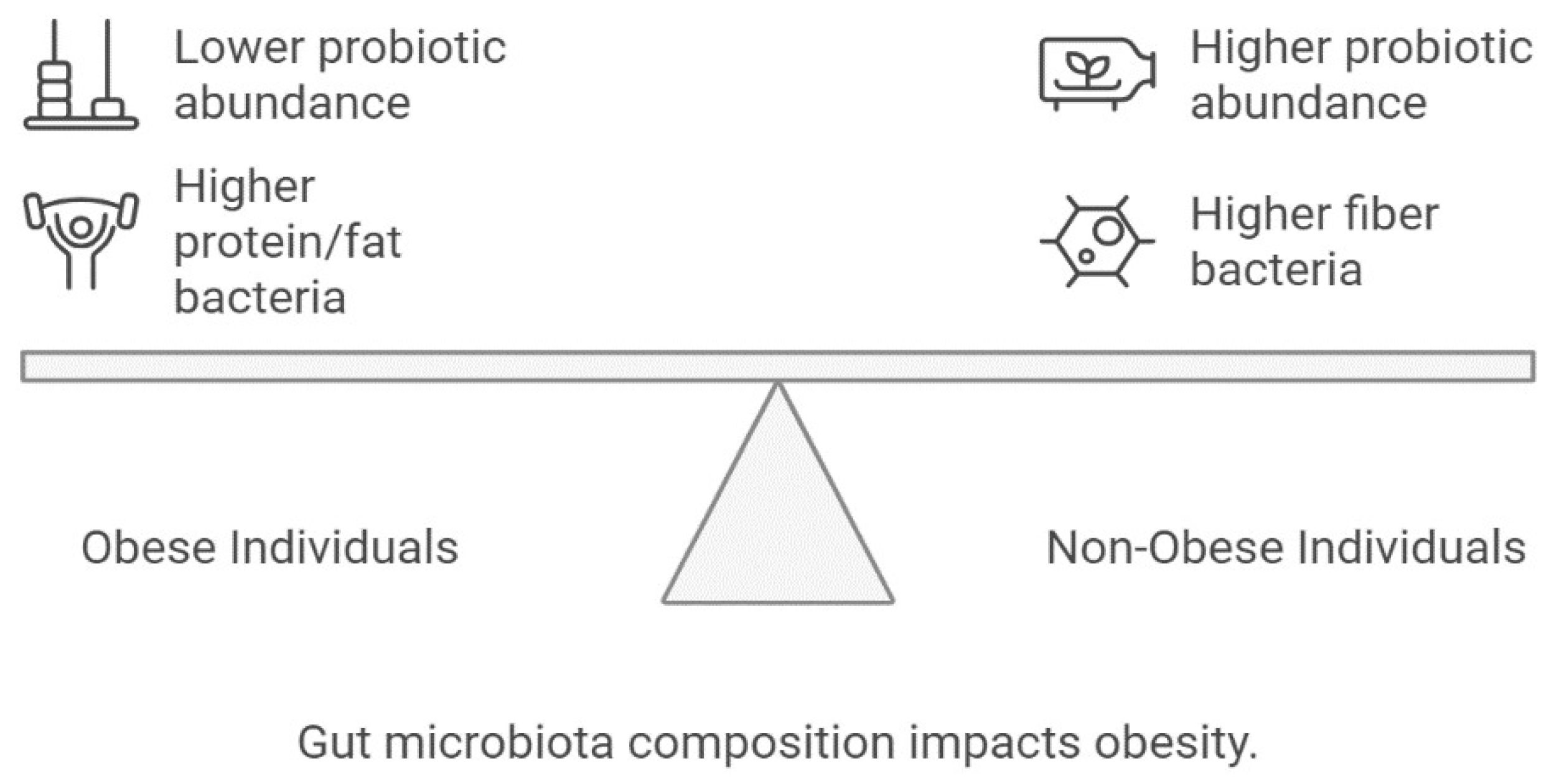
7. Role of Gut Microbiota in Obesity
8. Genetic and Clinical Implications for Future Directions
9. Limitations and Strengths
10. Conclusions and Future Genetic Perspectives in Obesity
- Functional characterization of obesity-associated loci;
- Cross-ancestry genomic mapping to address diversity gaps;
- Integrating multi-omics datasets with machine learning for risk prediction;
- Designing personalized interventions that optimize diet, physical activity, and metabolic health.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMI | Body Mass Index |
| GWAS | Genome-Wide Association Study |
| SNP | Single-Nucleotide Polymorphism |
| NGS | Next-Generation Sequencing |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic Acid |
| mRNA | Messenger RNA |
| miRNA | MicroRNA |
| lncRNA | Long Non-Coding RNA |
| CpG | Cytosine–Phosphate–Guanine (dinucleotide site) |
| ChIP-seq | Chromatin Immunoprecipitation Sequencing |
| ATAC-seq | Assay for Transposase-Accessible Chromatin Sequencing |
| eQTL | Expression Quantitative Trait Locus |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| FTO | Fat Mass and Obesity-Associated Gene |
| MC4R | Melanocortin 4 Receptor |
| PCOS | Polycystic Ovary Syndrome |
| NASH | Non-Alcoholic Steatohepatitis |
| T2D | Type 2 Diabetes |
| HDL | High-Density Lipoprotein |
| LDL | Low-Density Lipoprotein |
| GWAS-CNV | Genome-Wide Association Study–Copy Number Variant |
| MR | Mendelian Randomization |
| ACC | Acetyl-CoA Carboxylase |
| FAS | Fatty Acid Synthase |
| SCD-1 | Stearoyl-CoA Desaturase-1 |
| CPT | Carnitine Palmitoyltransferase |
| CoA | Coenzyme A |
References
- Albuquerque, D.; Stice, E.; Rodríguez-López, R.; Manco, L.; Nóbrega, C. Current review of genetics of human obesity: From molecular mechanisms to an evolutionary perspective. Mol. Genet. Genom. 2015, 290, 1191–1221. [Google Scholar] [CrossRef] [PubMed]
- Lingvay, I.; Cohen, R.V.; Le Roux, C.W.; Sumithran, P. Obesity in adults. Lancet 2024, 404, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Piché, M.-E.; Tchernof, A.; Després, J.-P. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ. Res. 2020, 126, 1477–1500. [Google Scholar] [CrossRef]
- Seravalle, G.; Grassi, G. Obesity and hypertension. Pharmacol. Res. 2017, 122, 1–7. [Google Scholar] [CrossRef]
- Mayoral, L.P.; Andrade, G.M.; Mayoral, E.P.; Huerta, T.H.; Canseco, S.P.; Canales, F.J.; Cabrera-Fuentes, H.A.; Cruz, M.M.; Santiago, A.D.; Alpuche, J.J.; et al. Obesity subtypes, related biomarkers & heterogeneity. Indian J. Med. Res. 2020, 151, 11–21. [Google Scholar] [CrossRef]
- Marrades, M.P.; Gonzalez-Muniesa, P.; Martínez, J.A.; Moreno-Aliaga, M.J. A dysregulation in CES1, APOE and other lipid metabolism-related genes is associated with cardiovascular risk factors linked to obesity. Obes. Facts 2010, 3, 312–318. [Google Scholar] [CrossRef]
- Bray, M.S. Genomics, genes, and environmental interaction: The role of exercise. J. Appl. Physiol. 2000, 88, 788–792. [Google Scholar] [CrossRef]
- Kopelman, P.G. Obesity as a medical problem. Nature 2000, 404, 635–643. [Google Scholar] [CrossRef]
- Hinney, A.; Vogel, C.I.; Hebebrand, J. From monogenic to polygenic obesity: Recent advances. Eur. Child Adolesc. Psychiatry 2010, 19, 297–310. [Google Scholar] [CrossRef]
- Leibel, R.L.; Chung, W.K.; Chua, S.C. The molecular genetics of rodent single gene obesities. J. Biol. Chem. 1997, 272, 31937–31940. [Google Scholar] [CrossRef] [PubMed]
- Barsh, G.S.; Farooqi, I.S.; O’Rahilly, S. Genetics of body-weight regulation. Nature 2000, 404, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Ozata, M.; Ozdemir, I.C.; Licinio, J. Human leptin deficiency caused by a missense mutation: Multiple endocrine defects, decreased sympathetic tone, and immune system dysfunction indicate new targets for leptin action, greater central than peripheral resistance to the effects of leptin, and spontaneous correction of leptin-mediated defects. J. Clin. Endocrinol. Metab. 1999, 84, 3686–3695. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.L. Obese and diabetes: Two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia 1978, 14, 141. [Google Scholar] [CrossRef]
- Fatima, W.; Shahid, A.; Imran, M.; Manzoor, J.; Hasnain, S.; Rana, S.; Mahmood, S. Leptin deficiency and leptin gene mutations in obese children from Pakistan. Int. J. Pediatr. Obes. 2011, 6, 419–427. [Google Scholar] [CrossRef]
- Clément, K.; Vaisse, C.; Lahlou, N.; Cabrol, S.; Pelloux, V.; Cassuto, D.; Gourmelen, M.; Dina, C.; Chambaz, J.; Lacorte, J.-M.; et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 1998, 392, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Kalinderi, K.; Goula, V.; Sapountzi, E.; Tsinopoulou, V.R.; Fidani, L. Syndromic and monogenic obesity: New opportunities due to genetic-based pharmacological treatment. Children 2024, 11, 153. [Google Scholar] [CrossRef]
- Stieg, M.R.; Sievers, C.; Farr, O.; Stalla, G.K.; Mantzoros, C.S. Leptin: A hormone linking activation of neuroendocrine axes with neuropathology. Psychoneuroendocrinology 2015, 51, 47–57. [Google Scholar] [CrossRef]
- Lan, J.; Lian, C.; Shao, Y.; Chen, S.; Lu, Y.; Zhu, L.; Mu, D.; Tang, Q. Genome-Wide Identification of Seven in Absentia E3 Ubiquitin Ligase Gene Family and Expression Profiles in Response to Different Hormones in Uncaria rhynchophylla. Int. J. Mol. Sci. 2024, 25, 7636. [Google Scholar] [CrossRef]
- Muller, G. Personalized strategies for the diagnosis and therapy of type II diabetes and obesity. Immunol. Endocr. Metab. Agents Med. Chem. (Former. Curr. Med. Chem.-Immunol. Endocr. Metab. Agents) 2012, 12, 80–109. [Google Scholar] [CrossRef]
- Bouchard, C.; Pérusse, L.; Dériaz, O.; Després, J.P.; Tremblay, A. Genetic influences on energy expenditure in humans. Crit. Rev. Food Sci. Nutr. 1993, 33, 345–350. [Google Scholar] [CrossRef]
- Tena-Sempere, M. Interaction between energy homeostasis and reproduction: Central effects of leptin and ghrelin on the reproductive axis. Horm. Metab. Res. 2013, 45, 919–927. [Google Scholar] [CrossRef]
- Saldanha, R.; Mohr, G.; Belfort, M.; Lambowitz, A.M. Group I and group II introns. FASEB J. 1993, 7, 15–24. [Google Scholar] [CrossRef]
- Farooqi, I.S. Monogenic human obesity syndromes. In Handbook of Clinical Neurology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 181, pp. 301–310. [Google Scholar] [CrossRef]
- Huszar, D.; Lynch, C.A.; Fairchild-Huntress, V.; Dunmore, J.H.; Fang, Q.; Berkemeier, L.R.; Gu, W.; Kesterson, R.A.; Boston, B.A.; Cone, R.D.; et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [Google Scholar] [CrossRef]
- Hinney, A.; Hebebrand, J. Polygenic obesity in humans. Obes. Facts 2008, 1, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.; Lindgren, C.M.; Li, S.; Wheeler, E.; Zhao, J.H.; Prokopenko, I.; Inouye, M.; Freathy, R.M.; Attwood, A.P.; Beckmann, J.S.; et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008, 40, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Loos, R.J. Obesity genomics: Assessing the transferability of susceptibility loci across diverse populations. Genome Med. 2013, 5, 55. [Google Scholar] [CrossRef]
- Granado-Lorencio, F.; Hernández-Alvarez, E. Functional foods and health effects: A nutritional biochemistry perspective. Curr. Med. Chem. 2016, 23, 2929–2957. [Google Scholar] [CrossRef]
- Herrera, B.M.; Keildson, S.; Lindgren, C.M. Genetics and epigenetics of obesity. Maturitas 2011, 69, 41–49. [Google Scholar] [CrossRef]
- Mühlhaus, J.; Pütter, C.; Brumm, H.; Grallert, H.; Illig, T.; Scherag, S.; Reinehr, T.; Pott, W.; Albayrak, Ö.; Wang, H.J.; et al. Do common variants separate between obese melanocortin-4 receptor gene mutation carriers and non-carriers? The impact of cryptic relatedness. Horm. Res. Paediatr. 2012, 77, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, J.H.; Luan, J.; Ekelund, U.; Luben, R.N.; Khaw, K.-T.; Wareham, N.J.; Loos, R.J.F. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from the EPIC-Norfolk prospective population study. PLoS Med. 2010, 7, e1000332. [Google Scholar] [CrossRef]
- Rampersaud, E.; Mitchell, B.D.; Pollin, T.I.; Fu, M.; Shen, H.; O’Connell, J.R.; Ducharme, J.L.; Hines, S.; Sack, P.; Naglieri, R.; et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch. Intern. Med. 2008, 168, 1791–1797. [Google Scholar] [CrossRef]
- Davis, C.D. The gut microbiome and its role in obesity. Nutr. Today 2016, 51, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-N.; Liu, X.-T.; Liang, Z.-H.; Wang, J.-H. Gut microbiota in obesity. World J. Gastroenterol. 2021, 27, 3837–3850. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Franzago, M.; Di Nicola, M.; Fraticelli, F.; Marchioni, M.; Stuppia, L.; Vitacolonna, E. Nutrigenetic variants and response to diet/lifestyle intervention in obese subjects: A pilot study. Acta Diabetol. 2022, 59, 69–81. [Google Scholar] [CrossRef]
- Smith, C.E.; Fullerton, S.M.; Dookeran, K.A.; Hampel, H.; Tin, A.; Maruthur, N.M.; Schisler, J.C.; Henderson, J.A.; Tucker, K.L.; Ordovás, J.M. Using genetic technologies to reduce, rather than widen, health disparities. Health Aff. 2016, 35, 1367–1373. [Google Scholar] [CrossRef]
- Ngowi, E.E.; Wang, Y.Z.; Khattak, S.; Khan, N.H.; Mahmoud, S.S.; Helmy, Y.A.; Jiang, Q.Y.; Li, T.; Duan, S.F.; Ji, X.Y.; et al. Impact of the factors shaping gut microbiota on obesity. J. Appl. Microbiol. 2021, 131, 2131–2147. [Google Scholar] [CrossRef]
- Angelidi, A.M.; Belanger, M.J.; Kokkinos, A.; Koliaki, C.C.; Mantzoros, C.S. Novel noninvasive approaches to the treatment of obesity: From pharmacotherapy to gene therapy. Endocr. Rev. 2022, 43, 507–557. [Google Scholar] [CrossRef]
- Andrews, L.; Fullarton, J.; Holtzman, N.; Motulsky, A. Social, legal, and ethical implications of genetic testing. In Assessing Genetic Risks: Implications for Health and Social Policy; National Academies Press: Washington, DC, USA, 1994. [Google Scholar] [CrossRef]
- Flint, H.J. Obesity and the gut microbiota. J. Clin. Gastroenterol. 2011, 45 (Suppl. S2), S128–S132. [Google Scholar] [CrossRef]
- Konarzewski, M.; Książek, A. Determinants of intra-specific variation in basal metabolic rate. J. Comp. Physiol. B 2013, 183, 27–41. [Google Scholar] [CrossRef]
- Lee, H.; Ash, G.I.; Angelopoulos, T.J.; Gordon, P.M.; Moyna, N.M.; Visich, P.S.; Zoeller, R.F.; Gordish-Dressman, H.; Deshpande, V.; Chen, M.H.; et al. Obesity-related genetic variants and their associations with physical activity. Sports Med. Open 2015, 1, 34. [Google Scholar] [CrossRef]
- Yang, W.; Kelly, T.; He, J. Genetic epidemiology of obesity. Epidemiol. Rev. 2007, 29, 49–61. [Google Scholar] [CrossRef]
- Ahmad, S.; Varga, T.V.; Franks, P.W. Gene × environment interactions in obesity: The state of the evidence. Hum. Hered. 2013, 75, 106–115. [Google Scholar] [CrossRef]
- Campion, J.; Milagro, F.; Martínez, J.A. Epigenetics and obesity. Prog. Mol. Biol. Transl. Sci. 2010, 94, 291–347. [Google Scholar] [CrossRef] [PubMed]
- Loid, P.; Mustila, T.; Mäkitie, R.E.; Viljakainen, H.; Kämpe, A.; Tossavainen, P.; Lipsanen-Nyman, M.; Pekkinen, M.; Mäkitie, O. Rare variants in genes linked to appetite control and hypothalamic development in early-onset severe obesity. Front. Endocrinol. 2020, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Arora, S. Role of neuropeptides in appetite regulation and obesity—A review. Neuropeptides 2006, 40, 375–401. [Google Scholar] [CrossRef]
- Clemente-Postigo, M.; Queipo-Ortuno, M.I.; Fernandez-Garcia, D.; Gomez-Huelgas, R.; Tinahones, F.J.; Cardona, F. Adipose tissue gene expression of factors related to lipid processing in obesity. PLoS ONE 2011, 6, e24783. [Google Scholar] [CrossRef]
- Malhotra, S.; Sivasubramanian, R.; Srivastava, G. Evaluation and management of early onset genetic obesity in childhood. J. Pediatr. Genet. 2021, 10, 194–204. [Google Scholar] [CrossRef]
- Chu, D.T.; Bui, N.L.; Thi, H.V.; Thi, Y.V. Role of DNA methylation in diabetes and obesity. Prog. Mol. Biol. Transl. Sci. 2023, 197, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Cho, Y.A. Gene–environment interaction and obesity. Nutr. Rev. 2008, 66, 684–694. [Google Scholar] [CrossRef]
- Antecol, H.; Bedard, K. Unhealthy assimilation: Why do immigrants converge to American health status levels? Demography 2006, 43, 337–360. [Google Scholar] [CrossRef]
- Ebrahim, S.; Kinra, S.; Bowen, L.; Andersen, E.; Ben-Shlomo, Y.; Lyngdoh, T.; Ramakrishnan, L.; Ahuja, R.C.; Joshi, P.; Das, S.M.; et al. The effect of rural-to-urban migration on obesity and diabetes in India: A cross-sectional study. PLoS Med. 2010, 7, e1000268. [Google Scholar] [CrossRef]
- Schulz, L.O.; Bennett, P.H.; Ravussin, E.; Kidd, J.R.; Kidd, K.K.; Esparza, J.; Valencia, M.E. Effects of traditional and western environments on prevalence of type 2 diabetes in Pima Indians in Mexico and the US. Diabetes Care 2006, 29, 1866–1871. [Google Scholar] [CrossRef]
- Schutz, Y.; Dulloo, A. Resting Metabolic Rate, Thermic Effect of Food, and Obesity. In Handbook of Obesity; CRC Press: Boca Raton, FL, USA, 2023; Volume 1, pp. 286–295. [Google Scholar]
- Brittain, E.L.; Han, L.; Annis, J.; Master, H.; Hughes, A.; Roden, D.M.; Harris, P.A.; Ruderfer, D.M. Physical activity and incident obesity across the spectrum of genetic risk for obesity. JAMA Netw. Open 2024, 7, e243821. [Google Scholar] [CrossRef]
- Knowler, W.C.; Barrett-Connor, E.; Fowler, S.E.; Hamman, R.F.; Lachin, J.M.; Walker, E.A.; Nathan, D.M.; Diabetes Prevention Program Research Group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002, 346, 393–403. [Google Scholar] [CrossRef]
- Pérusse, L.; Bouchard, C. Gene–diet interactions in obesity. Am. J. Clin. Nutr. 2000, 72, 1285S–1290S. [Google Scholar] [CrossRef][Green Version]
- Abbey, M. The influence of apolipoprotein polymorphism on the response to dietary fat and cholesterol. Curr. Opin. Lipidol. 1992, 3, 12–16. [Google Scholar] [CrossRef]
- Wright, L.; Davies, N.M.; Shireby, G.; Williams, D.M.; Morris, T.T.; Bann, D. Genetic Risk for High Body Mass Index Before and Amidst the Obesity Epidemic: Cross-Cohort Analysis of Four British Birth Cohort Studies. medRxiv 2024. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Luo, Z. Effect of Adiponectin Variant on Lipid Profile and Plasma Adiponectin Levels: A Multicenter Systematic Review and Meta-Analysis. Cardiovasc. Ther. 2022, 2022, 4395266. [Google Scholar] [CrossRef] [PubMed]
- Reda, D. Narrative review of metabolic syndrome and its relationships with non-alcoholic fatty liver disease, gonadal dysfunction and obstructive sleep apnea. Diabetol. Metab. Syndr. 2025, 17, 353. [Google Scholar] [CrossRef]
- Roberts, M.C.; Mensah, G.A.; Khoury, M.J. Leveraging implementation science to address health disparities in genomic medicine: Examples from the field. Ethn. Dis. 2019, 29 (Suppl. S1), 187. [Google Scholar] [CrossRef]
- Gérard, P. Gut microbiota and obesity. Cell. Mol. Life Sci. 2016, 73, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Airoldi, J.; Berghella, V. Hepatitis C and pregnancy. Obstet. Gynecol. Surv. 2006, 61, 666–672. [Google Scholar] [CrossRef]
- Cheung, W.W.; Mao, P. Recent advances in obesity: Genetics and beyond. ISRN Obes. 2012, 2012, 536905. [Google Scholar] [CrossRef] [PubMed]
- Masi, D.; Tozzi, R.; Watanabe, M. Genome editing and obesity. In Genome Editing in Cardiovascular and Metabolic Diseases; Springer Nature: Singapore, 2022; pp. 179–190. [Google Scholar] [CrossRef]
- Bouchard, C. Genetics of obesity: Overview and research directions. In The Genetics of Obesity; Academic Press: Cambridge, MA, USA, 2020; pp. 223–233. [Google Scholar]
- Moustafa, J.S.E.-S.; Froguel, P. From obesity genetics to the future of personalized obesity therapy. Nat. Rev. Endocrinol. 2013, 9, 402–413. [Google Scholar] [CrossRef]
- Viguerie, N.; Vidal, H.; Arner, P.; Holst, C.; Verdich, C.; Avizou, S.; Astrup, A.; Saris, W.H.; Macdonald, I.A.; Klimcakova, E.; et al. Adipose tissue gene expression in obese subjects during low-fat and high-fat hypocaloric diets. Diabetologia 2005, 48, 123–131. [Google Scholar] [CrossRef]
- Fansa, S.; Acosta, A. The melanocortin-4 receptor pathway and the emergence of precision medicine in obesity management. Diabetes Obes. Metab. 2024, 26, 46–63. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.J.; Orlando, R.A.; Garver, W.S. Gene-nutrient interactions and susceptibility to human obesity. Genes Nutr. 2017, 12, 29. [Google Scholar] [CrossRef]
- Janić, M.; Janež, A.; El-Tanani, M.; Rizzo, M. Obesity: Recent advances and future perspectives. Biomedicines 2025, 13, 368. [Google Scholar] [CrossRef]
- Sobalska-Kwapis, M.; Suchanecka, A.; Słomka, M.; Siewierska-Górska, A.; Kępka, E.; Strapagiel, D. Genetic association of FTO/IRX region with obesity and overweight in the Polish population. PLoS ONE 2017, 12, e0180295. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Ghosh, S.; Sathain, B.; Banerjee, I. Genetic basis of obesity: A review. J. Biomed. Sci. 2016, 3, 24–28. [Google Scholar] [CrossRef][Green Version]
- James, W.P.; Trayhurn, P. An integrated view of the metabolic and genetic basis for obesity. Lancet 1976, 308, 770–773. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M. An overview of epigenetics in obesity: The role of lifestyle and therapeutic interventions. Int. J. Mol. Sci. 2022, 23, 1341. [Google Scholar] [CrossRef] [PubMed]
- Speakman, J.R. Obesity: The integrated roles of environment and genetics. J. Nutr. 2004, 134, 2090S–2105S. [Google Scholar] [CrossRef]
- Johannsen, W. The genotype conception of heredity. Am. Nat. 1911, 45, 129–159. [Google Scholar] [CrossRef]
- Xia, Q.; Grant, S.F. The genetics of human obesity. Ann. N. Y. Acad. Sci. 2013, 1281, 178–190. [Google Scholar] [CrossRef]
- Bouchard, C. Genetics of obesity: What we have learned over decades of research. Obesity 2021, 29, 802–820. [Google Scholar] [CrossRef]
- Caballero, B. Humans against obesity: Who will win? Adv. Nutr. 2019, 10, S4–S9. [Google Scholar] [CrossRef]
- Bersten, D.C.; Bruning, J.B.; Peet, D.J.; Whitelaw, M.L. Human variants in the neuronal basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) transcription factor complex NPAS4/ARNT2 disrupt function. PLoS ONE 2014, 9, e85768. [Google Scholar] [CrossRef]
- Omeragić, E.; Imamović, B.; Bečić, E.; Dedić, M.; Hashemi, F. Modulating the human microbiome: The impact of xenobiotics on gut microbial composition and therapeutic strategies. In Human Microbiome: Techniques, Strategies, and Therapeutic Potential; Springer Nature: Singapore, 2024; pp. 587–623. [Google Scholar] [CrossRef]
- Mosbah, H.; Poitou, C.; Clément, K. Single-gene defects and obesity. In Handbook of Obesity; CRC Press: Boca Raton, FL, USA, 2023; Volume 1, pp. 123–132. [Google Scholar]
- Michaud, J.L.; Boucher, F.; Melnyk, A.; Gauthier, F.; Goshu, E.; Lévy, E.; Mitchell, G.A.; Himms-Hagen, J.; Fan, C.M. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum. Mol. Genet. 2001, 10, 1465–1473. [Google Scholar] [CrossRef]
- Plum, L.; Lin, H.V.; Dutia, R.; Tanaka, J.; Aizawa, K.S.; Matsumoto, M.; Kim, A.J.; Cawley, N.X.; Paik, J.H.; Loh, Y.P.; et al. The obesity susceptibility gene Cpe links FoxO1 signaling in hypothalamic pro-opiomelanocortin neurons with regulation of food intake. Nat. Med. 2009, 15, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, D.; Manco, L.; Nóbrega, C. Genetics of human obesity. In Obesity: A Practical Guide; Springer International Publishing: Cham, Switzerland, 2016; pp. 87–106. [Google Scholar] [CrossRef]
- Ali, H. Artificial intelligence in multi-omics data integration: Advancing precision medicine, biomarker discovery and genomic-driven disease interventions. Int. J. Sci. Res. Arch. 2023, 8, 1012–1030. [Google Scholar] [CrossRef]
- Meyre, D.; Delplanque, J.; Chèvre, J.C.; Lecoeur, C.; Lobbens, S.; Gallina, S.; Durand, E.; Vatin, V.; Degraeve, F.; Proença, C.; et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009, 41, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, G.S.; Chu, X.; Wood, G.C.; Gerhard, G.M.; Benotti, P.; Petrick, A.T.; Gabrielsen, J.; Strodel, W.E.; Still, C.D.; Argyropoulos, G. Next-generation sequence analysis of genes associated with obesity and nonalcoholic fatty liver disease-related cirrhosis in extreme obesity. Hum. Hered. 2013, 75, 144–151. [Google Scholar] [CrossRef]
- Manusov, E.G.; Diego, V.P.; Almeida, M.; Galan, J.A.; Bala, A.A.; Arriaga, M.A.; Garcia-Rodriguez, N.S.; Hernandez, R.; Kumar, S.; Blangero, J.; et al. Gene–environment interactions in nonalcoholic fatty liver disease: Insights from Mexican American populations. In A Comprehensive Guide to Non-Alcoholic Fatty Liver Disease IntechOpen Limited; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Heard-Costa, N.L.; Zillikens, M.C.; Monda, K.L.; Johansson, Å.; Harris, T.B.; Fu, M.; Haritunians, T.; Feitosa, M.F.; Aspelund, T.; Eiriksdottir, G.; et al. NRXN3 is a novel locus for waist circumference: A genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009, 5, e1000539. [Google Scholar] [CrossRef]



| Gene | Encoded Protein | Primary Function | Clinical Impact | References |
|---|---|---|---|---|
| LEP | Leptin | Regulates appetite & energy balance | Congenital leptin deficiency → severe hyperphagia | [14] |
| LEPR | Leptin receptor | Mediates leptin signaling | Impaired satiety, early-onset obesity | [15] |
| MC4R | Melanocortin-4 receptor | Controls food intake & energy expenditure | Accounts for ~5% of severe early-onset obesity | [12] |
| MC3R | Melanocortin-3 receptor | Regulates energy balance | Variants associated with obesity phenotypes | [20] |
| POMC | Pro-opiomelanocortin | Precursor for α-MSH, binds MC4R | Mutations cause extreme hyperphagia & adrenal dysfunction | [17] |
| PCSK1 | Prohormone convertase 1 | Activates appetite-regulating peptides | Loss-of-function → defective energy regulation | [17] |
| Gene/Locus | Chromosomal Location | Encoded Protein/Gene Name | Biological Function | Associated Trait/Relevance | Replication/References |
|---|---|---|---|---|---|
| FTO | 16q12 | Fat mass and obesity-associated protein | Epigenetic regulation of energy balance | BMI, fat deposition | Strong, multiple populations [31,32] |
| MC4R | 18q21 | Melanocortin-4 receptor | Regulates appetite & energy expenditure | BMI, obesity, and appetite control | Strong, replicated across ancestries [33] |
| MC3R | 20q13.2–13.3 | Melanocortin-3 receptor | Regulates energy homeostasis | BMI, obesity | Moderate, Caucasian and Hispanic populations [20,34] |
| TMEM18 | 2p25 | Transmembrane protein 18 | Body fat storage | BMI | Consistent replication [35] |
| NEGR1 | 1p31 | Neuronal growth regulator 1 | Regulation of adiposity and waist circumference | BMI, waist circumference | Strong [36] |
| SEC16B | 1q25 | SEC16 homolog B | Visceral fat regulation | BMI, fat distribution | Consistent [37] |
| BDNF | 11p14 | Brain-derived neurotrophic factor | Satiety and neuronal differentiation | BMI, extreme obesity | European cohorts [38] |
| GNPDA2 | 4p12 | Glucosamine-6-phosphate deaminase 2 | Appetite modulation and obesity risk | BMI | Moderate, replicated in Europeans [39] |
| MAP2K5–SKOR1 | 15q23 | MAP kinase pathway genes | BMI regulation | BMI | Strong in Europeans [40] |
| SLC6A14 | Xq23 | Solute carrier family 6, member 14 | Amino acid transport, nutrient sensing | Obesity | French/Finnish cohorts [41] |
| PCSK1 | 5q15–q21 | Proprotein convertase subtilisin/kexin type 1 | Activation of appetite-regulating peptides | BMI, obesity | East Asian replication [8] |
| APOA2 | 1q23 | Apolipoprotein A-II | Modifies lipid metabolism depending on diet | Diet–gene interaction (saturated fat) | Functional nutrigenomic evidence |
| LEP | 7q31.3 | Leptin | Regulates appetite & energy balance | Congenital leptin deficiency, hyperphagia | Disease-causing [16] |
| LEPR | 1p31 | Leptin receptor | Mediates leptin signaling | Impaired satiety, early-onset obesity | Disease-causing [20] |
| POMC | 2p23.3 | Pro-opiomelanocortin | Precursor for α-MSH, binds MC4R | Hyperphagia & adrenal dysfunction | Disease-causing [16] |
| ADCY3 | 2p23.3 | Adenylate cyclase 3 | Hypothalamic cAMP signaling | Severe obesity variants | Associated [20] |
| ARNT2 | 15q25 | Aryl hydrocarbon receptor nuclear translocator 2 | Hypothalamic neuronal differentiation | Developmental role | Animal models [42] |
| CPE | 4q32.3 | Carboxypeptidase E | Neuropeptide processing | Obesity-associated variants | [41] |
| GRPR | Xq22 | Gastrin-releasing peptide receptor | Satiety regulation | Obesity variants | [20] |
| ISL1 | 5q11.2 | ISL LIM home-box 1 | POMC expression, hypothalamic neuron differentiation | Developmental role | Linkage analysis |
| LRP2 | 2q31.1 | LDL receptor-related protein 2 | Enhances leptin-induced STAT3 | Obesity variants | [20] |
| MYT1L | 2p25.3 | Myelin transcription factor 1-like | Hypothalamic development | Obesity variants | [20] |
| NPY | 7p15.3 | Neuropeptide Y | Stimulates food intake | Obesity variants | [43] |
| NTRK2 | 9q22 | BDNF receptor | Hypothalamic differentiation | Disease-causing | [20] |
| OTP | 5q13.1 | Orthopedic home-box | Hypothalamic development | Animal model | [20] |
| OXT | 20p13 | Oxytocin | Appetite regulation | Hypothalamic circuit role | [20] |
| NEUROG3 | 10q21.3 | Neurogenin 3 | Hypothalamic transcription factor | Developmental role | Animal model [42] |
| POU3F2 | 6q16.1 | POU class 3 home-box 2 | Hypothalamic transcription factor | CNV studies | [20] |
| SH2B1 | 16p11.2 | Src homology 2 B adapter protein 1 | Modulates leptin & insulin signaling | Hyperphagia, obesity | Disease-causing [44] |
| SIM1 | 6q16.3 | Single-minded homolog 1 | Hypothalamic differentiation | Disease-causing | [45] |
| TUB | 11p15 | Tubby transcription factor | Hypothalamic neuropeptides | Syndromic obesity | [44] |
| Gene/Mutation | Function in the Development of Obesity Complications | References |
|---|---|---|
| Gene of adiponectin (variants rs1501299, rs2241766, rs266729 and rs17300539) | Marker of cardiometabolic risk | [62] |
| SREBF1 | Responsible for the increased risk of coronary heart disease in patients with obstructive sleep apnea | [63] |
| Deletion of the long arm of chromosome 15 | Prader–Willi syndrome-increased obstructive sleep apnea risk | [62] |
| Rs926198 variant of the gene encoding caveolin-1 | Increased risk of cardiovascular disease in obesity | [62] |
| Genes of transcription factor TCF7L2 and PPAR-γ2 receptor | Occurrence of type 2 diabetes mellitus in the obese | [55] |
| SLC16A11 gene variants | Development of type 2 diabetes in the inhabitants of Mexico and other Latin American countries | [55] |
| Gene encoding the amyloid A | The size of the adipocytes increased in obese people | [62] |
| PPP1R15A, HADHA, NR1P1, FOS, FOSB and JUN | Co-existence of osteoporosis, colon cancer, and Obesity | [62] |
| Polymorphism Ala55Val of UCP2 gene | Weight loss in obese patients undergoing laparoscopic adjustable gastric banding | [55] |
| Gene | Chromosomal Location | Phenotype | Population Studied | References |
|---|---|---|---|---|
| FTO | 16q12 | BMI; WC; Fat percentage; extreme obesity | European, African, Asian | [32] |
| MC4R | 18q21 | BMI; WC; extreme obesity | European: Indian Asian | [33] |
| MC3R | 20q13.2–13.3 | Obesity | The Caucasian population, the Hispanic population | [34] |
| SLC6A14 | Xq23 | Obesity | Finish, French | [41] |
| POMC | 2p23.3 | BMI | European | [65] |
| BDNF | 11p4 | BMI; extreme Obesity | European | [38] |
| TMEM18 | 2p25 | BMI; extreme Obesity | European | [35] |
| NEGR1 | 1p31 | BMI | European | [36] |
| PCSK1 | 5q15-q21 | BMI | East Asian | [39] |
| GNPDA2 | 4p12 | BMI | European | [66] |
| MAP2K5 | 15q23 | BMI | European | [40] |
| SEC16B | 1q25 | BMI | European | [37] |
| Gene/SNP | Dietary Modifier | Metabolic Effect | Phenotypic Impact | References |
|---|---|---|---|---|
| FTO rs9939609 | High protein vs. high carb | Alters IRX3/IRX5 expression | Protein-rich diets mitigate BMI risk | [75] |
| APOA2 CC genotype | Saturated fat | Modifies lipid storage | High saturated fat intake an increase (↑) BMI | [16] |
| PPARγ2 Pro12Ala | Dietary fats | Enhances insulin sensitivity | Improved glucose metabolism on high MUFA diets | [58] |
| TCF7L2 variants | Refined carbohydrate load | Influences β-cell function | Modulates diabetes & adiposity risk | [62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farzand, A.; Rohin, M.A.K.; Awan, S.J.; Ahmad, A.M.R.; Akram, H.; Saleem, T.; Imran, M.M. Nutrigenomics of Obesity: Integrating Genomics, Epigenetics, and Diet–Microbiome Interactions for Precision Nutrition. Life 2025, 15, 1658. https://doi.org/10.3390/life15111658
Farzand A, Rohin MAK, Awan SJ, Ahmad AMR, Akram H, Saleem T, Imran MM. Nutrigenomics of Obesity: Integrating Genomics, Epigenetics, and Diet–Microbiome Interactions for Precision Nutrition. Life. 2025; 15(11):1658. https://doi.org/10.3390/life15111658
Chicago/Turabian StyleFarzand, Anam, Mohd Adzim Khalili Rohin, Sana Javaid Awan, Abdul Momin Rizwan Ahmad, Hiba Akram, Talha Saleem, and Muhammad Mudassar Imran. 2025. "Nutrigenomics of Obesity: Integrating Genomics, Epigenetics, and Diet–Microbiome Interactions for Precision Nutrition" Life 15, no. 11: 1658. https://doi.org/10.3390/life15111658
APA StyleFarzand, A., Rohin, M. A. K., Awan, S. J., Ahmad, A. M. R., Akram, H., Saleem, T., & Imran, M. M. (2025). Nutrigenomics of Obesity: Integrating Genomics, Epigenetics, and Diet–Microbiome Interactions for Precision Nutrition. Life, 15(11), 1658. https://doi.org/10.3390/life15111658







